Open Access Article
Open Access ArticleTurn-on fluorescent probes that can light up endogenous RNA in nucleoli and cytoplasm of living cells under a two-photon microscope†
Ruiqing Feng‡
ab,
Longlong Li‡a,
Bing Lia,
Jinhui Lia,
Dan Penga,
Yitao Yua,
Qiuhong Mua,
Ning Zhao *a,
Xiaoqiang Yu*b and
Zhenhua Wang*c
*a,
Xiaoqiang Yu*b and
Zhenhua Wang*c
aShandong Key Laboratory for Adhesive Materials, Advanced Materials Institute, Shandong Academy of Sciences, Jinan 250014, Shandong, P. R. China. E-mail: zhaon@sdas.org
bCenter of Bio & Micro/Nano Functional Materials, State Key Laboratory of Crystal Materials, Shandong University, Jinan 250100, P. R. China. E-mail: yuxq@sdu.edu.cn; Tel: +86 53188366418
cShandong Analysis and Testing Center, Shandong Academy of Science, Jinan 250014, Shandong, P. R. China. E-mail: wzh312@mail.sdu.edu.cn
First published on 16th March 2017
Abstract
We have synthesized two-photon organic molecule probes (IMT-E and IMT-M), which selectively stain endogenous RNA in the nucleolus and cytoplasm of living cells in a short incubation time. Using these probes, bright two-photon excitation fluorescence (TPEF) images of living cells have been developed. Titrations and digest tests of ribonuclease indicate the markedly higher affinity of these probes for RNA, especially when using IMT-E. Upon binding to RNA, the fluorescence intensity of IMT-E increases by about 15-fold, showing that IMT-E is a turn-on probe for the detection of RNAs. MTT assays demonstrate that the mitochondria of cells maintain their electron mediating ability after being stained with the probes. These results demonstrate that IMT-E is an attractive two-photon turn-on fluorescent probe for visualizing RNA in living cells.
Introduction
Ribonucleic acid (RNA) plays an active role within living cells by catalyzing biological reactions, controlling gene expression, and sensing and communicating responses to cellular signals.1–4 Recently, researchers have found that the variations in nucleolus morphology are closely associated with disease phenotypes, and the relationship between the morphology of the nucleolus and cancerous status in tumor cells has attracted increasing attention.5,6 As is known, RNA is mainly distributed in the nucleolus and cytoplasm of living cells, and the nucleolus is the key site in the nucleus that synthesizes and assembles ribosomal RNAs (rRNA).7 Therefore, gaining a better understanding of the morphology of the nucleolus using RNA imaging in living cells could offer powerful help for the advancement of molecular biology, pathophysiology, and medical diagnostics.8–11 As one of the most powerful techniques for monitoring biomolecules in living systems, fluorescence imaging techniques are widely used to visualize morphological details of RNA in living cells.12 Consequently, it is significant and essential to design and develop RNA fluorescent probes for imaging the nucleolus of living cells.In the field of fluorescent probes for bio-imaging, two-photon turn-on fluorescence probes have gained tremendous attention.13 This is because two-photon fluorescence microscopy (TPM) has unique merits in imaging living specimen, such as high detection sensitivity, deep penetration, no image distortion, low photodamage and reduced photobleaching.14–17 Moreover, turn-on probes possess the advantages of low background noise and high sensitivity when binding to biomolecules.18 For imaging nucleolus RNA in living cells, the probe not only needs to penetrate the cell membrane, but also enter the nucleus. Thus, RNA probes have a high requirement for membrane permeability. Generally, large conjugated structures could help probes to achieve better two-photon properties.19,20 However, it is a contradiction that increasing the conjugate structure could reduce the membrane permeability.21 Recently, small-molecule fluorescent probes have become the most practical tool for imaging living cells, due to their good membrane permeability and chemical tractability.22 However, it is rather difficult to design small-molecule fluorescent probes with excellent two-photon properties and membrane permeability at the same time. In the field of two-photon probes for RNA detection, an appropriate probe is rarely available. Ohulchanskyy et al. found that the conventional nucleic acid dye cyan 40 can be used to image RNA in nucleoli using TPM. However, the two-photon action absorption cross section (δ × Φ) of the probe, a crucial parameter for TPEF probes, was not reported.23 Saini et al. synthesized a new trivalent aluminium metal complex (1) which could achieve nucleolus imaging in a variety of cell lines, but its two-photon properties were not reported.24 Our research group has also reported several carbazole- and indole-based two-photon RNA probes for cell imaging.25,26 However, because of the poor membrane permeability, living cell imaging using these probes is limited. Subsequently, in order to improve the permeability, we introduced a long alkyl chain into indole-based di-cationic salts, and synthesized two RNA probes that can stain living cells and have two-photon properties. Unfortunately, the incubation time of 1.5 hours demonstrated that their permeability was still less than satisfactory.27 Only Guo et al. synthesized an indole-based cyanine complex as a two-photon RNA-selective probe that realized successful imaging in living cells.28 Therefore, it is a significant challenge to develop two-photon turn-on probes with good membrane permeability for imaging RNA in living cells.
According to a variety of theoretical and experimental data, the introduction of terminal donor (D) and acceptor (A) moieties into small π-conjugates can result in a considerable two-photon absorption cross section (δ) value and a simultaneous red shift in emission.20,29,30 Based on previous work, two-photon RNA probes with good membrane permeability may be obtained by using various cationic salts with a particular chemical structure by appropriately increasing the donor–acceptor strength and intramolecular conjugation length. In this paper, we introduced indole as a donor group and benzothiazole iodized salt as an acceptor group into π-conjugates, and prepared two mono-cationic compounds (IMT-E: (E)-2-(2-(1H-indol-3-yl)vinyl)-3-(2-hydroxyethyl)benzo[d]thiazol-3-ium, MW: 448; IMT-M: (E)-2-(2-(1H-indol-3-yl)vinyl)-3-methylbenzo[d]thiazol-3-ium, MW: 418, see Fig. 1). Because of their smaller molecular structures and the appropriate electron donating ability of indole and electron withdrawing ability of benzimidazole iodized salt, IMT-E and IMT-M may be two-photon probes with good membrane permeability for imaging nucleolus RNA in living cells.
The subsequent testing of IMT-E and IMT-M gave us a pleasant surprise. The probes were found to possess a two-photon property, and can light up the RNA in the nucleolus and cytoplasm of living cells with excellent biological compatibility. After binding to RNA, the fluorescence intensity from IMT-E/IMT-M increased by about 15/10-fold, showing that the dyes are turn-on probes for the detection of RNAs.
Experimental
Synthesis
The synthetic details for IMT-E and IMT-M are shown in Scheme 1 and the ESI.† The dyes were synthesized via Knoevenagel condensation reactions between N-methyl-2-methylbenzothiazole iodine and 4-dimethylaminobenzaldehyde, using piperidine as the catalyst.Reagents and chemicals
Indole was purchased from J&K Chemical (Beijing, China). Iodomethane, iodoethanol and 2-aminobenzimidazole were purchased from Aladdin Co. All other chemicals were from commercial sources and of analytical reagent grade, unless indicated otherwise. Ultrapure water was used throughout. Calf thymus DNA and E. coli RNA, used as the models for DNA and RNA, and ribonuclease A (RNase) were obtained from Sigma. Tris and PBS were purchased from Seikagaku Corporation (Japan).Apparatus and general methods
Nuclear magnetic resonance spectra (1H and 13C) were obtained on a Bruker Avance 400 spectrometer. The HRMS spectra were recorded on an Agilent Technologies 6510 Q-TOF LC/MS. The UV-visible-near-IR absorption spectra of dilute solutions were recorded on a HITACHI U-2910 spectrophotometer using a quartz cuvette with a path length of 1 cm. One-photon fluorescence spectra were obtained on a HITACHI F-2700 spectrofluorimeter equipped with a 450 W Xe lamp. TPEF was measured on a SpectroPro300i and the pump laser beam came from a mode-locked Ti:sapphire laser system with a pulse duration of 220 fs, and a repetition rate of 76 MHz (Coherent Mira900-D). PBS buffer solution (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4·12H2O, 2 mM NaH2PO4·2H2O, pH = 7.40) and Tris-HCl buffer solution (10 mM Tris, 100 mM KCl, pH = 7.2) were used. All pH measurements were performed with a METTLER TOLEDO FE20-FiveEasy™ pH-meter. Cell viability was assayed using a cell proliferation Kit I, with the 492 nm absorbance being detected using a PerkinElmer Victor plate reader. Two-photon fluorescence images were obtained using a IX71 (Olympus) multiphoton laser scanning microscope with a Coherent Mira900-D. The dyeing rate experiment was carried out using flow cytometry (ImageStreamX MarkII). The data were obtained using INSPIRE software and analyzed using IDEAS Application v6.0 software.Spectroscopic measurements
The fluorescence quantum yields (Φ) can be calculated using eqn (1):
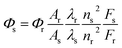 | (1) |
Two-photon absorption (TPA) cross sections were measured using the two-photon induced fluorescence method. Rhodamine B in MeOH (δ = 210 GM) was used as the standard,32 whose two-photon properties have been reported in the literature, and thus, cross sections can be calculated using eqn (2):
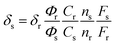 | (2) |
Cell culture and staining
Human cervical cancer cell lines (HeLa) were cultured in Dulbecco's modified Eagle's medium supplemented with penicillin/streptomycin and 10% bovine calf serum in a 5% CO2 incubator at 37 °C. IMT-E/IMT-M were dissolved in DMSO at a concentration of 1 mM.For living cell staining experiments, cultured cells grown on glass coverslips were stained with 5 μM IMT-E/IMT-M in culture medium for 30 min at 37 °C and then imaged using fluorescence microscopy.
For fixed cell staining experiments, cultured cells grown on glass coverslips were pretreated according to the following procedure: cells were first fixed by 4% paraformaldehyde for 30 min or pre-chilled methanol (−20 °C) for 15 min and were then permeabilized by 0.5% Triton X-100 for 2 min at ambient temperature.
For the RNase digest test, two sets of prefixed HeLa cells were stained with 5 μM IMT-E/IMT-M in PBS (pH = 7.4) for 30 min. After rinsing with PBS twice, a total of 1 mL PBS (as a control experiment) was added to one group of cells and 25 mg mL−1 DNase-free RNase (GE) was added to the other group of cells. Then the two sets of cells were incubated at 37 °C in 5% CO2 for 2 h. After rinsing with PBS twice, both two sets of cells were imaged using wide-field fluorescence microscopy.
Fluorescence imaging
Wide-field fluorescence microscopy images were acquired with an Olympus IX71 inverted microscope coupled with a CCD and display controller software. A confocal microscopic image and differential interference contrast (DIC) image were taken with a 488 nm Argon laser on an Olympus IX71 confocal microscope. TPEF imaging was obtained using an Olympus FV 300 Laser Confocal Microscope. In the two-photon experiments, the excitation wavelength was 800 nm from a Ti:sapphire femtosecond laser source (Coherent Chamelon Ultra). The incident power on the samples was modified by means of an attenuator and the samples were examined using a Power Monitor (Coherent). A multiphoton emission filter (FF01-750; Semrock) was used to block the IR laser. The differential interference contrast (DIC) images were taken with a 488 nm Ar ion laser.Results and discussion
Photophysical properties of IMT-E/IMT-M
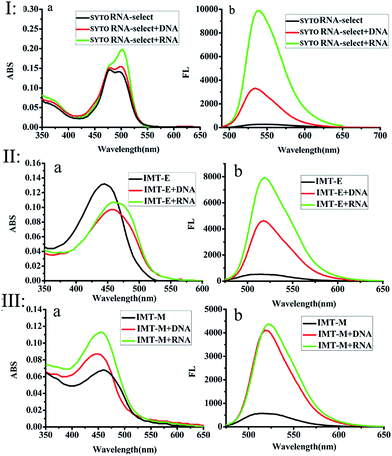
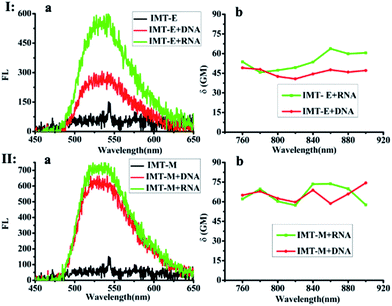
The photophysical properties of IMT-E/IMT-M in different solvents at room temperature were measured, as shown in Fig. S1.† Their one-photon absorption wavelengths in various solvents are similar, mainly in the range of 350–525 nm. Furthermore, these probes have appropriate molar extinction coefficients (ε: 6.62 × 104 cm−1 M−1 for IMT-E and 3.39 × 104 cm−1 M−1 for IMT-M in PBS), making them suitable for imaging applications. In addition, the emission maximum of the two probes in these solvents was around 520 nm. Such long-wavelength emission is also approved for cellular imaging, to efficiently avoid the background fluorescence of endogenous fluorophores.Subsequently, the photophysical properties of IMT-E/IMT-M in the presence of DNA and RNA were carefully studied (Fig. 1 and Table 1). In order to make an intuitive comparison, the commercial RNA stain “SYTO RNA-Select” was also studied. In the absorption and emission spectra, IMT-E and IMT-M display a tiny shift (Fig. 1a) when binding to DNA or RNA, while their emission fluorescence intensities show a large change. IMT-E and IMT-M are weakly fluorescent in Tris–HCl buffer, whereas their fluorescence intensities show an approximately 15-fold (IMT-E) and 10-fold (IMT-M) enhancement in the presence of RNA, and a 10-fold (IMT-E) and 10-fold (IMT-M) enhancement in the presence of DNA (Fig. 1b). As shown in Table 1, in Tris–HCl buffer, the fluorescence quantum yields (Φ) of IMT-E and IMT-M are 0.062% and 0.031%, but the values increase to 6.157% and 2.476% in the presence of RNA, respectively. The “turn-on” behaviors are similar to commercial SYTO RNA-Select (RNA: 35-fold, DNA: 12-fold). The obvious “turn-on” behavior can be attributed to the restraining of intramolecular motions when binding to DNA and RNA. Furthermore, as shown in Fig. 2, the TPEF properties of these dyes give rather similar results with excitation at 800 nm, an optimal excitation wavelength provided by a commercial Ti:sapphire laser source. The probes exhibit much brighter two-photon excited fluorescence in the presence of RNA, than that in the presence of DNA. The δ × Φ values of IMT-E and IMT-M in buffer are 0.07 GM and 0.04 GM, while in the presence of RNA, they increase to 5.95 GM and 4.9 GM (Table 1), respectively. The δ × Φ values at different wavelengths of the IMT-E and IMT-M in saturated RNA or DNA and Tris–HCl buffer are shown in Fig. 2b. From Table 1, the maximum δ values (in the presence of RNA) for IMT-E and IMT-M are 63.7 GM and 73.7 GM, respectively. These results indicate that these dyes show higher fluorescence responses to RNA than to DNA. It is expected that the compounds prefer to bind to RNA over DNA when entering cells. In addition, in the intracellular environment, acid compartments such as lysosomes change the intracellular pH dramatically. Therefore, the photophysical properties of an ideal probe in various intracellular environments should be stable. As shown in Fig. S2,† the peak positions of the fluorescence spectra of IMT-E and IMT-M at different pH values are unchanged, and their fluorescence intensities also show very little change. This finding demonstrates an important advantage.
| Sample | Solvent | λ1/nm | λ2/nm | λ3/nm | ε/M−1 cm−1 | Φ/% | δ/GM | δ × Φ/GM |
|---|---|---|---|---|---|---|---|---|
a λ1, λ2 and λ3 are the linear absorption, and one- and two-photon emission peaks, respectively; ε is molar absorptivity; Φ is the one-photon fluorescence quantum yield determined using fluorescein (Φ = 0.95) as the standard; δ is two-photon absorption cross section determined using of Rhodamine B in MeOH (δ = 210 GM) as the standard; δ × Φ is two-photon action absorption cross section. 1 GM = 10−50 cm4 s per photon; concentration of samples: 2 × 10−6 mol L−1; buffer: 10 mM Tris–HCl, 100 mM KCl, pH = 7.2. DNA/RNA: in Tris–HCl buffer in the presence of DNA/RNA. Ratio: (phosphate of NA/dye): 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for IMT-M and IMT-E. Error limits of Φ, δ and δ × Φ are 10%, 20% and 32%, respectively. 1 for IMT-M and IMT-E. Error limits of Φ, δ and δ × Φ are 10%, 20% and 32%, respectively. |
||||||||
| IMT-E | Tris | 446 | 517 | 543 | 6.62 × 104 | 0.177 | 39.5 | 0.07 |
| DNA | 460 | 519 | 533 | 4.83 × 104 | 5.691 | 49.1 | 2.79 | |
| RNA | 465 | 522 | 534 | 5.34 × 104 | 9.334 | 63.7 | 5.95 | |
| IMT-M | Tris | 460 | 520 | 541 | 3.39 × 104 | 0.092 | 47.6 | 0.04 |
| DNA | 447 | 518 | 535 | 4.39 × 104 | 6.151 | 74.4 | 4.58 | |
| RNA | 453 | 517 | 534 | 4.54 × 104 | 5.481 | 73.7 | 4.90 | |
RNA titrations
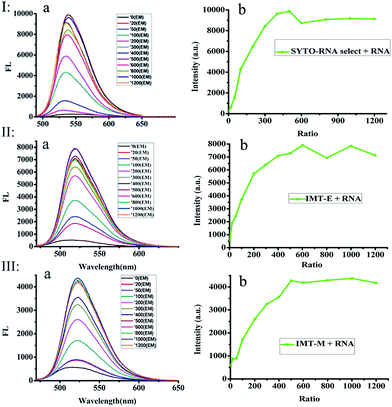
In order to further study the RNA binding characteristics of these dyes and check for the possibility of using them as RNA probes, fluorescence titration experiments of SYTO RNA-Select, IMT-E and IMT-M (Fig. 3) were carried out to further study the RNA binding characteristics. Two factors can be used to explain the mechanism of the light-up effect upon binding to RNA.33,34 Firstly, the torsional motion of vinyl groups, which can result in rapid non-radiative decay in water, could be restricted by the steric effect derived from static interactions between the cationic benzothiazole and anionic phosphate of nucleic acids, therefore, the fluorescence of the probes increased significantly. Secondly, intramolecular charge transfer (ICT) can occur between the electron donor of the indole moiety and the electron acceptor of the benzothiazole cation. In water, the larger dipole moment in the excited state than that in the ground state interacts strongly with the polar solvent, which could lead to charge separation and result in the formation of a twist intramolecular charge transfer (TICT) state. When the dyes are protected from water in the grooves of RNA, the formation of a TICT state is restricted, and dyes become brightly luminescent.More importantly, in Fig. 3, the fluorescence intensity evidently increases with the addition of RNA (especially the IMT-E probe). As shown in Table 1, the enhancement of δ × Φ induced by RNA causes this value to increase by about 100-fold. The saturation ratios ([RNA]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [dye]) for SYTO RNA-Select, IMT-E and IMT-M are 300
[dye]) for SYTO RNA-Select, IMT-E and IMT-M are 300![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 500
1, 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 500
1 and 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, respectively (Fig. 3). These results indicate that IMT-E prefers RNA, and its sensitivity is similar to the commercial probe SYTO RNA-Select. Thus, IMT-E should be a good RNA probe.
1, respectively (Fig. 3). These results indicate that IMT-E prefers RNA, and its sensitivity is similar to the commercial probe SYTO RNA-Select. Thus, IMT-E should be a good RNA probe.
TPEF/OPEF imaging
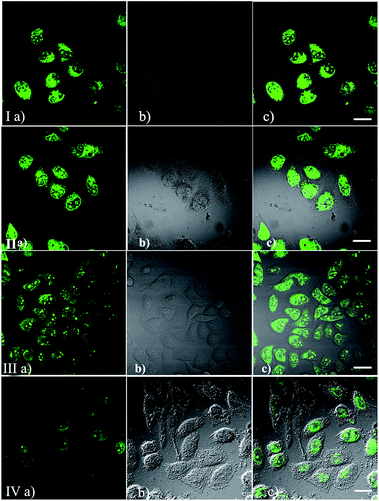
To demonstrate the permeability and one-photon imaging ability of IMT-E and IMT-M, cell imaging was performed using HeLa cells under a confocal microscope. The cells were incubated in culture medium with 5 μM of IMT-E or IMT-M for about 30 min before the imaging experiments. Using flow cytometry experiments it can be seen that the dyeing rates of IMT-E and IMT-M are 99.8 and 96.4% respectively (Fig. S3†). Thus, the two dyes show an excellent staining effect. As shown in Fig. 4(I and II), cells incubated with these probes show intense fluorescence inside the cytoplasm and nucleolus, while no fluorescence is found in the extracellular spaces. These results imply that these probes exhibit suitable permeability to plasma membranes, and entrance into the HeLa cells is enabled through simple incubation. Meanwhile, the bright intracellular fluorescence also proves that the probes can maintain their good optical properties when staining cells.After this experiment, TPEF images were also captured to examine the intracellular performance of these probes, as shown in Fig. 4(III and IV). The TPEF images, DIC pictures and merged images clearly demonstrate that cells stained with IMT-E and IMT-M show bright intracellular cytoplasma and nucleolus fluorescence when excited with 800 nm, a commonly used wavelength for TPM.
The RNase digest experiment
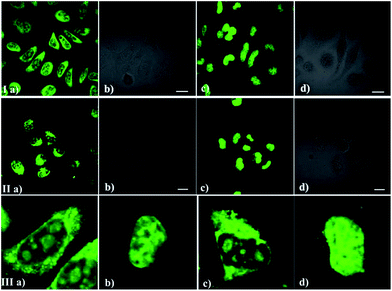
As we all know, the nucleolus contains abundant proteins and RNAs, especially ribosomal proteins and rRNA. To further confirm the selectivity of the dyes for RNA, a digest test of ribonuclease (RNase), which only hydrolyzes the RNA in the cell and does not influence the DNA and proteins, was performed (Fig. 5). Fixed-permeabilized HeLa cells (Fig. 5a) were used in this experiment. After treatment with RNase, the fluorescence in the cytoplasm and nucleoli dramatically diminished and tended to redistribute to the nucleus (Fig. 5c and d) in contrast to the untreated sample (Fig. 5a and b). The above results reconfirm that these probes prefer RNA to DNA in the complex internal environment of cells, although there was an interaction between these probes and NA in the solution fluorescence measurements (Fig. 2 and 3). This puzzling state is possibly due to the difference between the structure of RNA/DNA in vitro and their real states in cells. It may also be influenced by the specific interaction between the different probes and nucleic acid.Cytotoxicity of IMT-E and IMT-M
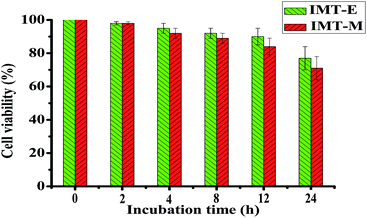
The cytotoxicity of IMT-E and IMT-M was also evaluated in HeLa cells using MTT assays. The cell viability assay data from HeLa cells treated with 5 μM IMT-E and IMT-M during different incubation time were quantified (Fig. 6). HeLa cells showed >70% viability within 24 hours. Thus, under the conditions of short treatment time (30 min) and at micromolar concentrations of IMT-E or IMT-M, these probes can be considered as having low cytotoxicity.Counterstain compatibility
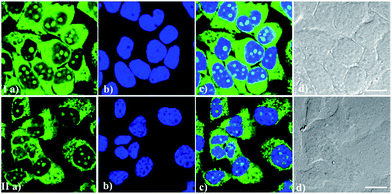
It is important to simultaneously stain one or more targets using two or more probes with different light-emitting ranges during cell imaging. Good compatibility of these RNA biosensors with other classic dyes would be helpful for these experiments. Particularly, good compatibility of RNA probes with the classic DNA-staining dye Hoechst 33342 can realize the imaging of DNA and RNA distribution simultaneously, probably revealing different patterns of RNA-DNA colocalization. From Fig. 7, it can be seen that IMT-E/IMT-M and Hoechst 33342 maintained their own labeling pattern and are not subject to any interference with each other during bioimaging. Therefore, IMT-E and IMT-M have good counterstain compatibility with Hoechst 33342 and are suitable for co-staining imaging.Photostability
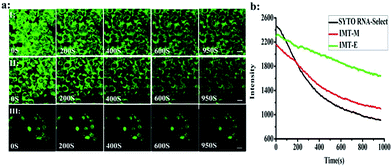
Photostability is one of the most important criteria for fluorescent imaging agents. Photostable fluorescent probes that can selectively illuminate cellular RNA are powerful tools for monitoring morphological changes in the nucleolus and studying these processes. Continuous scanning using a confocal microscope was used to quantitatively investigate the photostability of IMT-E, IMT-M and SYTO RNA-Select (Fig. 8). Within 950 s, IMT-E and IMT-M were still able to maintain a clear image, and the fluorescence signals of IMT-E and IMT-M decreased more slowly than SYTO RNA-Select. The results indicate that IMT-E and IMT-M display better photostability than SYTO RNA-Select.Conclusions
In summary, we have presented two-photon turn-on probes (IMT-E and IMT-M) for imaging intracellular RNA distribution. The probes possess a “turn-on” effect when binding to RNA, and brightness TPEF images of living cells have been obtained. Moreover the probes possess low cytotoxicity, excellent membrane permeability, good photostability and the ability to light up RNA in the nucleolus and cytoplasm of living cells. In addition, these probes show good counterstain compatibility with Hoechst 33342, and are suitable for DNA-RNA colocalization experiments. Therefore, IMT-E and IMT-M should be appropriate RNA probes for imaging in living cells using TPM.Acknowledgements
For financial support, we thank the National Natural Science Foundation of China (51303097 and 51273107), the Projects of National Natural Science Foundation of China (21277084), Natural Science Foundation of Shandong Province, China (ZR2012EMZ001), Open Project of State Key Laboratory for Supramolecular Structure and Materials (SKLSSM201524), and Shandong Province Key Research and Development Plan (2016GSF117015).Notes and references
- G. Bao, W. J. Rhee and A. Tsourkas, Annu. Rev. Biomed. Eng., 2009, 11, 25–47 CrossRef CAS PubMed.
- A. C. Peacock and C. W. Dingman, Biochemistry, 1968, 7, 668–674 CrossRef CAS PubMed.
- J. M. Chirgwin, A. E. Przybyla, R. J. MacDonald and W. J. Rutter, Biochemistry, 1979, 18, 5294–5299 CrossRef CAS PubMed.
- V. Glisin, R. Crkvenjakov and C. Byus, Biochemistry, 1974, 13, 2633–2637 CrossRef CAS PubMed.
- D. Zink, A. H. Fischer and J. A. Nickerson, Nat. Rev. Cancer, 2004, 4, 677–687 CrossRef CAS PubMed.
- A. I. Lamond and W. C. Earnshaw, Science, 1998, 280, 547–553 CrossRef CAS PubMed.
- M. Carmo-Fonseca, L. Mendes-Soares and I. Campos, Nat. Cell Biol., 2000, 2, E107–E112 CrossRef CAS PubMed.
- E. S. Lander, Nature, 2011, 470, 187–197 CrossRef CAS PubMed.
- N. N. Boustany, S. A. Boppart and V. Backman, Annu. Rev. Biomed. Eng., 2010, 12, 285 CrossRef CAS PubMed.
- L. Wei, Y. Shen, F. Xu, F. Hu, J. K. Harrington, K. L. Targoff and W. Min, ACS Chem. Biol., 2015, 10, 901–908 CrossRef CAS PubMed.
- A. J. Macario, Int. J. Clin. Lab. Res., 1995, 25, 59–70 CrossRef CAS PubMed.
- C. L. Evans and X. S. Xie, Annu. Rev. Anal. Chem., 2008, 1, 883–909 CrossRef CAS PubMed.
- L. Zeng, S. Chen, T. Xia, W. Hu, C. Li and Z. Liu, Anal. Chem., 2015, 87, 3004–3010 CrossRef CAS PubMed.
- P. T. So, C. Y. Dong, B. R. Masters and K. M. Berland, Annu. Rev. Biomed. Eng., 2000, 2, 399–429 CrossRef CAS PubMed.
- W. Denk, J. H. Strickler and W. W. Webb, Science, 1990, 248, 73–76 CAS.
- B. R. Masters and P. T. So, Microsc. Res. Tech., 2004, 63, 3–11 CrossRef CAS PubMed.
- J. A. Feijó and N. Moreno, Protoplasma, 2004, 223, 1–32 CrossRef PubMed.
- T. Terai and T. Nagano, Curr. Opin. Chem. Biol., 2008, 12, 515–521 CrossRef CAS PubMed.
- B. H. Cumpston, S. P. Ananthavel, S. Barlow, D. L. Dyer, J. E. Ehrlich, L. L. Erskine, A. A. Heikal, S. M. Kuebler, I. Y. S. Lee, D. McCord-Maughon, J. Qin, H. R. ckel, M. Rumi, X. Wu, S. R. Marder and J. W. Perry, Nature, 1999, 398, 51–53 CrossRef CAS.
- M. Pawlicki, H. A. Collins, R. G. Denning and H. L. nderson, Angew. Chem., Int. Ed., 2009, 48, 3244–3266 CrossRef CAS PubMed.
- H. Nikaido and M. Vaara, Microbiol. Rev., 1985, 49, 1–32 CAS.
- H. M. Kim and B. R. Cho, Chem. Rev., 2015, 115, 5014–5055 CrossRef CAS PubMed.
- T. Y. Ohulchanskyy, H. E. Pudavar, S. M. Yarmoluk, V. M. Yashchuk, E. J. Bergey and P. N. Prasad, Photochem. Photobiol., 2003, 77, 138–145 CrossRef CAS PubMed.
- A. K. Saini, V. Sharma, P. Mathur and M. M. Shaikh, Sci. Rep., 2016, 6, 34807 CrossRef CAS PubMed.
- X. Liu, Y. Sun, Y. Zhang, F. Miao, G. Wang, H. Zhao, X. Yu, H. Liu and W. Y. Wong, Org. Biomol. Chem., 2011, 9, 3615–3618 CAS.
- G. Song, F. Miao, Y. Sun, X. Yu, J. Z. Sun and W. Y. Wong, Sens. Actuators, B, 2012, 173, 329–337 CrossRef CAS.
- Y. Liu, W. Zhang, Y. Sun, G. Song, F. Miao, F. Guo, M. Tian, X. Yu and J. Sun, Dyes Pigm., 2014, 103, 191–201 CrossRef CAS.
- L. Guo, M. S. Chan, D. Xu, D. Y. Tam, F. Bolze, P. K. Lo and M. S. Wong, ACS Chem. Biol., 2015, 10, 1171–1175 CrossRef CAS PubMed.
- R. Kannan, G. S. He, L. Yuan, F. Xu, P. N. Prasad, A. G. Dombroskie and L. S. Tan, Chem. Mater., 2001, 13, 1896–1904 CrossRef CAS.
- R. Feng, Y. Sun, M. Tian, G. Zhang, R. Zhang, L. Guo and N. Zhao, J. Mater. Chem. B, 2015, 3, 8644–8649 RSC.
- G. A. Crosby and J. N. Demas, Review, J. Phys. Chem., 1971, 75, 991–1024 CrossRef CAS.
- C. Xu and W. W. Webb, J. Opt. Soc. Am. B, 1996, 13, 481–491 CrossRef CAS.
- Y. Zhang, J. Wang, P. Jia, X. Yu, H. Liu, X. Liu and B. Huang, Org. Biomol. Chem., 2010, 8, 4582–4588 CAS.
- H. Malak, F. N. Castellano, I. Gryczynski and J. R. Lakowicz, Biophys. Chem., 1997, 67, 35–41 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra28284a |
| ‡ Feng and Li made equal contributions to this work. |
| This journal is © The Royal Society of Chemistry 2017 |