Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Fluorescent analogs of cyclic and linear dinucleotides as phosphodiesterase and oligoribonuclease activity probes†
Jie Zhouab,
Yue Zhengac,
Benjamin T. Roembkec,
Sarah M. Robinson c,
Clement Opoku-Temengac,
David A. Sayrec and
Herman O. Sintim
c,
Clement Opoku-Temengac,
David A. Sayrec and
Herman O. Sintim *abcd
*abcd
aPurdue Institute for Drug Discovery, Purdue University, West Lafayette, IN 47907, USA. E-mail: hsintim@purdue.edu
bDepartment of Chemistry, Purdue University, West Lafayette, IN 47907, USA
cDepartment of Chemistry and Biochemistry, University of Maryland, College Park, Maryland 20742, USA
dPurdue Institute of Inflammation, Immunology and Infectious Disease, West Lafayette, IN 47907, USA
First published on 17th January 2017
Abstract
Cyclic dinucleotide-based second messengers, including c-di-GMP, c-di-AMP and cGAMP, are universal signaling molecules widely used by prokaryotes and eukaryotes. C-di-GMP and c-di-AMP play key roles in bacterial survival. Consequently, metabolism proteins that modulate cyclic dinucleotide concentrations, such as cyclic dinucleotide phosphodiesterases (PDE), are potential drug targets; thus, probes that report PDE activity could have great utility in high throughput screening for PDE inhibitors. However, there is a paucity of luminescent-based probes for monitoring cyclic dinucleotide PDE activity. In this study, we synthesized various fluorescent nucleobase (ethenoA and 2-AP) analogs of cyclic and linear dinucleotides. These analogs were readily cleaved by the cyclic dinucleotide PDE and oligoribonucleases (Orn). Cleavage of the fluorescent probes led to changes in fluorescence intensities. Our results suggest that these fluorescent analogs can be used to monitor the activity of cyclic dinucleotide PDEs in real time and that these probes could facilitate the identification of inhibitors of cyclic dinucleotide PDEs. Additionally these probes could be used to profile the activity of expressed PDEs and Orns.
Cyclic dinucleotides, such as c-di-GMP, c-di-AMP, and cGAMP, have come to the forefront of both chemical and biological investigations because they are involved in important signal transduction processes in both eukaryotes and prokaryotes.1 The intracellular concentrations of these second messengers fluctuate in response to changing environments, leading to the activation and/or repression of various cellular processes.1 Cyclic dinucleotides are synthesized from two molecules of purine triphosphates by synthases and their actions are terminated upon degradation by hydrolyases.2 The synthase for c-di-GMP is diguanylate cyclase (DGC), which is characterized by a GGDEF domain,3 whereas c-di-AMP is synthesized from ATP by diadenylate cyclase (DAC), characterized by a DisA_N domain.4 cGAMP, containing a non-canonical 2′,5′-phosphodiester linkage, is synthesized by cyclic GMP–AMP synthase (cGAS) in response to duplex DNA in the cytoplasm of immune cells of higher organisms.5 Both c-di-GMP and c-di-AMP are broken down into 5′-phosphoguanylyl/adenylyl-(3′-5′)-guanosine/adenosine (pGpG or pApA) and further into GMP or AMP, by phosphodiesterases (PDEs).6 C-di-GMP PDE-A (containing EAL domain) cleaves c-di-GMP in pGpG7 and further into GMP by PDE-B8 whereas PDEs containing the HD-GYP domain cleave c-di-GMP directly into GMP.9 Similarly, whereas some c-di-AMP PDEs cleave the cyclic dinucleotide to linear pApA, others can cleave into AMP.10–12 Recently it was revealed that oligoribonuclease (Orn), which had earlier been established as an essential enzyme in most bacteria involved in the recycling of RNA into ribonucleotides13,14 acts as c-di-GMP PDB-B in Pseudomonas aeruginosa.8,15 PDEs are now emerging as key regulators of bacterial fitness and virulence,14–18 and some could become potential antibacterial drug targets.
There is however a paucity of inhibitors that target cyclic dinucleotide signaling16–19 and no clinical drug is as yet in development for inhibiting cyclic dinucleotide signaling, but the case is different with signaling by cyclic mononucleotides such as cGMP20 and cAMP,21 which were identified in 1960s. For example, Sildenafil (Viagra®), a cGMP PDE5 inhibitor, is used to treat erectile dysfunction.22 There are also a handful of cyclic mononucleotide-related PDE inhibitors, such as GSK256066 (PDE4 inhibitor)23 and Milrinone (Primacor®, PDE3 inhibitor),24 which are currently in clinical use for the treatment of pulmonary hypertension and cardiovascular diseases. The successful identification of cyclic mononucleotide-related PDE inhibitors, such as Sildenafil, was facilitated by assays that could be used in high throughput manner to monitor the enzymatic cleavage of cGMP or cAMP by PDEs.25–27 Analogously, colorimetric or fluorimetric assays that could be used to monitor the cleavage of cyclic or linear dinucleotide in a high throughput manner would also facilitate the identification of inhibitors of cyclic dinucleotide metabolism enzymes. Thus far, cyclic dinucleotide detection methods, such as HPLC-MS,28 radiolabeled ribozyme29 or Spinach RNA,30 fluorescent or colorimetric intercalator assays31–35 and ELISA,36 have been reported. However these methods, albeit sensitive, have a few drawbacks. For example, the HPLC-MS method is not amenable for high throughput assays or real-time monitoring or is expensive to implement. The riboswitch method is also expensive to utilize in a high throughput way, notwithstanding the difficulty of working with RNA and some of the intercalator assays (such as the thiazole orange assay)31 require overnight incubation and hence not ideal for real-time monitoring. Others have also used p-nitrophenyl phosphate (pNPP) to monitor cyclic dinucleotide PDE activity37,38 but three limitations of this substrate are: (a) pNPP is not a good substrate for many cyclic dinucleotide PDEs or Orns; (b) background hydrolysis (PDE-independent cleavage) of pNPP is high and (c) the colorimetric readout of the cleavage product, para nitrophenol, overlaps with many compounds that are found in chemical libraries used for screening.
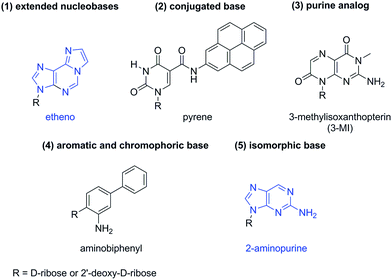
Fluorogenic analogs of cyclic dinucleotides are potential probes for profiling PDE activity. Fluorescent nucleobase analogs have been used in several applications,39–42 including the study of nucleic acid dynamics43 and structure.44 Several assays that are used to monitor nucleic acid processing enzymes have incorporated a fluorescent nucleobase into the nucleotide or oligonucleotide substrate.42 In general, fluorescent nucleobases can be categorized into five classes, namely (1) extended nucleobases; (2) conjugated base analogs; (3) purine analogs; (4) aromatic and chromophoric base analogs and (5) isomorphic base analogs45 (see Fig. 1). 2-Aminopurine (2-AP) and ethenoadenosine (ethenoA) are two widely utilized fluorescent nucleobases and are commercially available.46,47
2-AP is considered as “isomorphic” fluorescent nucleobase and closely resembles adenine. In fact it can form strong base pairs with thymine and uracil.46,47 As far back as 1969, Ward et al. showed that the fluorescence emission of 2-AP is reduced when placed within an oligonucleotide.48 Later, others demonstrated that the nature of the neighboring nucleobase also affected the fluorescence profile of 2-AP.47 EthenoA was developed by Secrist to study adenosine-containing coenzymes.49 Both the excitation (345 nm) and emission (435 nm) of ethenoA are more red shifted than those of 2-AP (ex. = 310 nm and em. = 375 nm), and this is due to the extended pi-system of etheno. Consequently, ethenoA is more suitable for biological assays than 2-AP as it suffers less from interference of biological fluorophores.41,42
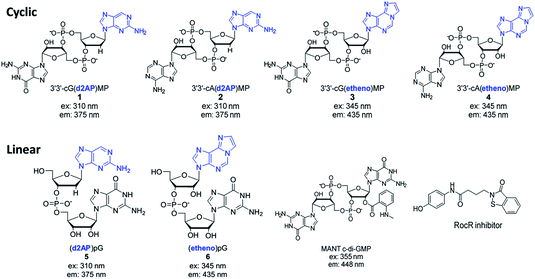
Chatterji et al.50 and the group of Banin15 reported that the fluorescence of MANT-c-di-GMP (see Fig. 2), slightly decreased upon cleavage by RocR,15 a c-di-GMP PDE that we have been interested in developing inhibitors against.19 However, MANT-c-di-GMP has a bulky fluorophore group appended to 2′-position of c-di-GMP and this steric block could account for the poor substrate profile for some PDEs. We recently reported that the replacement of one of the native nucleobases of c-di-GMP with 2-AP afforded a fluorescent analog (compound 1, Fig. 2) that could be used to monitor the activity of PDEs.33 Motivated by our initial success in developing a fluorescent probe that can monitor PDE activity we decided to develop a palette of fluorescent dinucleotide probes that could complement MANT-c-di-GMP. For the AP probes, we proceeded to investigate how the nature of the second nucleotide (adenine vs. guanine) in the 2-AP probe affected the fluorescence response to PDE cleavage (compare compound 1 vs. compound 2). Additionally, we explored other fluorescent nucleobases, such as etheno analog 3 and 4, hoping to increase the repertoire of fluorescent analogs of cyclic dinucleotides, which could facilitate high throughput screening for PDE inhibitors (Fig. 2). Also, in this new study we have explored fluorescent linear dinucleotides (compounds 5 and 6) as activity probes for oligoribonucleases.
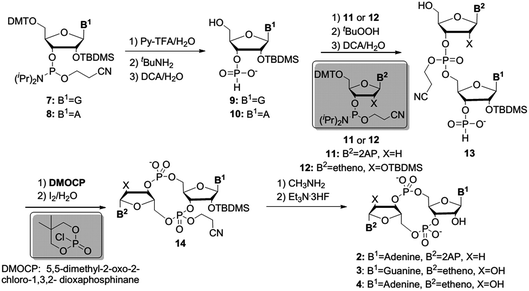
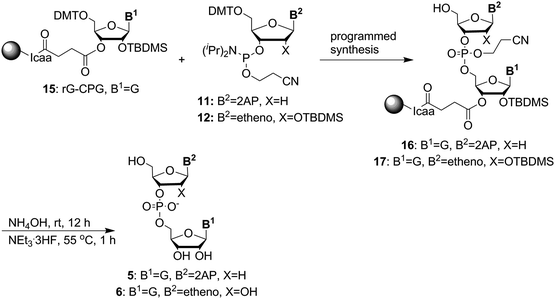
We first prepared the fluorescent probes 2 to 4 following Jones' strategy outlined in Scheme 1.51 For this strategy one portion of guanosine or adenosine phosphoramidite was first converted into H-phosphonate, followed by the coupling with another portion of deoxyribose 2-AP or ribose etheno phosphoramidite to achieve the linear dimer 13. The cyclization reagent 5,5-dimethyl-2-oxo-2-chloro-1,3,2-dioxaphosphinane (DMOCP), was used to convert the linear dimer 13 into a cyclized form 14 in the same flask. After deprotecting all of the protecting groups on the nucleobase exocyclic amine and 2′-OH, the crude product was subjected to HPLC purification. The fluorescent linear dinucleotides (5 and 6) were synthesized on solid support as shown in Scheme 2, using a DNA synthesizer.
With these new fluorescent probes in hand, we then proceeded to test if they were cyclic dinucleotide PDE substrates and if so, whether the fluorescence of these probes changed upon PDE cleavage. The fluorescence of cyclic probes (compounds 1 to 4) is quenched via electron transfer between adenine (A) or guanine (G) to the fluorescent base (see Fig. 3, S1 and S2†).52,53 Upon cleavage by PDE-A, the resulting linear form is less constrained, with more options of projecting the positions of the fluorescent base and A or G base. Compared to the parent cyclic probes, the distance between fluorescent and A/G base could be more, or less than or about the same as the distance in cyclic probes (see Fig. 3). Thus, overall fluorescence change upon cleavage of PDE-A could be either “turn-on” or “turn-off”. Upon PDE-B or oligoribonuclease cleavage, the resulting mononucleotides are far from each other (see Fig. 3, X4 ⋙ X1, X2 and X3), and therefore a fluorescence turn-on is expected. Similarly, when the cyclic probes are cleaved with SVPD (a control promiscuous phosphodiesterase that cleaves most cyclic and linear di- or polynucleotides into mononucleotides), a fluorescence turn-on would be expected (see Fig. 3).
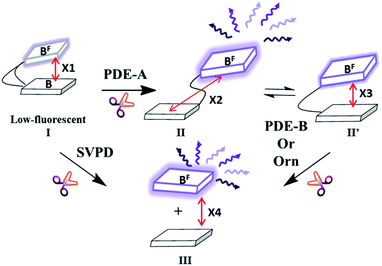 | ||
| Fig. 3 Our working model for fluorescence change upon PDE cleavage. Rectangular block indicates either fluorescent base (2-AP or ethenoA) or G or A. Black curved line represents phospho-linkage between bases. Purple arrows represent fluorescence. X indicates the distance between fluorescent base and normal base (X1 > X2 or X1 < X2 or X1 ∼ X2; X1 and X2 > X3 and X4 ⋙ X1, X2 and X3). See ESI, Fig. S1† for optimized structures of compounds 1 to 4. | ||
We incubated compounds 1 and 2 with RocR and YybT. RocR is c-di-GMP PDE, containing EAL domain, and was first identified in P. aeruginosa.54 RocR breaks down c-di-GMP first into linear form pGpG and then into GMP.54 On the other hand, YybT, a c-di-AMP PDE, is able to cleave c-di-AMP into its linear form, 5′-pApA.10 We therefore expected RocR to cleave compound 1 better than compound 2 and YybT to cleave 2 better than 1 due to the preferences of RocR and YybT for guanine-containing and adenine-containing cyclic dinucleotides respectively. Upon cleavage by RocR, both compounds 1 and 2 showed decrease in fluorescence (Fig. 4) but compound 1 showed a greater fluorescence decrease, confirming our expectation that RocR would prefer the guanine-containing probe. Compound 1 was cleaved as fast as the natural substrate, c-di-GMP (see Fig. S3†). Similarly, YybT also cleaved both compounds 1 and 2, resulting in a decrease in fluorescence (Fig. 4). In contrast to RocR, the magnitude of fluorescence decrease upon cleavage of compound 2 (adenine-containing) by YybT was larger than upon cleavage of compound 1 (guanine-containing) by YybT. Here too this was expected because YybT preferentially cleaves c-di-AMP over c-di-GMP.10 The degree of cleavage of compound 2 (10 μM) by YybT (1 μM) was similar to the cleavage of c-di-AMP by YybT (see Fig. S4†). Of note, as stated, upon cleavage by RocR or YybT, the linearized probes showed lower fluorescence than the cyclized form. As proposed in Fig. 3, the distance between fluorescent and normal base after cleavage by RocR or YybT would change (see Fig. 3) and the decrease in fluorescence upon cleavage implies that the proportion of conformers whereby the nucleobases are close enough for photoelectron transfer is higher in the linear dinucleotide than in the cyclized form. To ascertain if these cyclic probes (compounds 1 to 4) could be used for high throughput screening assays, we investigated if a known inhibitor of RocR or YybT could affect the fluorescence of the assays. We therefore chose EDTA as it chelates divalent metals that are essential for the PDE activity (Mg2+ for RocR and Mn2+ for YybT). We also chose a known inhibitor of RocR as a second control. Upon cleavage of compound 1 by RocR in the presence of EDTA, the fluorescence intensity decreased by only 20% (versus 60% without EDTA) (Fig. 4A). Also, in the presence of a RocR benzoisothiazolinone inhibitor,19 the change in fluorescence intensity was minimal (see Fig. 4A). The same trend was observed for the cleavage of compound 2 by YybT. Upon cleavage of compound 2 by YybT in the presence of EDTA, the fluorescence intensity decreased by only 10% (versus 50% without EDTA) (Fig. 4B). The fluorescence of the cyclic ethenoA analogs, compounds 3 and 4, are too low (guanine or adenine seem to efficiently quench the fluorescence of etheno adenine in both the cyclic and linear dinucleotide) to be used as activity probe for PDE-A (see ESI, Fig. S2†) but these probes were useful for monitoring PDE-B or Orn activity, vide infra.
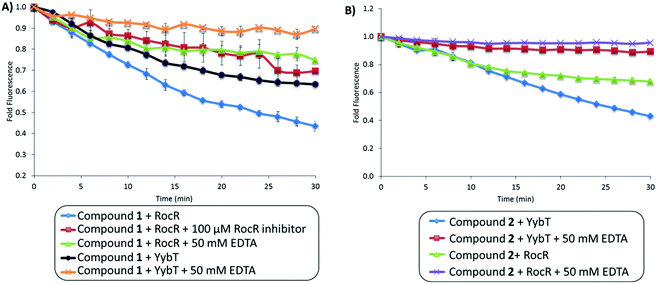 | ||
| Fig. 4 Fluorescence changes upon cleavage of compounds 1 and 2 by PDEs. (A) PDE cleavage assay of compound 1 by RocR and YybT in the absence or presence of 50 mM EDTA or RocR inhibitor.19 λex = 310 nm and λem = 375 nm. (B) PDE cleavage assay of compound 2 by YybT and RocR, in the absence or presence of 50 mM EDTA. λex = 310 nm and λem = 375 nm. [RocR or YybT] = 0.25 μM, [compound 1 or 2] = 2.5 μM, [RocR inhibitor] = 100 μM. Enzymatic assays were carried out at 37 °C. | ||
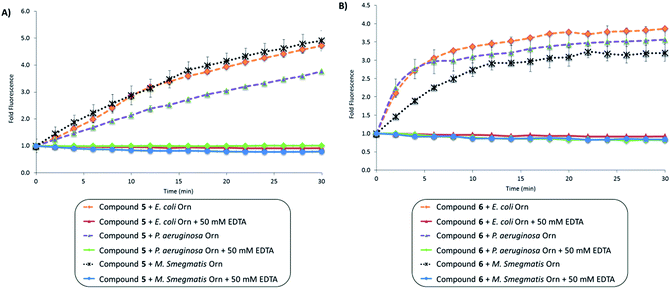
PDE-B or Orn cleave linear dinucleotides into mononucleotides. As shown in Fig. 3, the distance between the nucleobases becomes greatest upon PDE-B or Orn cleavage (compared to the distances in the cyclic or linear dinucleotides). Therefore we expected that dinucleotides containing either 2-AP or etheno adenine would be excellent turn-on probes for monitoring the activities of PDE-B or Orn. To confirm our hypothesis, we incubated linear probes (compounds 5 and 6) with different Orns (E. coli, P. aeruginosa and M. smegmatis Orns) (see Fig. 5). Upon cleavage of compounds 5 and 6 by all three different Orns, an increase in fluorescence intensity was observed (Fig. 5). To confirm that the increase in fluorescence was due to the enzymatic cleavage of the compounds, we again performed the enzymatic reactions in the presence of 50 mM EDTA, which would inhibit the enzymatic cleavage. As expected, there was no change in fluorescence intensity with the linear probes (compounds 5 and 6) when EDTA was present. Compound 5 gave the highest fold change in fluorescence. E. coli Orn cleaved compound 5 as well as the natural pGpG (see Fig. S5†) and we recommend 5 as the ideal substrate to screen for inhibitors of oligoribonuclease or to assay the activity of Orns.
Conclusion
In summary, we disclose new fluorescent linear and cyclic dinucleotide probes, which could facilitate cyclic dinucleotide PDE research and drug discovery. Recently several reports have documented the roles of cyclic dinucleotide (c-di-GMP, c-di-AMP and cGAMP) phosphodiesterases in bacterial virulence1 as well in innate immunity (cleavage of 2′3′-cGAMP) and probes,55,56 such as the ones reported in this manuscript could be used to confirm the activity of expressed PDE enzymes or used to screen for inhibitors of dinucleotide phosphodiesterases.19,57Acknowledgements
We sincerely thank Dr Zhao-Xun Liang for providing RocR and YybT plasmids. We also thank Dr Ehud Banin (P. aeruginosa Orn) and Dr Nicholas E. Dixon (E. coli and M. smegmatis Orns) for providing plasmids. Funding for this work was provided by the National Science Foundation (CHE1307218 and CHE1636752), Purdue University, University of Maryland Graduate Dean's Dissertation Fellowship (JZ), University of Maryland Graduate Dean's Dissertation Fellowship (YZ), Millard and Lee Alexander Fellowship in Chemistry (BTR) and Kraybill Biochemistry Fellowship (YZ).Notes and references
- D. Kalia, G. Merey, S. Nakayama, Y. Zheng, J. Zhou, Y. Luo, M. Guo, B. T. Roembke and H. O. Sintim, Chem. Soc. Rev., 2013, 42, 305–341 RSC.
- U. Römling, M. Y. Galperin and M. Gomelsky, Microbiol. Mol. Biol. Rev., 2013, 77, 1–52 CrossRef PubMed.
- U. Römling, M. Gomelsky and M. Y. Galperin, Mol. Microbiol., 2005, 57, 629–639 CrossRef PubMed.
- R. M. Corrigan and A. Gründling, Nat. Rev. Microbiol., 2013, 11, 513–524 CrossRef CAS PubMed.
- V. Hornung, R. Hartmann, A. Ablasser and K. P. Hopfner, Nat. Rev. Immunol., 2014, 14, 521–528 CrossRef CAS PubMed.
- P. Schaap, IUBMB Life, 2013, 65, 897–903 CrossRef CAS PubMed.
- A. J. Schmidt, D. A. Ryjenkov and M. Gomelsky, J. Bacteriol., 2005, 187, 4774–4781 CrossRef CAS PubMed.
- M. W. Orr, G. P. Donaldson, G. B. Severin, J. Wang, H. O. Sintim, C. M. Waters and V. T. Lee, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, E5048–E5057 CrossRef CAS PubMed.
- R. P. Ryan, Y. Fouhy, J. F. Lucey, L. C. Crossman, S. Spiro, Y. W. He, L. H. Zhang, S. Heeb, M. Camara, P. Williams and J. M. Dow, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 6712–6717 CrossRef CAS PubMed.
- F. Rao, R. Y. See, D. W. Zhang, D. C. Toh, Q. Ji and Z. X. Liang, J. Biol. Chem., 2010, 285, 473–482 CrossRef CAS PubMed.
- K. Manikandan, V. Sabareesh, N. Singh, K. Saigal, U. Mechold and K. M. Sinha, PLoS One, 2014, 9, e86096 Search PubMed.
- R. M. Corrigan, J. C. Abbott, H. Burhenne, V. Kaever and A. Gründling, PLoS Pathog., 2011, 7, e1002217 CAS.
- S. Ghosh and M. P. Deutscher, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 4372–4377 CrossRef CAS.
- A. Young Park, C. M. Elvin, S. M. Hamdan, R. J. Wood, N. E. Liyou, T. E. Hamwood, P. A. Jennings and N. E. Dixon, Protein Expression Purif., 2008, 57, 180–187 CrossRef CAS PubMed.
- D. Cohen, U. Mechold, H. Nevenzal, Y. Yarmiyhu, T. E. Randall, D. C. Bay, J. D. Rich, M. R. Parsek, V. Kaever, J. J. Harrison and E. Banin, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 11359–11364 CrossRef CAS PubMed.
- S. Nakayama, J. Zhou, Y. Zheng, H. Szmacinski and H. O. Sintim, Future Sci. OA, 2016, 2 DOI:10.4155/fso.4115.4193.
- C. Opoku-Temeng and H. O. Sintim, Sci. Rep., 2016, 6, 25445 CrossRef CAS PubMed.
- C. Opoku-Temeng and H. O. Sintim, Chem. Commun., 2016, 52, 3754–3757 RSC.
- Y. Zheng, G. Tsuji, C. Opoku-Temeng and H. O. Sintim, Chem. Sci., 2016, 7, 6238–6244 RSC.
- R. W. Bernlohr, M. K. Haddox and N. D. Goldberg, J. Biol. Chem., 1974, 249, 4329–4331 CAS.
- R. S. Makman and E. W. Sutherland, J. Biol. Chem., 1965, 240, 1309–1314 CAS.
- A. T. Bender and J. A. Beavo, Pharmacol. Rev., 2006, 58, 488–520 CrossRef CAS PubMed.
- C. J. Tralau-Stewart, R. A. Williamson, A. T. Nials, M. Gascoigne, J. Dawson, G. J. Hart, A. D. Angell, Y. E. Solanke, F. S. Lucas, J. Wiseman, P. Ward, L. E. Ranshaw and R. G. Knowles, J. Pharmacol. Exp. Ther., 2011, 337, 145–154 CrossRef CAS PubMed.
- D. H. Maurice, H. Ke, F. Ahmad, Y. Wang, J. Chung and V. C. Manganiello, Nat. Rev. Drug Discovery, 2014, 13, 290–314 CrossRef CAS PubMed.
- J. A. Beavo and L. L. Brunton, Nat. Rev. Mol. Cell Biol., 2002, 3, 710–718 CrossRef CAS PubMed.
- T. Hiratsuka, J. Biol. Chem., 1982, 257, 13354–13358 CAS.
- R. L. Kincaid and V. C. Manganiello, Methods Enzymol., 1988, 159, 457–470 CAS.
- R. Simm, M. Morr, U. Rerriminghorst, M. Andersson and U. Romling, Anal. Biochem., 2009, 386, 53–58 CrossRef CAS PubMed.
- H. Gu, K. Furukawa and R. R. Breaker, Anal. Chem., 2012, 84, 4935–4941 CrossRef CAS PubMed.
- S. Nakayama, Y. Luo, J. Zhou, T. K. Dayie and H. O. Sintim, Chem. Commun., 2012, 48, 9059–9061 RSC.
- J. Wang, J. Zhou, G. P. Donaldson, S. Nakayama, L. Yan, Y. F. Lam, V. T. Lee and H. O. Sintim, J. Am. Chem. Soc., 2011, 133, 9320–9330 CrossRef CAS PubMed.
- S. Nakayama, K. Roelofs, V. T. Lee and H. O. Sintim, Mol. BioSyst., 2012, 8, 726–729 RSC.
- B. T. Roembke, J. Zhou, Y. Zheng, D. Sayre, A. Lizardo, L. Bernard and H. O. Sintim, Mol. BioSyst., 2014, 10, 1568–1575 RSC.
- J. Zhou, D. A. Sayre, Y. Zheng, H. Szmacinski and H. O. Sintim, Anal. Chem., 2014, 86, 2412–2420 CrossRef CAS PubMed.
- S. Nakayama, I. Kelsey, J. X. Wang and H. O. Sintim, Chem. Commun., 2011, 47, 4766–4768 RSC.
- A. J. Underwood, Y. Zhang, D. W. Metzger and G. Bai, J. Microbiol. Methods, 2014, 107, 58–62 CrossRef CAS PubMed.
- F. Rao, Y. Qi, H. S. Chong, M. Kotaka, B. Li, J. Li, J. Lescar, K. Tang and Z. X. Liang, J. Bacteriol., 2009, 191, 4722–4731 CrossRef CAS PubMed.
- T. N. Huynh, S. Luo, D. Pensinger, J. D. Sauer, L. Tong and J. J. Woodward, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, E747–E756 CrossRef CAS PubMed.
- K. Börjesson, P. Sandin, B. Albinsson, J. Mårtensson, T. Brown and L. M. Wilhelmsson, Nucleic Acids Symp. Ser., 2008, 3–4 CrossRef PubMed.
- D. Shin, P. Lönn, S. F. Dowdy and Y. Tor, Chem. Commun., 2015, 51, 1662–1665 RSC.
- S. F. Singleton, A. I. Roca, A. M. Lee and J. Xiao, Tetrahedron, 2007, 63, 3553–3566 CrossRef CAS PubMed.
- R. W. Sinkeldam, N. J. Greco and Y. Tor, Chem. Rev., 2010, 110, 2579–2619 CrossRef CAS PubMed.
- N. Venkatesan, Y. J. Seo and B. H. Kim, Chem. Soc. Rev., 2008, 37, 648–663 RSC.
- K. Börjesson, S. Preus, A. H. El-Sagheer, T. Brown, B. Albinsson and L. M. Wilhelmsson, J. Am. Chem. Soc., 2009, 131, 4288–4293 CrossRef PubMed.
- Y. Tor, S. Del Valle, D. Jaramillo, S. Srivatsan, A. Rios and H. Weizman, Tetrahedron, 2007, 63, 3608–3614 CrossRef CAS.
- L. M. Wilhelmsson, Q. Rev. Biophys., 2010, 43, 159–183 CrossRef CAS PubMed.
- J. N. Wilson, Y. Cho, S. Tan, A. Cuppoletti and E. T. Kool, ChemBioChem, 2008, 9, 279–285 CrossRef CAS PubMed.
- D. C. Ward, E. Reich and L. Stryer, J. Biol. Chem., 1969, 244, 1228–1237 CAS.
- J. A. Secrist, J. R. Barrio, N. J. Leonard and G. Weber, Biochemistry, 1972, 11, 3499–3506 CrossRef CAS PubMed.
- I. M. Sharma, T. Dhanaraman, R. Mathew and D. Chatterji, Biochemistry, 2012, 51, 5443–5453 CrossRef CAS PubMed.
- B. L. Gaffney, E. Veliath, J. Zhao and R. A. Jones, Org. Lett., 2010, 12, 3269–3271 CrossRef CAS PubMed.
- O. Somsen, V. Hoek and V. Amerongen, Chem. Phys. Lett., 2005, 402, 61–65 CrossRef CAS.
- N. J. Leonard, CRC Crit. Rev. Biochem., 1984, 15, 125–199 CrossRef CAS PubMed.
- M. W. Chen, M. Kotaka, C. Vonrhein, G. Bricogne, F. Rao, M. L. Chuah, D. Svergun, G. Schneider, Z. X. Liang and J. Lescar, J. Bacteriol., 2012, 194, 4837–4846 CrossRef CAS PubMed.
- Q. Chen, L. Sun and Z. J. Chen, Nat. Immunol., 2016, 17, 1142–1149 CrossRef CAS PubMed.
- L. Li, Q. Yin, P. Kuss, Z. Maliga, J. L. Millán, H. Wu and T. J. Mitchison, Nat. Chem. Biol., 2014, 10, 1043–1048 CrossRef CAS PubMed.
- C. Opoku-Temeng, J. Zhou, Y. Zheng, J. Su and H. O. Sintim, Chem. Commun., 2016, 52, 9327–9342 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra25394f |
| This journal is © The Royal Society of Chemistry 2017 |