Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
An electrochemical cytosensor for ultrasensitive detection of cancer cells using modified graphene–gold nanostructures†
Amir Yadegariad,
Meisam Omidi*b,
Fatemeh Yazdianc,
Hakimeh Zalia and
Lobat Tayebide
aDepartment of Tissue Engineering and Regenerative Medicine, School of Advanced Technologies in Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran
bProtein Research Centre, Shahid Beheshti University, GC, Velenjak, Tehran, Iran. E-mail: m_omidi@sbmu.ac.ir
cFaculty of New Science and Technology, University of Tehran, Tehran, Iran
dDepartment of Developmental Sciences, Marquette University School of Dentistry, Milwaukee, WI 53233, USA
eDepartment of Engineering Science, University of Oxford, Oxford OX1 3PJ, UK
First published on 5th December 2016
Abstract
The ultrasensitive detection of human prostate metastatic cancer cells (Du-145) was investigated through a novel electrochemical cytosensor. The proposed biosensor was simultaneously developed via two approaches: multivalent identification and signal amplification. Herein, anti-CD166 monoclonal antibody-modified gold electrode was applied to capture and recognize target cells (Du-145). Also, a graphene (G)/gold nanoparticle (GNP)/horseradish peroxidase (HRP)-conjugated trastuzumab antibody (G/GNP/Ab-HRP) hybrid nanostructure was designed as a nanoprobe for accurate recognition of target cells and efficient amplification of enzymatic signals simultaneously. The performance of the cytosensor could be significantly improved by utilizing this novel signal-amplification strategy. The cytosensor described here exhibited an appropriate cell-capture ability, broad range of detection, and exceptional sensitivity with a low limit of detection (20 cells). The fabricated cytosensor showed high sensitivity and selectivity for detection of Du-145 cancer cells while keeping an extended linear range from 102 to 106 cells per ml, and a conveniently low limit of detection of 20 cells per ml. The extraordinary analytical performance of this cytosensor indicates that it has a great potential for the detection of cancer cells and cancer stem cells.
1. Introduction
Developing highly sensitive nanobiosensors for the detection of bacteria, fungal pathogens, viruses, and cancer cells is essential in biomedical, clinical, and food-industry applications.1,2 A rapid and sensitive approach for early detection or diagnosis of cancer is absolutely crucial for more effective therapy. In particular, detection of metastatic cancer cells is one of the critical steps in cancer diagnosis. Hence, introducing a simple strategy for this intention is very important.3 While the techniques available for detection of cancer cells have their own benefits and drawbacks, the absence of developing an unsophisticated, inexpensive, rapid and sensitive procedure for cell detection is tangible. Electrochemical cytosensing methods play an important part in cell detection because of inherent advantages such as rapid response, miniaturization, affordability, and easy operation.4–7 Recently, electrochemical techniques such as differential pulse voltammetry (DPV),8–11 cyclic voltammetry (CV),12 stripping voltammetry (SV)13,14 and electrochemical luminescence (ECL)15,16 have been used for detection of cell number and type. However, the low selectivity and sensitivity of these electrochemical techniques may hinder their potential application. To improve the sensitivity of detection in electrochemical methods, electrocatalysts for signal amplification have been proposed.8 Signal amplification based on enzymatic reactions is one of the most prevalent approaches used in electrochemical cytosensors for improving sensitivity.9 Although significant progress has been made to improve the sensitivity of electrochemical biosensors, they still suffer from low sensitivity in comparison with other cell-sensing approaches. For instance, Gehring et al. reported the electrochemical detection of Salmonella typhimurium cells through capture of magnetic bead-conjugated antibodies.17 Briefly, they deposited magnetic beads on a graphite electrode and employed an alkalinephosphatase-labelled goat anti-Salmonella antibody to catalyse the production of para-aminophenol from the substrate. The produced para-aminophenyl phosphate was electrothermally detected, and the number of captured cells was proportioned to the generated signal with a sensitivity of 8 × 103 cells per ml. Similarly, Radke et al. designed an electrochemical-based impedimetric platform for detection of Escherichia coli bacteria with a sensitivity of 104 to 107 cfu ml−1 using goat anti-IgG antibody.18 Dill et al. developed light-addressable potentiometric sensors for high-accuracy detection of S. typhimurium (119 cfu). They utilized fluorescein-labelled anti-Salmonella antibodies and biotinylated antibodies as the recognition biomarker for highly sensitive monitoring of Salmonella cells.19 Hnaiein et al. fabricated a conductimetric immunosensor for high-accuracy detection of E. coli through capture of a biotinylated polyclonal antibody on magnetic nanoparticles.20 The conjugation of biomarkers on the conductimetric electrode was completed using glutaraldehyde. The magnetic nanoparticles could increase conductivity and facilitate the highly sensitive detection of S. epidermis cells (0.5 cfu ml−1). Nevertheless, selectivity is still a major problem for developing high-accuracy immunosensors, so a more selective and sensitive electrochemical cytosensor needs to be developed for detection of cancer cells.21 In the last decade, graphene (G) and its derivatives have been widely used for fabrication of different types of sensors,22,23 biosensors,24 and cytosensors25 due to their excellent properties. Sharma et al. reported an electrochemical preparation of a functionalized G-graphene-oxide (GO) nanocomposite electrode as a highly sensitive immunosensor. They believed that the functionalized GO noticeably improved the electrical properties of the nanocomposite due to high conductivity of G sheets. The results showed that the presence of G sheets significantly amplified the electrochemical signals for detection of diuron.26 Also, the immobilization and hybridization of GO with metallic nanoparticles have been suggested as an appropriate candidate for cancer cells detection because it can enhance the rate of electron transfer and amplification of electrochemical signals.27 In particular, gold nanoparticles (GNPs) show excellent biocompatibility, poor adsorption of biomolecules, high conductivity, and high catalytic behavior towards electrochemical reactions. Thus, combination of GNPs with G sheets significantly improves the rate of electron transfer between biomarkers and the electrode surface by increasing the rate of electrochemical reactions.28 Huang et al. designed a sensitive label-free amperometric immunosensor based on the hybridization of functionalized G and GNPs. The fabricated immunosensor was immobilized with α-fetoprotein antibody (anti-AFP) for detection of α-fetoprotein. The results showed that the combination of G sheets and GNPs considerably improved sensitivity, long-term bioactivity and stability of the immunosensor owing to high electron transfer, large surface area, and appropriate electrostatic adsorption.29 In the present study, a novel electrochemical cytosensor working upon multivalent recognition and signal amplification was developed for ultrasensitive detection of human prostate cancer cells (Du-145). Du-145 is a cell line of prostatic cancer with metastatic potential.30 Immunohistologic studies have revealed overexpression of cluster of differentiation 166 (CD166)31 and human epidermal growth factor receptor 2 (HER2)32 in Du-145 cells. Herein, an anti-CD166 antibody-modified gold electrode was used to capture and recognize target cells (Du-145). Also, a G/GNP/horseradish peroxidase (HRP) conjugated anti-HER2 antibody (G/GNP/Ab-HRP) hybrid nanostructure was designed as a nanoprobe for accurate recognition of target cells and efficient amplification of enzymatic signals. The electrochemical cytosensor that we developed exhibited a unique performance for the detection of cancer cells.2. Experimental
2.1. Chemicals and materials
All chemical materials and reagents in this study were supplied from Sigma-Aldrich (Saint Louis, MO, USA). GO was prepared according to our previous work.33 All chemical materials and reagents were used in their initial form without further purification or manipulation. HRP–HER2 and CD166 monoclonal antibody were purchased from Abcam (Cambridge, MA, USA). Deionized water (DI) with a resistivity of ∼18 MΩ cm was achieved using a Millipore (Bedford, MA, USA) system and used throughout all experiments. 293T, L02, HepG2 and MCF-7 lines were purchased from American Type Culture Collection (Manassas, VA, USA).2.2. Synthesis of gold/reduced GO composite
GNPs were decorated on GO sheets through direct chemical reduction of chloroauric acid (HAuCl4) by sodium borohydride (NaBH4). Briefly, 200 μl of HAuCl4 (0.02 M) was gradually added to GO solution (1 mg ml−1) and the mixture was stirred for 2 h at room temperature. Then, 0.1 ml (0.1 M) NaBH4 was gently poured into the mixture and stirring maintained for 1 h at room temperature. Finally, the obtained sample was repeatedly washed with DI water thrice.2.3. Preparation of G/GNP/Ab-HRP nanoprobes
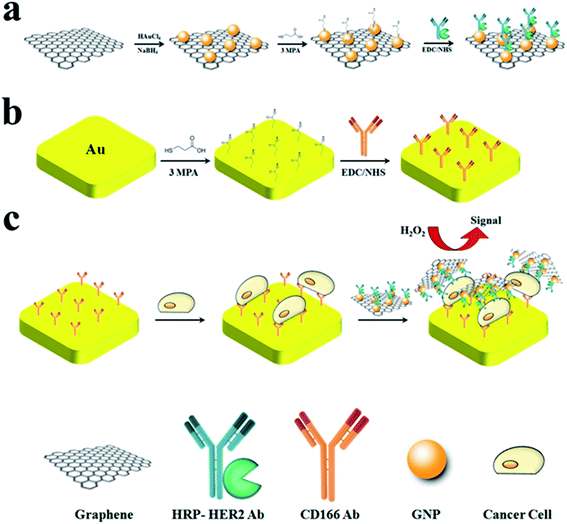
G/GNP/Ab-HRP nanoprobes were achieved by introducing 3-mercaptopropionic acid (3MPA) to G/GNP (Scheme 1a). Typically, 2.5 ml 3MPA solution in phosphate-buffered saline (PBS; 0.1 mM, pH = 7, with 0.1 mg ml−1 Tween 20) was added to 2.5 ml G/GNP (1 mg ml−1) and the mixture was stirred for 5 h at room temperature. Subsequently, the obtained mixture was centrifuged and repeatedly washed with PBS to remove excess reagent. Afterwards, 3MPA-modified G/GNP was incubated with 100 mM 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide and N-hydroxysuccinimide (EDC/NHS) for 10 min. Meanwhile, 0.4 mg ml−1 monoclonal HRP–HER2 antibody was injected into the mixture and maintained in 4 °C under an inert atmosphere for 2 h. The resulting G/GNP/Ab-HRP nanoprobes were collected and washed by centrifugation, and finally re-dispersed in PBS (0.1 mM, pH = 7, with 0.1 mg ml−1 Tween 20).2.4. Cell culture
Human prostate cancer cell line Du-145 was obtained from American Type Culture Collection. Cells were maintained in RPMI 1640 supplemented with 10% fetal bovine serum, 100 IU ml−1 penicillin, and 100 μg ml−1 streptomycin at 37 °C and 5% CO2. Trypsinization was used to obtain cell suspensions and a blood counting chamber was applied for determination of cell number.2.5. Flow cytometry
Flow cytometry was employed to consider the formation of binding between Du-145 cells and nanoprobes. In a typical experiment, G/GNP and G/GNP/Ab-HRP nanoprobes were labeled with fluorescein isothiocyanate. Subsequently, 10 μl of the labeled nanoprobes were incubated with Du-145 cells for 1 h.34 After that, cells were thoroughly washed with PBS. Finally, they were trypsinized and analyzed using a flow cytometry system (Sysmex Partec, Görlitz, Germany).2.6. Cytosensor fabrication
First, a gold working electrode (diameter, 2 mm) was polished with alumina slurry (0.3 μm) to obtain a mirror-like surface. Afterwards, the electrode was successively immersed in ethanol, acetone, piranha solution (H2SO4/H2O2 = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and DI water and sonicated for 3 min in each solution. The clean electrode was immediately immersed in 3MPA solution (0.5 mM, pH = 11) for 2 h at room temperature. Subsequently, the 3MPA-modified electrode was washed with DI water and immersed in 0.1 mg ml−1 CD166 antibody solution in the presence of EDC/NHS (100 mM) for 24 h under an inert atmosphere. Afterwards, the CD166-modified electrode was washed with DI water to remove excess reagent (Scheme 1b). Then, the electrode was immersed in different concentrations of cell suspension for 1 h at 37 °C. The resulting Du-145 cell/antibody/gold electrode was washed and incubated with G/GNP/Ab-HRP nanoprobes for 1 h. Eventually, the modified electrode was used for subsequent electrochemical measurements (Scheme 1c).
1) and DI water and sonicated for 3 min in each solution. The clean electrode was immediately immersed in 3MPA solution (0.5 mM, pH = 11) for 2 h at room temperature. Subsequently, the 3MPA-modified electrode was washed with DI water and immersed in 0.1 mg ml−1 CD166 antibody solution in the presence of EDC/NHS (100 mM) for 24 h under an inert atmosphere. Afterwards, the CD166-modified electrode was washed with DI water to remove excess reagent (Scheme 1b). Then, the electrode was immersed in different concentrations of cell suspension for 1 h at 37 °C. The resulting Du-145 cell/antibody/gold electrode was washed and incubated with G/GNP/Ab-HRP nanoprobes for 1 h. Eventually, the modified electrode was used for subsequent electrochemical measurements (Scheme 1c).
2.7. Characterization
Electrochemical measurements were carried out using an electrochemical interface and impedance analyzer (IVIUMSTAT.XR; Ivium Technologies, Eindhoven, the Netherlands). The morphology of samples was thoroughly investigated by transmission electron microscopy (TEM; EM208 transmission electron microscope; Philips, Amsterdam, the Netherlands) and atomic force microscopy (AFM; 0201/A atomic force microscope; Ara Research, Tehran, Iran). Ultraviolet-Visible (UV-Vis) spectra measurements were carried out on an AvaSpec 2048TEC spectrophotometer (Avantes, Apeldoorn, the Netherlands). X-ray diffraction (XRD) spectroscopy was performed with a step size of 0.02(2θ)/s with Cu Kα radiation using an X'Pert PRO MPD diffractometer (PANalytical, Almelo, the Netherlands).3. Results and discussion
3.1. Working principle of the electrochemical cytosensor
The electrochemical cytosensor based on a signal-amplification and dual-recognition strategy is presented in Scheme 1. The gold/reduced GO composite was prepared first owing to its catalytic properties and excellent biocompatibility. The nanoprobes were fabricated by introducing HRP–HER2 antibody to G/GNP (Scheme 1a). Then, CD166 antibody was attached to the surface of the working electrode for cell capture (Scheme 1b). Afterwards, the resulting cell/antibody/gold electrode was incubated with G/GNP/Ab-HRP nanoprobes to form a “sandwich-like” system. Due to the specific recognition and high binding affinity of the antibody, the sandwich-like system could be exclusively formed in the presence of the target cells (Scheme 1c). Electrochemical signals can be enhanced by hydroquinone (HQ), which is generated by the enzymatic reaction of HRP in the presence of hydrogen peroxide (H2O2). The electrochemical signals (DPV peaks) are directly related to the enzymatic reaction of HRP present on the electrode surface. As a result, the number of captured target cells can be estimated. Because of loading of a large number of HRP–HER2 conjugates, as well as the specific recognition and high binding affinity of antibody, the selectivity and sensitivity of this cytosensor was greatly enhanced.3.2. Characterization of nanoprobes
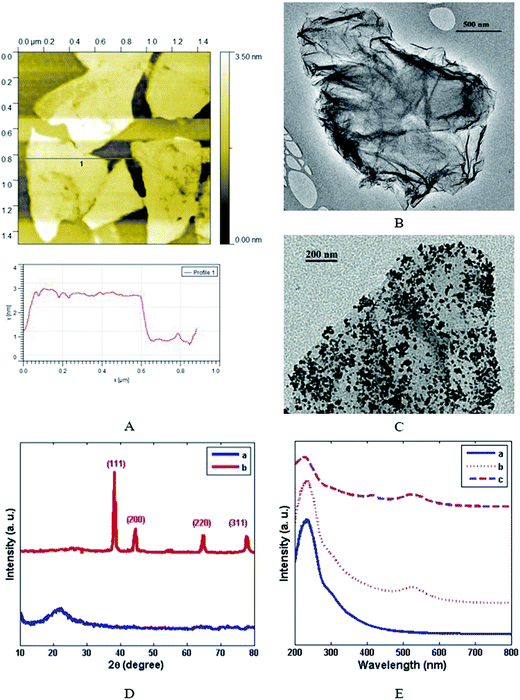
AFM and TEM images of the prepared GO are shown in Fig. 1A and B, respectively. As can be observed, the pristine graphite sheets were thoroughly exfoliated into single-layer GO nanosheets. In addition, the thickness of the obtained GO nanosheets was found to be ∼2 nm by AFM. With regard to the lateral width of G sheets, the size distribution was estimated to be 100 nm to 800 nm. Fig. 2C shows the TEM image of GNPs and GO composite: GNPs were uniformly deposited on the reduced GO sheets without obvious agglomeration. The average size of the deposited GNPs was found to be about 10 ± 1 nm. The XRD patterns of GO and G/GNP are shown in Fig. 1D. The distinctive peak at 2θ ∼ 26.01° is related to the (002) plane of graphite (curve a). As the XRD pattern of G/GNP clearly shows, the appearance of additional peaks at 2θ ∼ 38.3, 44.4, 64.7 and 77.7° correspond to the (111), (200), (220) and (311) planes of the GNPs, respectively (curve b).35 The UV-Vis absorption spectra of GO, G/GNP and G/GNP/Ab-HRP are shown in Fig. 1E. GO showed two absorption peaks: a strong absorption peak at ∼230 nm and a shoulder at ∼300 nm corresponding to the π–π* transition of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and n–π* transitions of C
C and n–π* transitions of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, respectively (curve a).36 G/GNP showed a new band at ∼520 nm (curve b), which was ascribed to the LSPR peak of GNP.37 In addition, due to the partial reduction of GO, the absorption peaks at ∼230 and ∼300 nm disappeared. The absorption spectra of G/GNP/Ab-HRP showed a LSPR peak at ∼525 nm and low-intensity band at ∼410 nm (curve c). These results confirmed that Ab-HRP was effectively conjugated onto G/GNP. In accordance with its excellent affinity, appropriate biocompatibility and large surface area, G/GNP was favored for increasing the loading of Ab-HRP, which improves amplification of enzymatic signals.
O, respectively (curve a).36 G/GNP showed a new band at ∼520 nm (curve b), which was ascribed to the LSPR peak of GNP.37 In addition, due to the partial reduction of GO, the absorption peaks at ∼230 and ∼300 nm disappeared. The absorption spectra of G/GNP/Ab-HRP showed a LSPR peak at ∼525 nm and low-intensity band at ∼410 nm (curve c). These results confirmed that Ab-HRP was effectively conjugated onto G/GNP. In accordance with its excellent affinity, appropriate biocompatibility and large surface area, G/GNP was favored for increasing the loading of Ab-HRP, which improves amplification of enzymatic signals.
Flow cytometry was also employed to consider the formation of binding between Du-145 cells and nanoprobes. As presented in Fig. S1,† the results of flow cytometry show that the connection between G/GNP and Du-145 cells was estimated to be ∼5% (Fig. S1A†); however, the connection between Du-145 cells and G/GNP/Ab-HRP was >90% (Fig. S1B†), indicating high affinity between Du-145 cells and the designed nanoprobes. The high affinity between Du-145 cells and G/GNP/Ab-HRP nanoprobes can be explained by the membrane overexpression of HER2 on the surface of Du-145 cells that has been reported in many studies.32
3.3. Electrochemical behavior of the biosensor
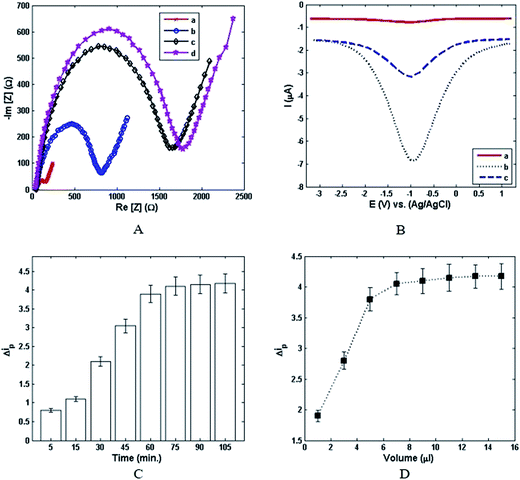
The stepwise assembly processes of the modified electrode were confirmed by electrochemical impedance spectroscopy (EIS). Fig. 2A shows the Nyquist plots of the gold electrode which was gradually modified step by step according to the Experimental section. A minuscule semicircle domain for the bare gold electrode was observed (curve a), implying a fast electron-transfer process. After that, CD166 was assembled on the gold electrode. A conspicuous escalation of the semicircle diameter was also observed (curve b), indicating a noticeable augmentation of electrochemical impedance. Moreover, the apparent increase in the resistance with sequential assembly of cancer cells (curve c) could be discerned. Eventually, as can be seen in Fig. 2A (curve d), increasing the electrochemical impedance after incubation of nanoprobes verified stepwise alteration in impedance. These results revealed accomplishment of an antibody–cell–nanoprobe sandwich-like architecture. Thus, EIS is an appropriate technique for highly sensitive detection of cancer cells. Apart from electrochemical impedance, DPV was employed as a supplementary technique for further investigation of the signal-amplification strategy. As shown in Fig. 2B, a very small and weak DPV current peak was observed when the G/GNP/Ab-HRP/Du-145 cell/CD166/gold electrode was placed in PBS (pH 7.0, 0.1 M) as an electrolyte (curve a). Whereas, by immersing the G/GNP/Ab-HRP/Du-145 cell/CD166/gold electrode in PBS in the presence of 1.5 mM H2O2, the intensification of the DPV current density was sharply increased (curve b). This phenomenon implies that the initiation of this reaction completely depends on the signal-catalysis of HRP in the presence of H2O2. The electrochemical characteristics of the proposed cytosensor were further examined in 100 mM PBS containing 1.5 mM H2O2. The obvious reduction of current density (curve c) was due to the incubation of CD166/gold electrode with G/GNP/Ab-HRP nanoprobes in the absence of Du-145 cells. Also, the DPV signal was determined by the total number of G/GNP/Ab-HRP nanoprobes attached to captured cancer cells. Thus, it can be concluded that the nanoprobes have a pivotal role not only for amplification of current density, but also for increasing the sensitivity of detection of Du-145 cells. It was also found that recognition of the specific nanoprobes on targeted cells as well as capture of cells on the CD166-modified gold surface could be affected by incubation time. The alteration of peak current density (Δip) with incubation time is shown in Fig. 2C. It can be seen that a minor increase in the incubation time led to a significant increase in Δip. However, the current signal reached a plateau after 75 min even if a longer incubation time was employed. Therefore, the desired incubation time for cell capture and nanoprobe incubation was found to be ∼75 min. In addition to the results mentioned above, the impact of the volume of nanoprobe solution was further investigated for the proposed cytosensor. Fig. 2D shows the relationship between Δip and the volume of nanoprobe solution. Accordingly, the intensity of peak current density linearly increased with increasing the volume of nanoprobe solution until it reached ∼7 μl. Then, from 7 μl to 15 μl, the plot revealed that the intensity of peak current density was approximately independent from the volume of nanoprobe solution. With regard to this behavior, the optimum volume solution for the nanoprobe is estimated to be ∼7 μl for the designed cytosensor.3.4. Detection of target cells
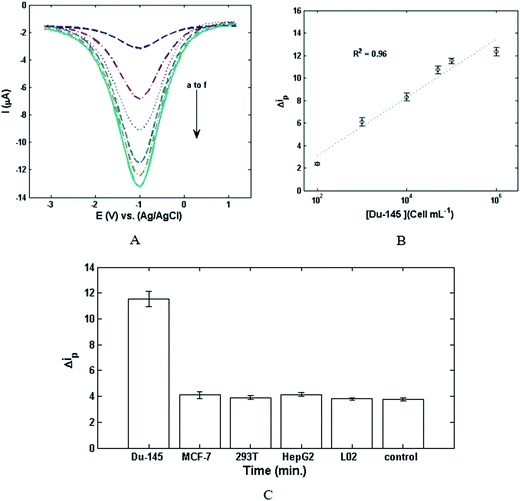
The responses of DPV and the linear calibration of Δip versus the concentration of Du-145 cells are illustrated in Fig. 3A and B. Fig. 3A shows that DPV responses increase with augmentation of the concentration of cancer cells. Fig. 3B exhibits an excellent linear relationship between Δip and the logarithmic concentration of the cancer cells ranging from 102 cells per ml to 106 cells per ml. The related linear regression equation is Δip (μA) = 2.0652 + 1.1213![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C cells (cells per ml) with a correlation coefficient of 0.96 (n = 5). Limit of detection (LOD) is defined as the lowest possible concentration of analyte which can be distinguished without any assurance about the imprecision of the obtained results. Herein, LOD was calculated from the following equation:37
C cells (cells per ml) with a correlation coefficient of 0.96 (n = 5). Limit of detection (LOD) is defined as the lowest possible concentration of analyte which can be distinguished without any assurance about the imprecision of the obtained results. Herein, LOD was calculated from the following equation:37| LOD = 3σ/b | (1) |
4. Conclusion
In this study, an electrochemical-based cytosensor via a dual functionality (i.e., specific recognition and signal amplification for Du-145 human prostate cancer cells) was successfully developed. The proposed cytosensor demonstrated high sensitivity and selectivity toward Du-145 cancer cells, a conveniently low LOD of 20 cells per ml with decreasing repeatability, and an extended linear range from 102 to 106 cells per ml. Compared to cytosensors reported previously, the designed electrochemical biosensor showed remarkable performance for detection of cancer cells. Owing to the above-mentioned considerations, our electrochemical cytosensor is a significant development for detection of cancer cells, and has great potential for the early diagnosis of cancer. Nevertheless, this electrochemical-based biosensor is still in the development phase. Thus, many more comprehensive studies should be done to implement a sensitive and selective electrochemical approach for commercial laboratories and real-world medical and biomedical applications.Abbreviations
| AFM | Atomic force microscopy |
| GO | Graphene oxide |
| Du-145 | Human prostatic metastatic cancer cell |
| HRP | Horseradish peroxidase |
| LOD | Limit of detection |
| GNP | Gold nanoparticle |
| DPV | Differential pulse voltammetry |
| EIS | Electrochemical impedance spectroscopy |
| HQ | Hydroquinone |
| MCF-7 | Human breast cancer cell |
| 293T | Human embryonic kidney cell |
| L02 | Human normal hepatocyte cell |
| HepG2 | Human liver hepatocellular carcinoma |
| TEM | Transmission electron microscopy |
| XRD | X-ray diffraction |
| LSPR | Localized surface plasmon resonance |
Acknowledgements
This work was supported by the Shahid Beheshti University of Medical Sciences (Grant No. 7655).Notes and references
- T. K. Sharma, R. Ramanathan, R. Rakwal, G. K. Agrawal and V. Bansal, Proteomics, 2015, 15, 1680–1692 CrossRef CAS PubMed.
- B. Byrne, E. Stack, N. Gilmartin and R. O'Kennedy, Sensors, 2009, 9, 4407–4445 CrossRef CAS PubMed.
- M. Omidi, M. Malakoutian, M. Choolaei, F. Oroojalian, F. Haghiralsadat and F. Yazdian, Chin. Phys. Lett., 2013, 30, 068701 CrossRef.
- J.-J. Zhang, M.-M. Gu, T.-T. Zheng and J.-J. Zhu, Anal. Chem., 2009, 81, 6641–6648 CrossRef CAS PubMed.
- T.-T. Zheng, R. Zhang, L. Zou and J.-J. Zhu, Analyst, 2012, 137, 1316–1318 RSC.
- C. Hu, D.-P. Yang, Z. Wang, P. Huang, X. Wang, D. Chen, D. Cui, M. Yang and N. Jia, Biosens. Bioelectron., 2013, 41, 656–662 CrossRef CAS PubMed.
- S. Zhang, L. Zhang, X. Zhang, P. Yang and J. Cai, Analyst, 2014, 139, 3629–3635 RSC.
- W. Cheng, L. Ding, J. Lei, S. Ding and H. Ju, Anal. Chem., 2008, 80, 3867–3872 CrossRef CAS PubMed.
- L. Ding, Q. Ji, R. Qian, W. Cheng and H. Ju, Anal. Chem., 2010, 82, 1292–1298 CrossRef CAS PubMed.
- X. Chen, Y. Wang, Y. Zhang, Z. Chen, Y. Liu, Z. Li and J. Li, Anal. Chem., 2014, 86, 4278–4286 CrossRef CAS PubMed.
- X. Wang, J. Ju, J. Li, J. Li, Q. Qian, C. Mao and J. Shen, Electrochim. Acta, 2014, 123, 511–517 CrossRef CAS.
- X. Zhu, J. Yang, M. Liu, Y. Wu, Z. Shen and G. Li, Anal. Chim. Acta, 2013, 764, 59–63 CrossRef CAS PubMed.
- L. Ding, W. Cheng, X. Wang, S. Ding and H. Ju, J. Am. Chem. Soc., 2008, 130, 7224–7225 CrossRef CAS PubMed.
- H. Liu, S. Xu, Z. He, A. Deng and J.-J. Zhu, Anal. Chem., 2013, 85, 3385–3392 CrossRef CAS PubMed.
- C. Ding, S. Wei and H. Liu, Chem.–Eur. J., 2012, 18, 7263–7268 CrossRef CAS PubMed.
- H.-W. Shi, M.-S. Wu, Y. Du, J.-J. Xu and H.-Y. Chen, Biosens. Bioelectron., 2014, 55, 459–463 CrossRef CAS PubMed.
- A. G. Gehring, C. G. Crawford, R. S. Mazenko, L. J. Van Houten and J. D. Brewster, J. Immunol. Methods, 1996, 195, 15–25 CrossRef CAS PubMed.
- S. M. Radke and E. C. Alocilja, Biosens. Bioelectron., 2005, 20, 1662–1667 CrossRef CAS PubMed.
- K. Dill, L. H. Stanker and C. R. Young, J. Biochem. Biophys. Methods, 1999, 41, 61–67 CrossRef CAS PubMed.
- M. Hnaiein, W. Hassen, A. Abdelghani, C. Fournier-Wirth, J. Coste, F. Bessueille, D. Leonard and N. Jaffrezic-Renault, Electrochem. Commun., 2008, 10, 1152–1154 CrossRef CAS.
- D. Sun, J. Lu, Z. Chen, Y. Yu and M. Mo, Anal. Chim. Acta, 2015, 885, 166–173 CrossRef CAS PubMed.
- M. S. Artiles, C. S. Rout and T. S. Fisher, Adv. Drug Delivery Rev., 2011, 63, 1352–1360 CrossRef CAS PubMed.
- A. Yadegari, M. Omidi, M. Choolaei, F. Haghiralsadat and F. Yazdian, Procedia Eng., 2014, 87, 967–970 CrossRef CAS.
- M. Omidi, A. Fathinia, M. Farahani, Z. Niknam, A. Yadegari, M. Hashemi, H. Jazayeri, H. Zali, M. Zahedinik and L. Tayebi, Advanced 2D Materials, 2016, pp. 433–471 Search PubMed.
- G. Yang, J. Cao, L. Li, R. K. Rana and J.-J. Zhu, Carbon, 2013, 51, 124–133 CrossRef CAS.
- P. Sharma, S. K. Tuteja, V. Bhalla, G. Shekhawat, V. P. Dravid and C. R. Suri, Biosens. Bioelectron., 2013, 39, 99–105 CrossRef CAS PubMed.
- X. Luo, A. Morrin, A. J. Killard and M. R. Smyth, Electroanalysis, 2006, 18, 319–326 CrossRef CAS.
- W. Hong, H. Bai, Y. Xu, Z. Yao, Z. Gu and G. Shi, J. Phys. Chem. C, 2010, 114, 1822–1826 CAS.
- K.-J. Huang, D.-J. Niu, J.-Y. Sun and J.-J. Zhu, J. Electroanal. Chem., 2011, 656, 72–77 CrossRef CAS.
- S. M. Pulukuri, C. S. Gondi, S. S. Lakka, A. Jutla, N. Estes, M. Gujrati and J. S. Rao, J. Biol. Chem., 2005, 280, 36529–36540 CrossRef CAS PubMed.
- A. Roth, D. C. Drummond, F. Conrad, M. E. Hayes, D. B. Kirpotin, C. C. Benz, J. D. Marks and B. Liu, Mol. Cancer Ther., 2007, 6, 2737–2746 CrossRef CAS PubMed.
- D. Carrión-Salip, C. Panosa, J. A. Menendez, T. Puig, G. Oliveras, A. Pandiella, R. De Llorens and A. Massaguer, Int. J. Oncol., 2012, 41, 1128–1138 Search PubMed.
- M. Hashemi, A. Yadegari, G. Yazdanpanah, S. Jabbehdari, M. Omidi and L. Tayebi, RSC Adv., 2016, 6, 74072–74084 RSC.
- Q. Mu, G. Su, L. Li, B. O. Gilbertson, L. H. Yu, Q. Zhang, Y.-P. Sun and B. Yan, ACS Appl. Mater. Interfaces, 2012, 4, 2259–2266 CAS.
- J. Zhang, J. Lei, R. Pan, C. Leng, Z. Hu and H. Ju, Chem. Commun., 2011, 47, 668–670 RSC.
- M. Hashemi, A. Yadegari, G. Yazdanpanah, M. Omidi, S. Jabbehdari, F. Haghiralsadat, F. Yazdian and L. Tayebi, Biotechnol. Appl. Biochem., 2016 DOI:10.1002/bab.1487.
- M. Omidi, G. Amoabediny, F. Yazdian and M. Habibi-Rezaei, EPL, 2015, 109, 18005 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra25938c |
| This journal is © The Royal Society of Chemistry 2017 |