Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
B. amyloliquefaciens TCCC 11319, a new Cr(III)-tolerant bacterium for chromium-tanned leather shaving disposal
Shan Cao abc,
Qinglong Xina,
Shiting Zhouc,
Bin Xuea,
Bing Liud,
Fuping Lua,
Yanping Wangb and
Yu Li*a
abc,
Qinglong Xina,
Shiting Zhouc,
Bin Xuea,
Bing Liud,
Fuping Lua,
Yanping Wangb and
Yu Li*a
aKey Laboratory of Industrial Fermentation Microbiology, College of Biotechnology, Tianjin University of Science and Technology, 415#, Tianjin, 300457, P. R. China. E-mail: cs1988@foxmail.com; liyu@tust.edu.cn; Tel: +86 02260601173
bCollege of Food Engineering and Biotechnology, Tianjin University of Science and Technology, Tianjin, 300457, P. R. China
cCollege of Material Science and Chemical Engineering, Tianjin University of Science and Technology, Tianjin, 300457, P. R. China
dRoom DM3.13 Gorlaeus Laboratory, Leiden, The Netherlands
First published on 15th February 2017
Abstract
A strain of B. amyloliquefaciens, TCCC11319, was isolated from tannery soil and identified by 16S rRNA gene sequencing. The enzyme produced by this strain was successfully applied to the treatment of chromium-tanned leather shavings (CTLS). Furthermore, the species and characteristics of the enzyme, including collagenase, alkaline protease, neutral protease, and keratinase, were analyzed. Under optimized conditions (5% inoculum, pH 8, 37 °C, 32 h), the collagenase activity of the crude broth reached 900 U mL−1 and exhibited relatively high tolerance toward Cr(III) (≤900 mg L−1). Moreover, the conditions and effects of enzyme-treated CTLS disposal were studied. The results showed that more than 95% CTLS was successfully liquefied at 10% enzyme concentration, pH 8, and 35 °C within 24 h. The enzyme has the potential to be reused in leather production processes at an industrial scale to generate value-added products.
1 Introduction
Leather products, such as shoes, bags, and clothes, are widely used. However, the leather production process causes pollution and resource consumption.1 The tanning process involves the conversion of putrescible biological material into leather, which is more resistant to microbial attack and has higher thermal stability. Accordingly, tanning is the most important process in leather production.2 Trivalent chromium salts with sulfate are currently the most important tanning agents, resulting in heavy metal pollution from the leather industry. Every metric ton of raw hide processed generates 200 kg of the final chrome-containing leather product, and 250 kg of chrome-containing tanned waste.3 In other words, the leather yield from raw hides is 25%, while the waste yield can be up to 75%. In China, over 700![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 metric tons of chromium-tanned leather shavings (CTLS) are produced each year. CTLS are composed of collagen cross-linked with small particles of Cr(III) complexes.4 As reported,5 the major disposal routes for tanned wastes are landfill and incineration. Therefore, the recovery and reuse of CTLS is urgently needed, with the impact on cost and environment also requiring consideration.6
000 metric tons of chromium-tanned leather shavings (CTLS) are produced each year. CTLS are composed of collagen cross-linked with small particles of Cr(III) complexes.4 As reported,5 the major disposal routes for tanned wastes are landfill and incineration. Therefore, the recovery and reuse of CTLS is urgently needed, with the impact on cost and environment also requiring consideration.6
Due to their high efficiency, high selectivity, and mild processing conditions, enzymes have received increasing attention for their applications to biodegradation in the leather industry. Waste disposal through enzymatic hydrolysis has environmentally friendly characteristics. The enzymatic hydrolysis of Cr-free solid waste has been studied since 1998.7 Song Jian reported that enzymatic hydrolysis can be accelerated using ultrasound, improving the final conversion ratio of non-tanned shavings to 84.1% at 40 °C.8 Karel Kolomaznik successfully converted chrome shavings into a reducing agent in a laboratory scale trial at a digestion temperature of 70 °C.9 CTLS usually requires pretreatment with alkali or lime (5–6%) at over 60 °C to digest collagen before enzymatic hydrolysis,10 and the mean molecular mass of the hydrolysates being about one-tenth of current gelatins (usually 15–30 kDa).11
Collagenase is an enzyme usually produced by animals or microorganisms, and is highly specific for natural or denatured collagen. The stable triple helix structure of collagen makes it difficult to hydrolyze using other proteases.12 However, denatured gelatin and collagen hydrolysates are easily digested by proteinases. Furthermore, most enzymes are deactivated by heavy metals. Chromium use in the leather tanning process decreases collagenase activity in CTLS waste treatment.13 As a solution, Katsifas identified a chromium-tolerant strain of Aspergillus carbonarius that successfully degraded chromium-containing solid wastes in a solid state fermentation process. The application of this strain to CTLS disposal was proposed to recover Cr(III) for reuse in tanning.14 Further screening studies and research into applications for new types of stain and enzymes due to the chromium-tolerant property are of great importance.
The characteristics of a new chromium-tolerant bacterial strain isolated from tannery soil are elaborated in this paper, and the enzyme it produced was analyzed. Furthermore, the effects of CTLS hydrolysis are evaluated. In the CTLS treatment process, biodegradation is expected to be a promising method for Cr(III) recovery. Furthermore, the chromium-containing regeneration products might be useful in retanning steps and fatliquoring steps. This research helps to both protect the environment and reduce tannery costs.
2 Materials and methods
2.1 Preliminary screening of Cr(III)-tolerant bacterial strain
Chromium-tanned leather shavings (CTLS) and soil samples were collected from a tannery (Dongming Leath. Co., Ltd, Hebei, China). Chromium-tolerant bacteria were isolated from soil samples using a dilution plate technique. A soil sample (1 g) was dissolved in normal saline (9 mL, 0.9%). An aliquot of the diluted suspension (1 mL) was added to a flask containing enrichment medium (50 mL/250 mL). The enrichment medium was composed of gelatin (10 g L−1), gelatin (5 g L−1), and NaCl (5 g L−1) in distilled water at pH 7.2–7.5. The bacteria were primarily cultured using shake flasks at 37 °C for 24 h. The broth was spread on nutrient agar plates comprising beef extract (10 g L−1), yeast (5 g L−1), and NaCl (10 g L−1), and then incubated on Petri plates at 37 °C for 24 h. The growing bacterial colonies were spread on plates and screened. The chemicals used for the culture were all chemically pure reagents (Jiangtian Chem. Co., Ltd, Tianjin, China).2.2 Classification of chromium-tolerant bacterial strain
After incubating the bacteria for 48 h at 37 °C, colony morphologies were observed on plates using the Gram straining method described by Bailey and Scott.15 Biochemicals and physiology were analyzed according to the work of Bergey.16 Moreover, characterization and species identification of the isolated chromium-tolerant bacterial strain was carried out by 16S rRNA gene sequencing. The standard method presented by Sambrook was used to extract bacterial DNA,17 and designed PCR primers 27F (AGAGTTTGATCCTGGCTCAG) and 1492R (TACGG(C/T)TACCTTGT TACGACTT) were used to amplify the 16S rRNA gene. Purification of the genomic DNA strain was carried out using a Wizard Genomic DNA Purification Kit (Promega, Madison, Wi, USA), and the purified DNA strain was used as a template for PCR amplification (denaturation, 30 cycles and 95 °C for 45 s; primer annealing, 56 °C for 30 s; and extension, 72 °C for 1.5 min). PCR products were evaluated on 0.8% agarose gel via amplification on a Primus 25 Advanced PCR machine (Peqlab Biotechnologie, GmbH, Germany). Strain gene sequences were identified by comparison with homologous sequences using the BLAST technique.182.3 Enzymatic assay
Enzyme production was optimized using the following parameters: inoculum ratios, 1%, 2%, 5%, 7%, and 10% (v/v); culture temperatures, 30–45 °C; pH 4–10; and fermentation time, 32 h. Optimum conditions were determined according to collagenase activity. All experiments were carried out in triplicate.The enzymes analyzed were collagenase, keratinase, neutral protease, and alkaline protease. Crude enzyme extract was concentrated by centrifugation (Eppendorf, 5480R, American) at 8000 rpm for 10 min. Collagenase activity was determined using ninhydrin colorimetric analysis for amino acids.19 One unit of collagenase was defined as the amount of enzyme that released 1 μg of glycine from collagen hydrolysis in 1 min, at pH 7.5 and 37 °C, as determined spectrophotometrically from the color intensity.20 Keratinase activity was determined using the method of Letourneau et al.21 One unit of keratinase activity was defined as the amount of enzyme that produced an absorbance change of 0.01 units (OD280). Furthermore, the neutral protease and alkaline protease activity assay was based on the method described by Huang.22 One unit of protease activity was defined as the amount of enzyme that releases 1 μg of tyrosine per minute at 40 °C and pH 10.5 for alkaline protease, and 30 °C and pH 7.5 for neutral protease. Chemicals used for analysis were all analytical grade (Jiangtian Chem. Co., Ltd, Tianjin, China).
2.4 Cr(III) tolerance of bacterial strain and enzyme
Chromium sulfate was used for Cr(III) tolerance analysis and leather production. To resist Cr(III), a bacterial colony was inoculated with 50 mL LB medium (yeast, 5 g L−1; peptone, 10 g L−1; NaCl, 5 g L−1), and grown for 12 h with shaking at 37 °C. After further inoculation (2%) of the LB medium with chromium sulfate (0, 1, 2, 3, 4, and 5 g mL−1), the broth was sampled every 2 h (2, 4, 6, 8, 10, and 12 h from the start) and evaluated by measuring the OD600 at 37 °C.23 To test the tolerance of the crude enzyme, chromium sulfate solutions with different Cr(III) concentrations (0, 0.2, 0.4, 0.6, 0.8, 1.0, 1.2, and 1.4 mg mL−1) were added to the broth and shaken for 24 h before determining the collagenase activity.2.5 Optimization of parameters and analytical procedures for CTLS degradation
Optimization of the crude enzyme involved changing parameters that obviously affected the CTLS degradation, namely temperature, pH, and enzyme dosage. To optimize CTLS enzymatic degradation efficiency, the temperature (30, 37, 45, 52, and 60 °C), pH (5, 6, 7, 8, and 9), and crude enzyme concentration (0.5%, 1%, 1.5%, 2%, and 2.5% (m/m)) were varied. The effects of final CTLS degradation were then investigated under optimized conditions. The hydrolyzed CTLS was characterized after 2 h using field emission scanning electron microscopy (FESEM, Hitachi, S4800, Japan) for microstructure evaluation.3 Results and discussions
3.1 Classification of Cr(III)-tolerant bacteria isolated from tannery
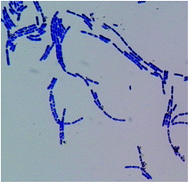
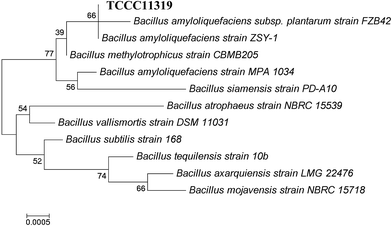
In total, fifty bacterial strains were isolated from the tannery soil samples using nutrient agar medium. Focusing on Cr(III) tolerance, an isolate was selected for further study in the degradation of CTLS, and denoted as TCCC 11319. The strain had an aerobic, Gram-positive, and rod-shaped nature, as shown in Fig. 1, and was a member of the genus Bacillus according to its morphophysiological and biochemical characterizations. From 16S rRNA gene sequencing and phylogenetic analysis, strain TCCC 11319 showed 99% sequence identity with the Bacillus amyloliquefaciens strain using the Genbank nucleotide database (Fig. 2).Bacteria with high Cr(VI)-reducing potential have been reported in recent years. Pal and Paul isolated a chromium-resistant bacterium from chromite-polluted soil that could recover chromate.24 Sasmita Das showed that CSB 9 (B. amyloliquefaciens) had a moderately high Cr(VI) ion tolerance and reduced Cr(VI) solid wastes with a rapid degradation rate.25 Both the bacteria-produced enzyme and enzyme metabolic products contributed to catalytic Cr(IV) reduction.26 Compared with chemical catalysis, biocatalysts are considered more moderate and eco-friendly for chromium pollutant disposal. Though a variety of soluble reductases from Cr(IV)-tolerance bacterium have been reported,27 few Cr(III) tolerant reductases have been presented.
3.2 Enzyme properties
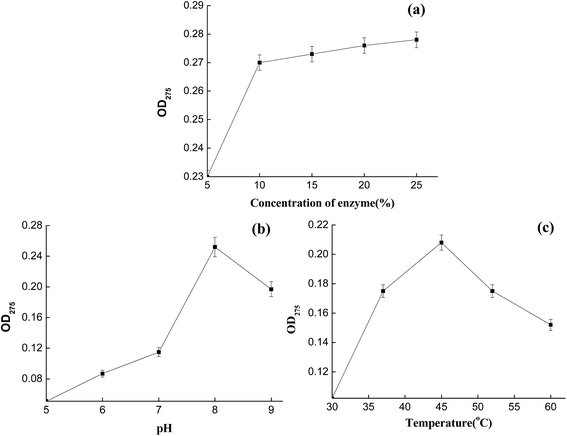
To determine optimum conditions for the collagenase, the activity was determined at different inoculum ratios, culture temperatures, and pH values (Fig. 3). In the assay, the inoculum ratio was an important parameter affecting collagenase activity. After fermentation for 32 h, optimum collagenase activity was observed to be around 5%. Studying the effect of temperature on collagenase showed that enzyme catalytic activity increased rapidly in the range 30–37 °C, with the same trend also present in the range 37–47 °C. Maximum collagenase activity was observed at 37 °C, with a further increase in incubation temperature drastically reducing collagenase activity (Fig. 3a). Meanwhile, collagenase showed optimum activity at pH 8.0 (Fig. 3b), and other variations in pH caused a reduction in activity. This variation in activity was more noticeable as the pH increased to pH 8.0. In leather production, the pH of CTLS from finished leather is usually in the range 6–8, with the temperature increasing with stacking storage. Under the optimized conditions, the enzyme acting on CTLS should have the desired effects.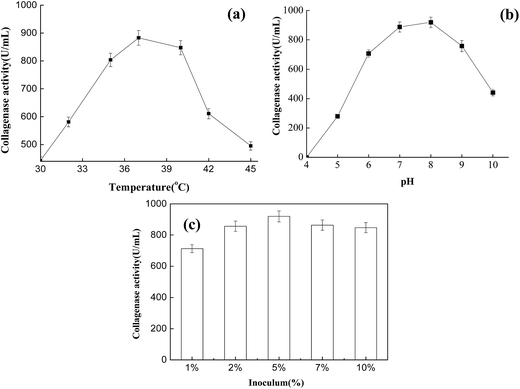 | ||
| Fig. 3 Collagenase activity at (a) different temperatures, (b) different pH values, and (c) with different inoculums. | ||
A summary of the crude enzyme from the culture medium of B. amyloliquefaciens TCCC 11319 is presented in Table 1. B. amyloliquefaciens is closely related to B. subtilis and can produce the majority of proteases, glucoamylase, and extracellular enzyme.28 In the crude broth of B. amyloliquefaciens TCCC 11319, there are at least four main types of protease, namely collagenase, alkaline protease, neutral protease, and keratinase. The main component of CTLS is collagen, with more than 70% content. The enzyme collagenase plays a major role in the degradation process. After degradation of the triple helix structures in the CTLS collagenous domain by collagenase, the alkaline protease and neutral protease would be helpful for further gelatin hydrolysis. Moreover, keratinase can act on hair in the solid waste simultaneously. In summary, all enzymes generated by the strain were useful in leather waste treatment. Strain TCCC 11319 has potential for waste treatment in the leather industry.
| Enzyme species | Enzyme activity (U mL−1) |
|---|---|
| Collagenase | 919.72 |
| Alkaline protease | 142.19 |
| Neutral protease | 90.33 |
| Keratinase | 53.47 |
3.3 Evaluation of Cr(III) tolerance of B. amyloliquefaciens TCCC11319 and the crude enzyme
The growth response curve of B. amyloliquefaciens TCCC11319 strain at various concentrations of Cr(III) (0.1, 0.2, 0.3, 0.4, and 0.5 mg L−1) showed that the optical density attained by the strains depended on the Cr(III) concentration in the solution (Fig. 4a).29 At Cr(III) concentrations of 0.3 mg L−1 and above, the maximal bacterial density reduced by about 40% compared with the initial bacteria. At Cr(III) concentrations of 0.4 and 0.5 mg mL−1, the B. amyloliquefaciens TCCC11319 strain showed no growth in the media. Furthermore, we investigated collagenase activity over a wide range of Cr(III) concentrations. Chromium sulfate was used as the substrate to determine the inhibitory effect of Cr(III) on collagenase activity. At a concentration of 1 mg mL−1, Cr(III) exhibited 80% inhibition of collagenase. Inhibition increased with increasing Cr(III) concentration, becoming obvious at a concentration of 0.6 mg mL−1. Collagenase was inactivated when the Cr(III) concentration was over 1.2 mg mL−1. Previously studied bacteria were easily inactivated at Cr(III) concentrations over 0.5 mg mL−1.30 Therefore, B. amyloliquefaciens TCCC11319 had the highest Cr(III) tolerance reported so far.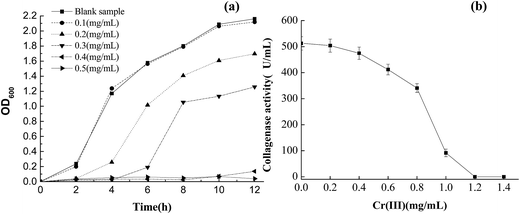 | ||
| Fig. 4 Tolerance curves of (a) B. amyloliquefaciens TCCC11319 for Cr(III) concentration and (b) collagenase for Cr(III) concentration. | ||
3.4 Optimization of CTLS degradation under different parameters
To obtain better CTLS degradation efficiency, the conditions for crude enzyme use were optimized. Compared with enzyme broth, enzyme powder is more convenient for use in industry. Therefore, the crude broth was centrifuged for 10 min at 8000 rpm and microorganisms were successfully removed. The concentrated solution was then mixed with 20% (w/w) maltodextrin and underwent spray-drying to afford enzyme powder.31 The collagenase activity of the enzyme powder was 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 U g−1. The CTLS were then treated, followed by trials to optimize the following parameters: enzyme concentration, pH, and temperature (Fig. 5).
000 U g−1. The CTLS were then treated, followed by trials to optimize the following parameters: enzyme concentration, pH, and temperature (Fig. 5).
Optical density measurements showed that a higher initial peptide concentration gave stronger absorption and stronger covalent peptide bonding.32 As collagen is the major component of CTLS, the degradation effect could be evaluated using optical behavior. Therefore, CTLS degradation effects at different concentrations, pH values, and temperatures were monitored by measuring optical density at 275 nm. The effect of enzyme concentration on CTLS degradation was studied in the range 5–25% (w/w). Substantial CTLS degradation occurred when the concentration was over 10% (Fig. 5a). The optimum pH range for CTLS degradation by collagenase was 7.5–9. The degradation effect increased in the pH range 5–8 and then decreased in the range 8–9 (Fig. 5b). Temperature also influenced the degradation effect, and was as important a factor as pH. The optimum temperature for CTLS degradation was 45 °C. Furthermore, the enzyme powder had a higher degradation rate at 35–60 °C, allowing it to be utilized for CTLS treatment more widely.
3.5 CTLS degradation studies
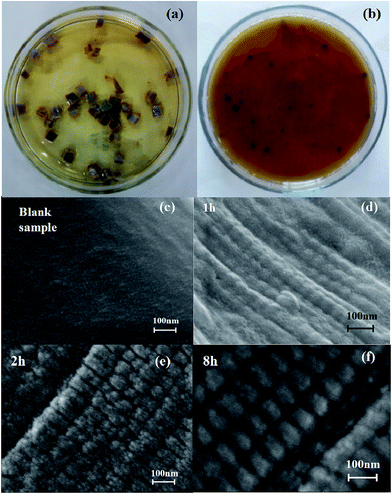
CTLS biodegradation was observed according to the hydrolysis effect. Under the optimum conditions, more than 95% weight loss occurred in 24 h, with the solid wastes being completely liquefied and the solution becoming more light blue due to increasing Cr(III) ions (Fig. 6). Fig. 6c and d show FESEM images of CTLS initially and after hydrolysis for 2 h with enzyme from B. amyloliquefaciens TCCC 11319. CTLS showed very smooth fiber surfaces with an average diameter of approximately 500 nm. Meanwhile, granular tissue appeared on the surface of CTLS fibers after enzymatic digestion (Fig. 6d). The formation of interior porosity was ascribed to enzymatic hydrolysis. Collagen cutting by the enzyme was orderly, causing the specific surface area to clearly expand. The increased specific surface area helped facilitate contact between the enzyme and collagen, accelerating the enzymatic reaction. The microstate generated rapid enzyme hydrolysis in the CTLS degradation process.4 Conclusion
Research on the Cr(III) tolerance of bacterial enzymes is valuable for the biodegradation of leather industry waste because most enzymes lose activity when exposed to high chrome concentrations. Herein, a strain, named TCCC11319, was isolated from tannery chromium-contaminated soil and identified as B. amyloliquefaciens. The enzyme produced by TCCC11319 tolerated a Cr(III) concentration of 900 mg L−1. Several factors, including inoculum, temperature, and pH, had a profound effect on the collagenase activity of the crude enzyme. Furthermore, the chromium-tolerant enzyme was able to hydrolyze CTLS for 24 h under optimum conditions. These findings could potentially be applied to solid waste disposal in the leather industry or other chromium-contaminated industries.References
- K. Kolomazník, M. Adámek and I. Andel, et al., Leather waste—potential threat to human health, and a new technology of its treatment, J. Hazard. Mater., 2008, 160(2), 514–520 CrossRef PubMed.
- A. D. Covington, Modern tanning chemistry, Chem. Soc. Rev., 1997, 26, 111–126 RSC.
- S. Hüffer and T. Taeger, Sustainable leather manufacturing: a topic with growing importance, J. Am. Leather Chem. Assoc., 2004, 99(10), 424–428 Search PubMed.
- L. F. Cabeza, M. M. Taylor and G. L. DiMaio, et al., Processing of leather waste: pilot scale studies on chrome shavings. Isolation of potentially valuable protein products and chromium, Waste Manag., 1998, 18(3), 211–218 CrossRef CAS.
- C. Mu, W. Lin and M. Zhang, Towards zero discharge of chromium-containing leather waste through improve alkali hydrolysis, Waste Manag., 2003, 23(9), 835–843 CrossRef CAS PubMed.
- A. Pati, R. Chaudhary and S. Subramani, A review on management of chrome-tanned leather shavings: a holistic paradigm to combat the environmental issues, Environ. Sci. Pollut. Res., 2014, 21(19), 11266–11282 CrossRef CAS PubMed.
- Z. Bajza and I. Markovi, Influence of enzyme concentration on leather waste hydrolysis kinetics, J. Soc. Leather Technol. Chem., 1998, 83(4), 172–176 Search PubMed.
- S. Jian, T. Wenyi and C. Wuyong, Ultrasound-accelerated enzymatic hydrolysis of solid leather waste, J. Cleaner Prod., 2008, 16(5), 591–597 CrossRef.
- D. Janacova, K. Kolomaznik and P. Mokrejs, et al., Optimization of enzymatic hydrolysis of leather waste, WSEAS Transactions on Systems, 2006, 5(11), 2651–2654 Search PubMed.
- V. J. Sundar, J. Raghava rao and C. Muralidharan, et al., Recovery and utilization of chromium-tanned proteinous wastes of leather making: a review, Crit. Rev. Environ. Sci. Technol., 2011, 41(22), 2048–2075 CrossRef CAS.
- F. Langmaier, P. Mokrejs and K. Kolomaznik, et al., Biodegradable packing materials from hydrolysates of collagen waste proteins, Waste Manag., 2008, 28(3), 549–556 CrossRef CAS PubMed.
- Z. Zhang, G. Li and B. Shi, Physicochemical properties of collagen, gelatin and collagen hydrolysate derived from bovine limed split wastes, J. Soc. Leather Technol. Chem., 2006, 90(1), 23 Search PubMed.
- R. Azmat and H. Akhter, Changes in some biophysical and biochemical parameters of mungbean (Vigna radiata L) Wilczek grown on chromium-contaminated soil treated with solid tea wastage, Pak. J. Bot., 2010, 42, 3065–3071 Search PubMed.
- E. A. Katsifas, E. Giannoutsou and M. Lambraki, et al., Chromium recycling of tannery waste through microbial fermentation, J. Ind. Microbiol. Biotechnol., 2004, 31(2), 57–62 CrossRef CAS PubMed.
- R. W. Bailey and E. G. Scott, Diagnostic Microbiology, The C.V. Mosby Company, Saint Louis, 2nd edn, 1996 Search PubMed.
- J. G. Holt, N. R. Krieg and P. H. A. Sneath, et al., Bergey's manual of determinative microbiology, Williams and Wilkins, Baltimore, MD, 1994, p. 527 Search PubMed.
- J. Sambrook and D. W. Russell, Molecular Cloning: A Laboratory Manual, 2001 Search PubMed.
- S. F. Altschul, T. L. Madden and A. A. Schäffer, et al., Gapped BLAST and PSI-BLAST: a new generation of protein database search programs, Nucleic Acids Res., 1997, 25(17), 3389–3402 CrossRef CAS PubMed.
- S. Moore and W. H. Stein, A modified ninhydrin reagent for the photometric determination of amino acids and related compounds, J. Biol. Chem., 1954, 211, 907–913 CAS.
- Y. Zhang, Y. Fu and S. Zhou, et al., A straightforward ninhydrin-based method for collagenase activity and inhibitor screening of collagenase using spectrophotometry, Anal. Biochem., 2013, 437(1), 46–48 CrossRef CAS PubMed.
- F. Letourneau, V. Soussotte and P. Bressollier, et al., Keratinolytic activity of Streptomyces sp. S. K1-02: a new isolated strain, Lett. Appl. Microbiol., 1998, 26(1), 77–80 CAS.
- X. Huang, B. Tian and Q. Niu, et al., An extracellular protease from Brevibacillus laterosporus G4 without parasporal crystals can serve as a pathogenic factor in infection of nematodes, Res. Microbiol., 2005, 156(5), 719–727 CrossRef CAS PubMed.
- S. P. Bachate, V. S. Nandre and N. S. Ghatpande, et al., Simultaneous reduction of Cr(VI) and oxidation of As(III) by Bacillus firmus TE7 isolated from tannery effluent, Chemosphere, 2013, 90(8), 2273–2278 CrossRef CAS PubMed.
- A. Pal and A. K. Paul, Aerobic chromate reduction by chromium-resistant bacteria isolated from serpentine soil, Microbiol. Res., 2004, 159(4), 347–354 CrossRef CAS PubMed.
- S. Das, J. Mishra and S. K. Das, et al., Investigation on mechanism of Cr(VI) reduction and removal by Bacillus amyloliquefaciens, a novel chromate tolerant bacterium isolated from chromite mine soil, Chemosphere, 2014, 96, 112–121 CrossRef CAS PubMed.
- I. Hwang, B. Batchelor and M. A. Schlautman, et al., Effects of ferrous iron and molecular oxygen on chromium(VI) redox kinetics in the presence of aquifer solids, J. Hazard. Mater., 2002, 92(2), 143–159 CrossRef CAS PubMed.
- H. Thatoi, S. Das and J. Mishra, et al., Bacterial chromate reductase, a potential enzyme for bioremediation of hexavalent chromium: a review, J. Environ. Manage., 2014, 146, 383–399 CrossRef CAS PubMed.
- D. Gangadharan, K. M. Nampoothiri and A. Pandey, α-Amylase production by Bacillus amyloliquefaciens using agro wastes as feed stock, Food Technol. Biotechnol., 2011, 49(3), 336–340 CAS.
- T. Srinath, T. Verma and P. W. Ramteke, et al., Chromium(VI) biosorption and bioaccumulation by chromate resistant bacteria, Chemosphere, 2002, 48(4), 427–435 CrossRef CAS PubMed.
- A. Singh, D. Vyas and P. Malaviya, Two-stage phyto-microremediation of tannery effluent by Spirodela polyrrhiza (L.) Schleid. and chromium resistant bacteria, Bioresour. Technol., 2016, 216, 883–893 CrossRef CAS PubMed.
- L. Cui, D. Zhang and L. Huang, et al., Stabilization of a new microbial transglutaminase from Streptomyces hygroscopicus WSH03-13 by spray drying, Process Biochem., 2006, 41(6), 1427–1431 CrossRef CAS.
- G. Marx, A. Hotovely-Salomon and L. Levdansky, et al., Haptide-coated collagen sponge as a bioactive matrix for tissue regeneration, J. Biomed. Mater. Res., Part B, 2008, 84(2), 571–583 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2017 |