Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Effective gene-silencing of siRNAs that contain functionalized spacer linkages within the central region†
Jean-Paul Desaulniers *,
Gordon Hagen,
Jocelyn Anderson,
Chris McKim and
Blake Roberts
*,
Gordon Hagen,
Jocelyn Anderson,
Chris McKim and
Blake Roberts
University of Ontario Institute of Technology, Faculty of Science, 2000 Simcoe Street North, Oshawa, ON L1H 7K4, Canada. E-mail: Jean-Paul.Desaulniers@uoit.ca
First published on 13th January 2017
Abstract
Short-interfering RNAs containing a variety of functional groups at the central region of the sense strand were synthesized and evaluated. The gene-silencing data suggest that these siRNAs are biocompatible and are very effective in cell-based assays.
Short-interfering RNAs are double-stranded RNAs that are capable of entering the endogenous RNA interference (RNAi) pathway to silence gene expression.1 This has enormous potential for modern medicine as several candidate siRNAs are showing promise, and some are progressing in clinical trials.2–5 Chemical modification of siRNAs is necessary to improve their pharmacokinetic properties, in addition to ameliorate stability.6–8 However, no universal chemical modification exists that is capable of addressing all the challenges of utilizing siRNAs as potential therapeutics.
Commonly utilized commercially-available chemical modifications within siRNAs include phosphorothioate backbone linkages, and sugar modifications such as the 2′-O-methyl and 2′-fluoro modification.6,7 Although many other modifications exist in the literature, several of them are not commercially available, and thus rely on in-house multi-step phosphoramidite synthesis.9–11
With this limitation in mind, we sought to generate modified siRNAs using easily synthesizable phosphoramidites. In our previous study, we identified a class of functional RNAs that contained simple carbon-based and ethylene glycol-based spacer linkages within the central region of the siRNA on the passenger strand.12 We noted that siRNAs bearing spacer linkages between eight or nine atoms (equivalent of two nucleosides) within the central region of siRNAs were particularly effective at gene-silencing. These siRNAs were thermally destabilizing, yet were able to function with a high degree of potency.
In this study, we explored new siRNA design, and report the straightforward synthesis of phosphoramidites bearing a variety of functional groups, followed by its incorporation within the central region area of the sense strand of siRNAs. These modified siRNAs were evaluated for their gene-silencing potential within the RNAi pathway. To the best of our knowledge, this is the first time that the effect of the described functionalized spacer modifications have been used within the central region of siRNAs.
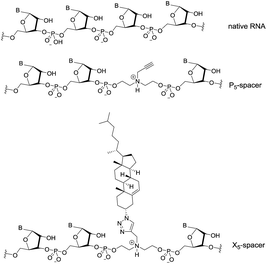
Based on prior reports, and the principle of thermodynamic asymmetry,13,14 a variety of abasic linkers bearing different functional groups were chosen to replace the central region of the siRNA. In Fig. 1, the P5- and X5-spacers contain five atoms between the oxygens of the phosphodiester linkages, which approximates the distance between a single nucleotide in native RNA. The P5-spacer contains a propargyl group, and the X5-spacer contains a cholesterol derivative bound covalently via a triazole linkage.
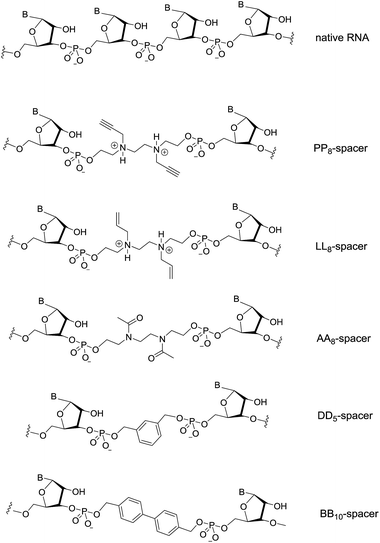
In addition, spacers bearing a variety of functional groups that span two nucleosides were also investigated and are illustrated in Fig. 2. The distance of atoms spanning two nucleosides in native RNA is nine atoms. Three of the spacers used in this study contain eight atoms, and contain either two propargyl groups (PP8-spacer), two allyl groups (LL8-spacer) or two acetyl groups (AA8-spacer). In addition, two aromatic groups were also used to determine whether conformationally rigid aromatic compounds would be useful scaffolds when incorporated within siRNAs (DD5- and BB10-spacer).
In order to synthesize various siRNAs bearing these modifications at the central region, a variety of phosphoramidites were synthesized that are compatible with solid-phase RNA synthesis.15 In general, the basic building blocks were inexpensive and commercially available.
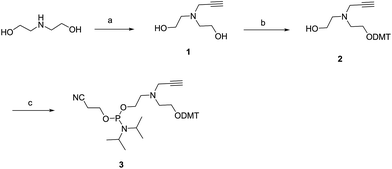
As described in Scheme 1, the nitrogen of diethanolamine was alkylated with propargyl bromide in the presence of K2CO3 to afford compound 1 in 32% yield. Subsequent reaction of the molecule with 4,4′-dimethoxytrityl chloride (DMT-Cl) afforded monoalcohol 2. Monoalcohol 2 was phosphytilated with 2-cyanoethyldiisopropylphosphoramidochloride to afford phosphoramidite 3.
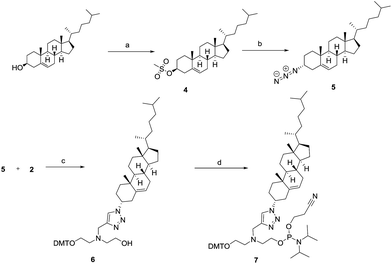
For the synthesis of phosphoramidite 7 (Scheme 2), cholesterol was first mesylated to afford compound 4 in excellent yield. Subsequent displacement of the mesyl group with the azido group afforded the azido-cholesterol compound 5.16 Finally, compound 5 reacted with alkyne 2 to afford triazole 6 in good yield. Furthermore, phosphitylation of compound 6 afforded phosphoramidite 7.
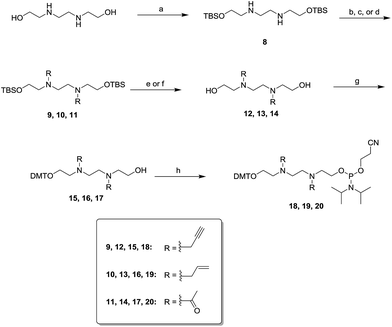
Scheme 3 describes the synthesis of spacer linkers spanning eight atoms. These spacer linkers were functionalized with alkynyl, allyl or acetyl groups. tert-Butyldimethylsilyl (TBS)-protected N,N′-bis(2-hydroxyethyl)ethylenediamine reacted with propargyl bromide, allyl bromide, or acetic anhydride to afford compounds 9, 10 and 11, respectively. Desilylation of compounds 9–11 was accomplished with tetrabutylammonium fluoride (TBAF) or hydrofluoric acid (HF) to afford diols 12–14. The diols were reacted with DMT-Cl to afford the mono-DMT protected alcohols 15–17. Phosphytilation of 15–17 with 2-cyanoethyldiisopropylchlorophosphoramidite in the presence of diisopropylethylamine (DIPEA) generated the phosphoramidites 18–20 in good yield.
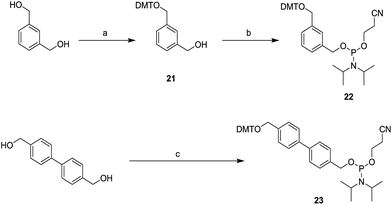
As is described in Scheme 4, to generate the aromatic phosphoramidites 22 and 23, 1,3-benzenedimethanol reacted with DMT-Cl to afford the monoalcohol 21. Phosphitylation of monoalcohol 21 with 2-cyanoethyldiisopropylchlorophosphoramidite generated phosphoramidite 22 in good yield. 4,4-bis(hydroxymethyl)biphenyl was also reacted with DMT-Cl, however, its monoalcohol was not stable. Therefore, the monoalcohol that was generated was immediately reacted with 2-cyanoethyldiisopropylchlorphosphoramidite to afford phosphoramidite 23 in good yield.
A library of seven different phosphoramidites were synthesized in relatively few steps. From these, various siRNAs were synthesized that contain a combination of the functional groups highlighted in the above schemes within the central region of siRNAs. These siRNAs were synthesized to target either firefly luciferase mRNA, or endogenous gene target BCL2 mRNA in mammalian cells. The siRNAs were gel purified and characterized by mass spectrometry (see ESI†). These siRNAs are highlighted in Table 1.
| RNA | siRNA duplex | Target | Tm | ΔTm |
|---|---|---|---|---|
| a P5 corresponds to the single propargyl modification; X5 corresponds to the single cholesterol modification; AA8 corresponds to the double acetyl modification; LL8 corresponds to the double allyl modification; PP8 corresponds to the double propargyl modification. DD5 corresponds to the benzene modification; BB10 corresponds to the biphenyl modification. The top strand corresponds to the sense strand; the bottom strand corresponds to the antisense strand. In all duplexes, the 5′-end of the bottom antisense strand contains a 5′-phosphate group. | ||||
| wtLuc | 5′-CUUACGCUGAGUACUUCGAtt-3′ | Luciferase | 72.7 | — |
| 3′-ttGAAUGCGACUCAUGAAGCU-5′ | ||||
| 24 | 5′-CUUACGCUP5AGUACUUCGAtt-3′ | Luciferase | 69.1 | −3.6 |
| 3′-ttGAAUGCGACUCAUGAAGCU-5′ | ||||
| 25 | 5′-CUUACGCUGP5GUACUUCGAtt-3′ | Luciferase | 70.2 | −2.5 |
| 3′-ttGAAUGCGACUCAUGAAGCU-5′ | ||||
| 26 | 5′-CUUACGCUX5AGUACUUCGAtt-3′ | Luciferase | 61.6 | −11.1 |
| 3′-ttGAAUGCGACUCAUGAAGCU-5′ | ||||
| 27 | 5′-CUUACGCUGX5GUACUUCGAtt-3′ | Luciferase | 62.5 | −10.2 |
| 3′-ttGAAUGCGACUCAUGAAGCU-5′ | ||||
| 28 | 5′-CUUACGCUAA8GUACUUCGAtt-3′ | Luciferase | 55.1 | −17.6 |
| 3′-ttGAAUGCGACUCAUGAAGCU-5′ | ||||
| 29 | 5′-CUUACGCULL8GUACUUCGAtt-3′ | Luciferase | 65.9 | −6.8 |
| 3′-ttGAAUGCGACUCAUGAAGCU-5′ | ||||
| 30 | 5′-CUUACGCUPP8GUACUUCGAtt-3′ | Luciferase | 64.6 | −8.1 |
| 3′-ttGAAUGCGACUCAUGAAGCU-5′ | ||||
| wtBcl2 | 5′-GGUCAUCCAUGACAACUUUtt-3′ | Bcl-2 | 72.5 | — |
| 3′-ttCCAGUAGGUACUGUUGAAA-5′ | ||||
| 31 | 5′-GGUCAUCCDD5GACAACUUUtt-3′ | Bcl-2 | 66.1 | −6.4 |
| 3′- ttCCAGUAGGUACUGUUGAAA-5′ | ||||
| 32 | 5′-GGUCAUCCBB10GACAACUUUtt-3′ | Bcl-2 | 57.2 | −15.3 |
| 3′-ttCCAGUAGGUACUGUUGAAA-5′ | ||||
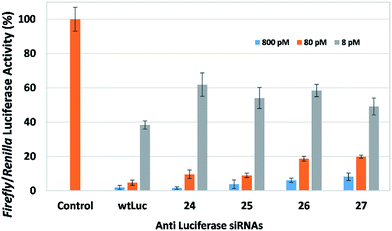
We first examined the effect of placing modifications that replace a single nucleotide within the polymeric RNA strand. In this study, we used the propargyl spacer (P5) and cholesterol-derivatized spacer (X5) within the central region of the sense strand of siRNAs that target firefly luciferase. Placing both of these modifications within functional siRNAs at either position 9 or 10 (counting from the 5′-end of the sense strand) resulted in efficient dose-dependent downregulation (Fig. 3). In fact, siRNAs 24 and 25 which contain the propargyl group downregulates firefly luciferase at levels comparable to wild-type siRNA wtLuc. The cholesterol-modified siRNAs 26 and 27 downregulate, however at a slightly reduced efficacy compared to wtLuc. Nevertheless, this result is exciting because it highlights the compatibility of employing simple small functional groups and molecules within this area. SiRNAs 24–27 are thermally destabilizing, as exhibited by their melting temperature (Tm) data highlighted in Table 1. The cholesterol-containing siRNAs (26 and 27) are much more destabilizing (ca. −10 °C) compared to siRNAs containing the propargyl group (ca. −2.5 °C) (24 and 25). This is not surprising as cholesterol is a relatively large bulky group. Prior work from our laboratory involving an siRNA bearing a cholesterol-derivatized triazole moiety also showed significant thermal destabilization, despite a favorable gene silencing biological response.17
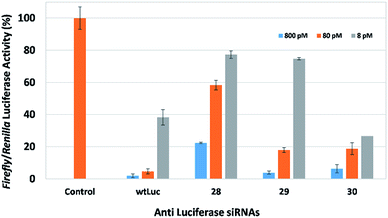
We next turned our attention to the derivatized spacer molecules that replace two consecutive nucleoside derivatives. With respect to flexible aliphatic spacers spanning positions 9 and 10 of the sense strand, we observed dose dependent gene-silencing with siRNAs 28, 29, and 30, which contain two acetyl groups, two propargyl groups, and two allyl groups, respectively (Fig. 4). SiRNA 28, was the least effective compared to wtLuc, whereas siRNAs 29 and 30 were quite effective, exhibiting gene-silencing comparable to wtLuc. Examination of the structural differences between the acetyl-modified siRNAs and both the propargyl- and allyl-modified siRNAs (29 and 30) reveal changes in their overall charge at physiological pH (Fig. 2). Both siRNAs 29 and 30 have two nitrogens which will be protonated, and thus positively charged. Whereas the acetyl-modified siRNA 28 will not be protonated due to the properties of the amide-bond. In addition, amide-bonds are capable of forming rotamers,18 which can allow for molecules to exhibit more than one conformational state. As a consequence, this may cause the siRNAs to adopt a variety of conformationally distinct structures. Melting temperature data for siRNAs 28–30 show destabilization. However, in the case of acetyl-modified siRNA 28, destabilization of −17.6 °C is observed, which is significantly more destabilizing compared to siRNAs 29 and 30 (ca. −7 °C to −8 °C). In general though, despite this loss in thermal stability, the addition of all functionalized spacer linkages spanning two nucleosides, the data suggests that the siRNAs act as suitable substrates for the RNAi machinery. In our previous study with siRNAs containing carbon- and ethylene-based spacers at the central region of the sense strand, the change in Tm data compared to wtLuc for these siRNAs were similar to the acetyl-modified siRNA 28.12 Given the protonation of the amine at physiological pH, it is likely that the positive charge of the modified backbone spacer of siRNAs 29 and 30 assist in increasing the Tm, when compared to the comparable modified siRNAs used in our previous study and that of siRNA 28.12
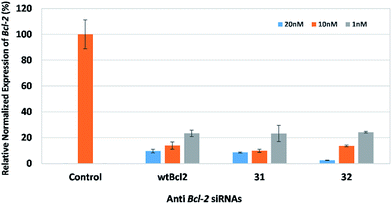
Following these results, we investigated the effect of using aromatic spacer molecules within this region, to see if restricting the degrees of freedom around the core functional group would allow for activity (Fig. 5). In this experiment, we examined the effect of a 1,3-dibenzenemethanol derivative, and a 4,4-biphenyl derivative containing siRNAs (32 and 33) that target the endogenous gene BCL2. BCL2 encodes the Bcl-2 protein, which is important for both type 1 and 2 programmed cell death (apoptosis and autophagy).19,20 BCL2 is viewed as an important endogenous target, and is associated with several clinical cancer types.21,22 Prior work from our research group identified siRNAs bearing triazole-backbone modifications were suitable gene silencing substrates for the endogenous gene BCL2.23 To our delight, siRNAs containing either of these aromatic groups were well accommodated within the central region of the siRNA, and exhibited gene-silencing potency comparable to wild-type wtBcl2.
Conclusions
In conclusion, this study provides evidence to suggest that siRNAs bearing non-cleavable aromatic and alkyl spacers derivatized with a variety of functionality are suitable gene silencing substrates in plasmid-borne and endogenous gene targets. These spacer linkers are particularly attractive due to their ease of synthesis, and their biocompatibility within the RNAi pathway. These modified siRNAs all exhibit classic A-type helices, which is the form that is recognized by the RNA-induced silencing complex (RISC).24 In addition, a reduction in negative charge occurs with several of these polyanionic siRNAs, which may be assisting the siRNA favourably. Furthermore, placing cholesterol at the central region also functions efficiency, and may offer a relatively new method to install cholesterol small molecule centrally within siRNAs. This could have important use in liposome or other related delivery systems.25 For example, cholesterol-labeled siRNAs for liposome packaging are often labelled at one end of the siRNA. An siRNA labelled with a cholesterol moiety at the central region could potentially offer altered and improved packaging into liposomes and/or lipid nanoparticles. Similarly, by altering the hydrophobicity and positive charge of the various linkers used in this study, changes in the physical properties of the siRNA duplex are possible. As such, these changes can potentially improve the packaging of the siRNA into delivery vehicles. Therefore, the use of new functional groups and small molecules at the central region allows for the creation of a new diverse class of biofunctionally active modified siRNAs. Future investigation involves modifying this area with other moieties such as selective tunable molecules, or cell-targeting molecules.Acknowledgements
We acknowledge the National Sciences and Engineering Research Council (NSERC) and the Canada Foundation for Innovation (CFI) for funding.Notes and references
- A. Fire, S. Q. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver and C. C. Mello, Nature, 1998, 391, 806–811 CrossRef CAS PubMed
.
- A. Bouchie, Nat. Biotechnol., 2014, 32, 203–204 CrossRef CAS PubMed
.
- J. K. Nair, J. L. S. Willoughby, A. Chan, K. Charisse, M. R. Alam, Q. F. Wang, M. Hoekstra, P. Kandasamy, A. V. Kel'in, S. Milstein, N. Taneja, J. O'Shea, S. Shaikh, L. G. Zhang, R. J. van der Sluis, M. E. Jung, A. Akinc, R. Hutabarat, S. Kuchimanchi, K. Fitzgerald, T. Zimmermann, T. J. C. van Berkel, M. A. Maier, K. G. Rajeev and M. Manoharan, J. Am. Chem. Soc., 2014, 136, 16958–16961 CrossRef CAS PubMed
.
- S. H. Lee, Y. Y. Kang, H. E. Jang and H. Mok, Adv. Drug Delivery Rev., 2016, 104, 78–92 CrossRef CAS PubMed
.
- T. Coelho, D. Adams, A. Silva, P. Lozeron, P. N. Hawkins, T. Mant, J. Perez, J. Chiesa, S. Warrington, E. Tranter, M. Munisamy, R. Falzone, J. Harrop, J. Cehelsky, B. R. Bettencourt, M. Geissler, J. S. Butler, A. Sehgal, R. E. Meyers, Q. Chen, T. Borland, R. M. Hutabarat, V. A. Clausen, R. Alvarez, K. Fitzgerald, C. Gamba-Vitalo, S. V. Nochur, A. K. Vaishnaw, D. W. Sah, J. A. Gollob and O. B. Suhr, N. Engl. J. Med., 2013, 369, 819–829 CrossRef CAS PubMed
.
- D. R. Corey, J. Clin. Invest., 2007, 117, 3615–3622 CrossRef CAS PubMed
.
- G. F. Deleavey and M. J. Damha, Chem. Biol., 2012, 19, 937–954 CrossRef CAS PubMed
.
- D. A. Braasch, S. Jensen, Y. Liu, K. Kaur, K. Arar, M. A. White and D. R. Corey, Biochemistry, 2003, 42, 7967–7975 CrossRef CAS PubMed
.
- T. Dowler, D. Bergeron, A. L. Tedeschi, L. Paquet, N. Ferrari and M. J. Damha, Nucleic Acids Res., 2006, 34, 1669–1675 CrossRef CAS PubMed
.
- D. Mutisya, C. Selvam, B. D. Lunstad, P. S. Pallan, A. Haas, D. Leake, M. Egli and E. Rozners, Nucleic Acids Res., 2014, 42, 6542–6551 CrossRef CAS PubMed
.
- T. C. Efthymiou, H. Vanthi, J. Oentoro, B. Peel and J.-P. Desaulniers, Bioorg. Med. Chem. Lett., 2012, 22, 1722–1726 CrossRef CAS PubMed
.
- T. C. Efthymiou, B. Peel, V. Huynh and J. P. Desaulniers, Bioorg. Med. Chem. Lett., 2012, 22, 5590–5594 CrossRef CAS PubMed
.
- R. C. Wilson and J. A. Doudna, Ann. Rev. Biophys., ed. K. A. Dill, 2013, vol. 42, pp. 217–239 Search PubMed
.
- D. S. Schwarz, G. Hutvagner, T. Du, Z. Xu, N. Aronin and P. D. Zamore, Cell, 2003, 115, 199–208 CrossRef CAS PubMed
.
- M. H. Caruthers, Science, 1985, 230, 281–285 CAS
.
- D. Neshchadin, F. Palumbo, M. S. Sinicropi, I. Andreu, G. Gescheidt and M.
A. Miranda, Chem. Sci., 2013, 4, 1608–1614 RSC
.
- B. J. Peel, G. Hagen, K. Krishnamurthy and J. P. Desaulniers, ACS Med. Chem. Lett., 2015, 6, 117–122 CrossRef CAS PubMed
.
- A. M. Watkins, R. Bonneau and P. S. Arora, J. Am. Chem. Soc., 2016, 138, 10386–10389 CrossRef CAS PubMed
.
- M. L. Cleary, S. D. Smith and J. Sklar, Cell, 1986, 47, 19–28 CrossRef CAS PubMed
.
- Y. Tsujimoto, J. Cossman, E. Jaffe and C. M. Croce, Science, 1985, 228, 1440–1443 CAS
.
- J. C. Reed, Toxicol. Lett., 1995, 82–83, 155–158 CrossRef CAS PubMed
.
- J. C. Reed, Curr. Opin. Oncol., 1995, 7, 541–546 CrossRef CAS PubMed
.
- G. Hagen, B. J. Peel, J. Samis and J. P. Desaulniers, MedChemComm, 2015, 6, 1210–1215 RSC
.
- H. Kobayashi and Y. Tomari, Biochim. Biophys. Acta, 2016, 1859, 71–81 CrossRef CAS PubMed
.
- S. H. Lee, Y. Y. Kang, H. E. Jang and H. Mok, Adv. Drug Delivery Rev., 2016, 104, 78–92 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra27701b |
| This journal is © The Royal Society of Chemistry 2017 |

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
: