Open Access Article
Open Access ArticleSelf-assembled nanostructures of a di-carbonitrile molecule on copper single-crystal surfaces†
Honghong Ma‡
,
Meiling Bao‡,
Liangliang Cai,
Qiang Sun and
Chunxue Yuan *
*
Tongji-Aarhus Joint Research Center for Nanostructures and Functional Nanomaterials, College of Materials Science and Engineering, Tongji University, Caoan Road 4800, Shanghai 201804, P. R. China. E-mail: cxyuan@tongji.edu.cn
First published on 9th January 2017
Abstract
Through high-resolution UHV-STM imaging and DFT calculations, we find that a di-carbonitrile molecule unexpectedly prefers to form a hydrogen-bonded nanostructure rather than coordination bonded ones on both Cu(110) and Cu(100) at room temperature. These findings highlight the importance of the surface symmetry and molecule–surface interactions in controllable fabrication of metal–ligand coordination nanostructures.
Introduction
On-surface self-assembly is a promising strategy for constructing low-dimensional nanostructures with well-controlled arrangements and functionalities. Weak noncovalent intermolecular interactions including van der Waals (vdW) forces, dipole–dipole interactions, hydrogen bonding, and metal–ligand interactions have been widely introduced and studied to form large-scale and well-ordered self-assembled nanostructures on different metal surfaces.1–9 Among these non-covalent intermolecular bonds, metal–ligand coordination is one of the mostly adopted strategies in fabricating nanostructures due to its relatively high strength and directionality.10–13 Moreover, a variety of choices of ligands and different metals give rise to the versatility of metal–ligand coordination in forming different kinds of novel nanostructures. Thus, exotic nanoarchitectures including Sierpiński triangle, quasicrystallinity, kagome lattice and Archimedean tessellation have been successfully achieved on surfaces.14–17In most cases, employed ligand molecules are terminated with pyridyl, carboxyl, hydroxyl or carbonitrile functional group, which have a tendency to coordinate with transition metals. Besides, it is noteworthy that the involvement of the elements with large electronegativity such as nitrogen and oxygen could also yield hydrogen bond.18,19 Therefore, a delicate competition between the metal-coordination bonds and hydrogen bonds is important to determine specific surface nanostructures in certain adsorbed systems and is also of fundamental interest.20
In this work, we have investigated the self-assembled behaviours of an organic molecule ([1,1′:3′,1′′-terphenyl]-4,4′′-dicarbonitrile, named TPDCN) functionalized with carbonitrile end groups on two copper surfaces with two-fold and four-fold symmetries, respectively, i.e. Cu(110) and Cu(100). It has been previously reported that the carbonitrile groups tend to coordinate with transition atoms (Cu, Ni, Co, Fe) on surfaces with three-fold symmetry such as Cu(111), Ag(111) and Au(111).15,21,22 Herein, we introduce the surfaces with two-fold and four-fold symmetries to study the influences of the surface symmetry on the formation of hydrogen bonded and metal–ligand coordinated nanostructure. By utilizing scanning tunneling microscopy (STM) and density functional theory (DFT) calculations we have surprisingly found a hydrogen-bonded chain structure formed by TPDCN with CH⋯N weak intermolecular interactions rather than metal–ligand coordination on both surfaces at room temperature (RT), which is in contrast to the coordinated structure on Cu(111) prepared at RT (cf. Fig. S1†). Further raising the sample temperature leads to metal–ligand bonding, while, in comparison with the Cu(111) surface, no extended coordinated structures are observed. These findings highlight the role of the surface symmetry for constructing coordinated nanostructure. This system could serve as a prototype for investigating the competition between hydrogen bond and coordination bond in forming intermolecular interactions.
Experimental section
The STM experiments were performed in a UHV chamber (base pressure 1 × 10−10 mbar) equipped with a variable temperature “Aarhus-type” STM purchased from SPECS,23,24 a molecular evaporator, and standard facilities for sample preparation. After the system was thoroughly degassed, the TPDCN molecule which was prepared according to the literature25 was deposited by thermal sublimation at ∼350 K onto the Cu(110) and Cu(100) surfaces, respectively. The sample was thereafter transferred from the UHV chamber to the microscope, where measurements were carried out at about 100–150 K unless specified, and the typical scanning parameters: It = 0.5–1.0 nA, Vt = ±1000–2000 mV.All of the calculations were carried out in the framework of DFT by using the Vienna Ab initio Simulation Package (VASP).26,27 The projector augmented wave method was used to describe the interaction between ions and electrons.28,29 We employed the Perdew–Burke–Ernzerhof generalized gradient approximation exchange–correlation functional,30 and van der Waals (vdW) interactions were included using the dispersion corrected DFT-D2 method of Grimme.31 The atomic structures were relaxed using the conjugate gradient algorithm scheme as implemented in VASP until the forces on all unconstrained atoms were ≤0.03 eV Å−1.
Results and discussion
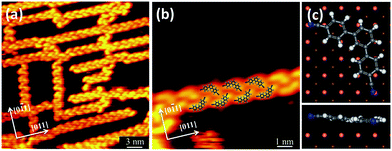
As shown in Fig. 1a, self-assembled chain structures were observed after deposition of the TPDCN molecules on Cu(110) at room temperature, and the chain structures aligned with a degree of 17° deviated from the close-packed direction of the surface. Fig. 1b shows the close-up STM image of the self-assembled molecular nanostructure of the TPDCN chain, where the single-molecule appearance and arrangement could be clearly resolved in the high-resolution STM image. The chain structure is formed by two columns of TPDCN molecules arranged in a staggered fashion, which is very similar to the hydrogen-bonded one observed on Ag(111) by Zhang et al.32 We conclude that TPDCN molecules are interconnected by hydrogen bonds between the phenyl rings and carbonitrile end groups, which is in agreement with the self-assembled chain structure on Au(111).15 Moreover, the DFT-optimized model of the hydrogen-bonded chain structure which is superimposed on the STM image in Fig. 1b is also consistent with the experimental result. However, we find that the carbonitrile molecules prefer to form hydrogen-bonded nanostructures on Cu(110) at room temperature, rather than its thermodynamically more stable coordination bonded counterpart prepared on Cu(111) at room temperature (Fig S1†). So, it is clearly shown that the symmetry of the surface has significant influence on the intermolecular interactions. However, it should also be noted that the geometrical reason cannot be totally excluded as one of the cause for the formation of the chains.Apart from the chain structure, isolated TPDCN molecules are also observed on the terrace. A closer inspection of the STM image of the isolated TPDCN molecule allows us to distinguish the depressions at the end of the carbonitrile groups as shown in Fig. 1c, which implies a strong interaction between the carbonitrile groups and the copper surface. DFT calculation has been conducted to get further insight. To position the TPDCN molecule on the surfaces, we chose extended supercells that had been checked to be big enough to avoid spurious interactions between the molecules with the neighboring supercells (cf. Fig. S2†). The DFT optimized models of the TPDCN molecule adsorbed on Cu(110) are shown in Fig. 1d and g, which clearly indicate that the molecule is stabilized by the formation of N–Cu bonds with the substrate due to the high affinity between the N and Cu atoms. As a result, the N atoms are located below the molecule plane and with a height of ∼1.80 Å above the Cu(110) substrate, in contrast to a 2.10 Å height of the molecular plane above the surface. Note that Cu adatom concentration should not be the limitation for the formation of coordination bonds since there are enough Cu adatoms at RT.33,34 Furthermore, we calculate the direction of the chain structure on the Cu(110) surface. As shown in Fig. 1e and f, the direction of the chain structure deviates from the [1−10] direction of the surface by 17°, which is in good agreement with the experiment. Moreover, the charge density difference map has also been plotted (cf. Fig. S3†), where we can clearly distinguish the hydrogen bonds between the phenyl rings and carbonitrile end groups within the chain structure.
Further annealing the sample to ∼400 K results in aggregates of the TPDCN molecules as shown in Fig. 2a. A close-up STM image in Fig. 2b indicates that the aggregates are attributed to divergent coordination assemblies because of their head-to-head binding modes. In particularly, two typical coordination motifs are clearly resolved in the STM image. Fig. 2c–f show the corresponding high-resolution STM images and DFT optimized gas-phase models of the nanostructure of the metal–ligand motifs. One of them is constituted by the two-fold symmetrical coordination motif (Fig. 2c). The three-fold symmetrical coordination motif constitutes a windmill structure, while no extended coordinated structures are observed due to the coexistence of divergent coordinated motifs including the chain structure and windmill structure. Here, we conclude that TPDCN molecules are interconnected by coordination bonds between the carbonitrile groups and the copper adatoms because they are thermodynamically more stable than hydrogen bonds. Note that the copper atoms are not resolved by STM, in conformity to most other experiments on copper-coordinated nanostructures.35
To further explore the influence of the symmetry of the surface, we have moved to the Cu(100) surface which has four-fold symmetry. As shown in Fig. 3a, after deposition of the TPDCN molecule on Cu(100) at RT, we also observe a chain structure, which is in analogy to the staggered chain structures formed on Cu(110). Thus, the same chain structures are formed on two surfaces with different chemical activities, which implies that the chain structures are independent of the molecule–substrate interactions. This finding also manifests that the intermolecular hydrogen bonds within the chain structure determine the formation of the chain structure. Moreover, there is no evidence of the metal–ligand coordinated structure, either. This result further demonstrates that metal–ligand coordination structures are not favored on incommensurate surfaces. As shown in Fig. 3c, to get further insight into the adsorption geometry of the TPDCN monomer on Cu(100), we have optimized the geometry of the TPDCN molecule on Cu(100), which shows that a flat-lying adsorption configuration on the substrate, while a strong N–Cu interaction is not found.
Fig. 4a displays a typical STM image by annealing the TPDCN covered surface at ∼400 K. The TPDCN molecules form aggregates after annealing. We could recognize the formation of a coordinated structure from the close-up STM image shown in Fig. 4b. However, due to the existence of multiple coordinated nodes, no extended ordered nanostructures observed. Fig. 4c shows the STM image of the three-fold metal–ligand coordinated nanostructure and its DFT optimized gas-phase model. These results further imply that the coordination bond is more stable than the hydrogen bond.34
It was previously shown that Fe-biphenolate coordinated network assembled in a three-fold symmetry on Cu(100),36 which indicates that the symmetry of the coordination bond is independent of the symmetry of the substrate. Also note that very recently, by introducing foreign Fe atoms on Ag(100) Zhang et al. observed geometrically appropriate metal–organic bonds on the symmetry-mismatched surface.37 While, in this system we show that an unfavored symmetry of the substrate might inhibit the formation of the preferred metal–ligand coordination.
Conclusions
In conclusion, two different Cu surfaces were utilized to study the impact of surface symmetry on the self-assembly of a di-carbonitrile molecule. Since the carbonitrile groups and the transition atoms prefer to form a three-fold coordination node, we introduce two unfavored surface symmetries i.e. Cu(110) and Cu(100). From a combination of high-resolution UHV-STM imaging and DFT calculations, we find that the carbonitrile molecule prefers to form hydrogen-bonded nanostructures on both Cu(110) and Cu(100) at room temperature, rather than its thermodynamically more stable coordination bonded counterpart prepared on Cu(111) at room temperature. Further annealing the hydrogen-bonded structures on both surfaces yield metal coordinated structures. However, divergent coordinated motifs have been established on the surfaces due to the incommensurability between the favored coordination symmetry and the surface symmetry. These findings highlight the importance of the surface symmetry and geometry of the surface in forming metal–ligand coordination nanostructures, which is crucial to the controllable construction of surface coordinated nanostructures.Acknowledgements
This work was funded by the National Natural Science Foundation of China (51403157), and the Fundamental Research Funds for the Central Universities (2014KJ061).Notes and references
- J. V. Barth, J. Weckesser, N. Lin, A. Dmitriev and K. Kern, Appl. Phys. A, 2003, 76, 645–652 CrossRef CAS.
- A. Mugarza, N. Lorente, P. Ordejon, C. Krull, S. Stepanow, M. L. Bocquet, J. Fraxedas, G. Ceballos and P. Gambardella, Phys. Rev. Lett., 2010, 105, 115702 CrossRef CAS PubMed.
- Y. Wei, W. Tong and M. B. Zimmt, J. Am. Chem. Soc., 2008, 130, 3399–3405 CrossRef CAS PubMed.
- W. Xu, M. Dong, H. Gersen, E. Rauls, S. Vázquez-Campos, M. Crego-Calama, D. N. Reinhoudt, I. Steensgaard, E. Lægsgaard, T. R. Linderoth and F. Besenbacher, Small, 2007, 3, 854–858 CrossRef CAS PubMed.
- W. Xu, J. Wang, M. Yu, E. Laegsgaard, I. Stensgaard, T. R. Linderoth, B. Hammer, C. Wang and F. Besenbacher, J. Am. Chem. Soc., 2010, 132, 15927–15929 CrossRef CAS PubMed.
- Q. Tan, Q. Sun, L. Cai, C. Zhang and L. Xie, J. Phys. Chem. C, 2015, 119, 8155–8159 CAS.
- Q. Tan, Q. Sun, L. Cai, J. Wang and Y. Ding, J. Phys. Chem. C, 2015, 119, 12935–12940 CAS.
- L. Dong, Z. Gao and N. Lin, Prog. Surf. Sci., 2016, 91, 101–135 CrossRef CAS.
- M. Shen, Z. Luo, S. Zhang, S. Wang, L. Cao, Y. Geng, K. Deng, D. Zhao, W. Duan and Q. Zeng, Nanoscale, 2016, 8, 11962–11968 RSC.
- A. Langner, S. L. Tait, N. Lin, C. Rajadurai, M. Ruben and K. Kern, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 17927–17930 CrossRef CAS PubMed.
- L. Wang, H. Kong, C. Zhang, Q. Sun, L. Cai, Q. Tan, F. Besenbacher and W. Xu, ACS Nano, 2014, 8, 11799–11805 CrossRef CAS PubMed.
- H. Kong, L. Wang, Q. Sun, C. Zhang, Q. Tan and W. Xu, Angew. Chem., Int. Ed., 2015, 54, 6526–6530 CrossRef CAS PubMed.
- H. Kong, Q. Sun, L. Wang, Q. Tan, C. Zhang, K. Sheng and W. Xu, ACS Nano, 2014, 8, 1804–1808 CrossRef CAS PubMed.
- J. I. Urgel, D. Ecija, G. Q. Lyu, R. Zhang, C. A. Palma, W. Auwarter, N. Lin and J. V. Barth, Nat. Chem., 2016, 8, 657–662 CrossRef CAS PubMed.
- Q. Sun, L. Cai, H. Ma, C. Yuan and W. Xu, Chem. Commun., 2015, 51, 14164–14166 RSC.
- Z. Shi and N. Lin, J. Am. Chem. Soc., 2009, 131, 5376–5377 CrossRef CAS PubMed.
- D. Ecija, J. I. Urgel, A. C. Papageorgiou, S. Joshi, W. Auwaerter, A. P. Seitsonen, S. Klyatskaya, M. Ruben, S. Fischer and S. Vijayaraghavan, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 6678–6681 CrossRef CAS PubMed.
- S. Clair, S. Pons, A. P. Seitsonen, H. Brune, K. Kern and J. V. Barth, J. Phys. Chem. B, 2004, 108, 14585–14590 CrossRef CAS.
- J. V. Barth, J. Weckesser, G. Trimarchi, M. Vladimirova, A. D. Vita, C. Cai, H. Brune, P. Gunter and K. Kern, J. Am. Chem. Soc., 2002, 124, 7991–8000 CrossRef CAS PubMed.
- S. Vijayaraghavan, D. Ecija, W. Auwarter, S. Joshi, K. Seufert, M. Drach, D. Nieckarz, P. Szabelski, C. Aurisicchio, D. Bonifazi and J. V. Barth, Chem.–Eur. J., 2013, 19, 14143–14150 CrossRef CAS PubMed.
- U. Schlickum, R. Decker, F. Klappenberger, G. Zoppellaro, S. Klyatskaya, M. Ruben, I. Silanes, A. Arnau, K. Kern, H. Brune and J. V. Barth, Nano Lett., 2007, 7, 3813–3817 CrossRef CAS PubMed.
- M. Pivetta, G. E. Pacchioni, E. Fernandes and H. Brune, J. Chem. Phys., 2015, 142, 101928 CrossRef PubMed.
- F. Besenbacher, Rep. Prog. Phys., 1996, 59, 1737–1802 CrossRef CAS.
- E. Laegsgaard, L. Osterlund, P. Thostrup, P. B. Rasmussen, I. Stensgaard and F. Besenbacher, Rev. Sci. Instrum., 2001, 72, 3537–3542 CrossRef CAS.
- D. A. Patrick, M. A. Ismail, R. K. Arafa, T. Wenzler, X. Zhu, T. Pandharkar, S. K. Jones, K. A. Werbovetz, R. Brun, D. W. Boykin and R. R. Tidwell, J. Med. Chem., 2013, 56, 5473–5494 CrossRef CAS PubMed.
- G. Kresse and J. Hafner, Phys. Rev. B: Condens. Matter Mater. Phys., 1993, 48, 13115–13118 CrossRef CAS.
- G. Kresse and J. Furthmuller, Phys. Rev. B: Condens. Matter Mater. Phys., 1996, 54, 11169–11186 CrossRef CAS.
- P. E. Blochl, Phys. Rev. B: Condens. Matter Mater. Phys., 1994, 50, 17953–17979 CrossRef.
- G. Kresse and D. Joubert, Phys. Rev. B: Condens. Matter Mater. Phys., 1999, 59, 1758–1775 CrossRef CAS.
- J. P. Perdew, K. Burke and M. Ernzerhof, Phys. Rev. Lett., 1996, 77, 3865–3868 CrossRef CAS PubMed.
- S. Grimme, J. Comput. Chem., 2006, 27, 1787–1799 CrossRef CAS PubMed.
- X. Zhang, N. Li, G. Gu, H. Wang, D. Nieckarz, P. Szabelski, Y. He, Y. Wang, C. Xie, Z. Shen, J. Lu, H. Tang, L. Peng, S. Hou, K. Wu and Y. Wang, ACS Nano, 2015, 9, 11909–11915 CrossRef CAS PubMed.
- T. Classen, G. Fratesi, G. Costantini, S. Fabris, F. L. Stadler, C. Kim, S. de Gironcoli, S. Baroni and K. Kern, Angew. Chem., Int. Ed., 2005, 44, 6142–6145 CrossRef CAS PubMed.
- S. L. Tait, A. Langner, N. Lin, S. Stepanow, C. Rajadurai, M. Ruben and K. Kern, J. Phys. Chem. C, 2007, 111, 10982–10987 CAS.
- T. Sirtl, S. Schlogl, A. Rastgoo-Lahrood, J. Jelic, S. Neogi, M. Schmittel, W. M. Heckl, K. Reuter and M. Lackinger, J. Am. Chem. Soc., 2013, 135, 691–695 CrossRef CAS PubMed.
- S. Stepanow, N. Lin, D. Payer, U. Schlickum, F. Klappenberger, G. Zoppellaro, M. Ruben, H. Brune, J. V. Barth and K. Kern, Angew. Chem., Int. Ed., 2007, 46, 710–713 CrossRef CAS PubMed.
- X. Zhang, N. Li, L. Liu, G. Gu, C. Li, H. Tang, L. Peng, S. Hou and Y. Wang, Chem. Commun., 2016, 52, 10578–10581 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available: Fig. S1–S3. See DOI: 10.1039/c6ra28157e |
| ‡ These authors contribute equally to this work. |
| This journal is © The Royal Society of Chemistry 2017 |