Open Access Article
Open Access ArticlePolydopamine assisted functionalization of boronic acid on magnetic nanoparticles for the selective extraction of ribosylated metabolites from urine†
Abrar Mohyuddin,
Dilshad Hussain and
Muhammad Najam-ul-Haq *
*
Division of Analytical Chemistry, Institute of Chemical Sciences, Bahauddin Zakariya University, Multan 60800, Pakistan. E-mail: najamulhaq@bzu.edu.pk; Tel: +92 306 7552653
First published on 31st January 2017
Abstract
A novel strategy for the rapid and selective extraction of ribosylated metabolites by dopamine assisted functionalization of boronic acid on magnetic (Fe3O4@PDA-FPBA) nanoparticles has been demonstrated under optimized conditions. The study is designed to overcome the drawbacks of dendrimer assisted functionalization of low molecular weight cis-diol compounds, in combination with the advantages of boronic acid affinity towards nucleosides. Magnetic particles provide rapid extraction processing, polydopamine contributes in achieving better functionalization of boronic acid on magnetic particles because of the presence of multiple active sites at terminal groups and boronic acid selectively captures cis-diol compounds via reversible covalent bonding. Scanning electron microscopy (SEM) and transmission electron microscopy (TEM) confirm the particle size (80–120 nm). Energy dispersive X-ray spectroscopy (EDS) justifies the elemental composition of the material. The BET surface area (64 m2 g−1) is obtained from nitrogen adsorption porosimetry and coating of polydopamine and boronic acid is confirmed by Fourier transform infra-red (FT-IR) spectroscopy. Fe3O4@PDA-FPBA is highly selective with better adsorption capacity (197.3, 183.9, 163.1 and 186.5 μg g−1 for adenosine, cytidine, guanidine and uridine respectively) and recovery for ribosylated metabolites ranging from 87 to 133% for different standards. The magnetic sorbent also extracts 47 endogenous nucleoside metabolites from urine samples of healthy and lung cancer patients. Magnetic separation, combined with superior performance as compared to previously reported materials, makes Fe3O4@PDA-FPBA a promising addition to targeted metabolites analysis.
1 Introduction
Metabolites, the reactants, intermediates or products of bodily metabolic processes, have been reported as potential biomarkers for several types of disease including cancers. Metabolites are mainly classified on the basis of their functional groups and modifications. Nucleosides, a class of metabolites originating from RNA, belong to the family of cis-diol compounds.1 This family also includes glycoproteins, catechol, saccharides and a few other drugs. During inflammation or malignant conditions, the rate of RNA turn over increases, which results in a higher level of nucleosides released into the body fluids including urine and blood. The abnormal levels of such metabolites during disease is associated with higher methyl transferase activity and RNA turnover activity.2 Nucleosides are also reported as the biomarkers of cancers including lung cancer,3 colorectal cancer,4,5 thyroid cancer,6 bladder cancer,7 cervical cancer8 and breast cancer.9 Nucleosides are abundantly found in urine under disease conditions as they are directly excreted out however in the healthy state they are decomposed into other forms.10Several approaches have been developed for the analysis of nucleosides which include immunoassay,11,12 capillary electrophoresis,13 high performance liquid chromatography and mass spectrometry.14 Highly selective sample preparation methods are necessary in minimizing the background interference prior to the analysis and for accurate quantification of nucleosides.
Over the past few years, the focus is placed on the targeted metabolite analysis because of the emergence of modern mass spectrometry tools and selective enrichment methodologies.15–19 Affinity chromatography in combination with the modern mass spectrometric techniques is used in the targeted profiling of metabolites to find the disease biomarkers.20 Affinity materials like the boronic acid functionalized have been reported for the extraction of ribonucleosides through the reversible covalent bond formation of boronic acid with cis-diol moieties.21–23 Metal oxides like ZrO2, TiO2, CeO2 and their composites have also been explored with varying success.24–28 Hybrid materials, monoliths and MOFs have also been used for the cis-diol capture.29–31 These materials lack specificity to be applied at the clinical level.
Magnetic solid phase extraction (MSPE) has low cast, high efficiency, rapid extraction and numerous functionalization options.32 Magnetic sorbents have been reported in MSPE for the extraction of biological compounds like proteins, peptides and metabolites.33–35 The surface functionalization of magnetic support material helps in the targeted analysis of biological samples.36 Previously, magnetic material with PEI dendrimer coating has been reported for the nucleoside extraction.37 The dendrimers with branched and multiple chain structures have advantages in the glycopeptides enrichment, however drawbacks exist in nucleoside extractions. The rigid structure unfavorable for quick equilibration and extraction and synergic effect of branched structures only suits to the analytes with more than one cis-diol moiety.38
In this study, polydopamine assisted functionalization of boronic acid on magnetic nanoparticles (Fe3O4@PDA-FPBA) is made for the fast and selective extraction of ribosylated metabolites. Magnetic material offers rapid extraction, polydopamine provides biocompatibility and efficient coating, and reversible covalent interaction of boronic acid towards cis-diol compounds results in the selective extraction of nucleosides. Extraction parameters are optimized with standard nucleosides and the material shows high selectivity, adsorption capacity and recovery. Finally the material is applied for the profiling of endogenous nucleosides from urine samples of healthy and lung cancer patients.
2 Experimental details
2.1 Chemicals and reagents
Ferric chloride, ferrous chloride, ammonium formate (NH4HCO3), ethylene glycol, polyethylene glycol, sodium acetate, sodium cyanoborohydride, 4-formylphenyl boronic acid, creatinine, adenosine (rA), cytidine (rC), uridine (rU), guanosine (rG), 1-methyladenosine (m1A), 5-methyluridine (5mU), 2′-deoxyadenosine (dA), 2′-deoxycytidine (dC), 2′-deoxyguanidine (dG), and Thymine (T) were obtained from Sigma-Aldrich (St. Louis, MO, USA). Chromatographic grade ammonia and methanol were purchased from TEDIA Co. Inc. (Ohio, USA). Milli-Q water was obtained from Millipore, Bedford, MA.2.2 Synthesis of Fe3O4@PDA-FPBA
Iron oxide nanoparticles were prepared according to the reported procedure with slight modifications.39 Briefly 3.24 g of ferric chloride and 1.2 g of ferrous chloride were dissolved in 200 mL of ethylene glycol and stirred for 30 min. Ten grams of sodium acetate and 4 g of polyethylene glycol were added to the mixture and stirred for 30 min. Twenty millilitres of ammonia solution was added to the mixture and heated in Teflon lined stainless steel autoclave at 250 °C for 24 hours. Material was washed with ethanol followed by the deionized water and dried under vacuum at 70 °C.PDA coating was carried out on magnetic nanoparticles by self-polymerization of dopamine under basic pH.40 Two grams of magnetic nanoparticles were dispersed in 10 mM Tris Buffer (pH 8) and sonicated for 15 min. After the addition of 1.2 g of PDA, mixture was stirred at room temperature for 24 hours. The PDA coated magnetic nanoparticles were washed with ethanol and dried at room temperature.
Finally the functionalization with 4-formylphenyl boronic acid was carried out. Twenty milligrams of 4-formylphenyl boronic acid were dissolved in anhydrous ethanol. Two grams of magnetic nanoparticles were dispersed in 0.5 mL of the solution and stirred for 24 hours at room temperature. Two hundred milligrams of sodium cyanoborohydride were added after every four hours. The mixture was separated by external magnet and washed with ethanol and water. The magnetic sorbent was then dried and stored for further use.
2.3 Characterization of Fe3O4@PDA-FPBA
The morphology and particle size of synthesized material was observed with scanning electron microscopy (Quanta 200 FEI, Holand) and transmission electron microscopy (TEM, JEOL, Kyoto, Japan). The elemental composition was determined by Shimadzu EDX-720 energy-dispersive X-ray analysis (EDX, Kyoto, Japan). FTIR analysis was carried out by FTIR-8400 Shimadzu Japan. Surface area was calculated by BET equation at P/P0 between 0.05 and 0.2 through nitrogen adsorption porosimetry and all the measurements were performed at 77 K using JW-BK surface area and pore size analyzer (JWGB Sci. & Tech., Beijing, China).2.4 Extraction of cis-diol compounds by Fe3O4@PDA-FPBA
Optimization with standard nucleosides was carried out using 20 μg mL−1 of standard nucleosides solution in water. pH of standard solutions was adjusted by ammonia solution. Different amounts of material (1–10 mg) were incubated with 1 mL of standards solution for different time durations (5–60 min). After washing with water, the content was eluted with 2% formic acid. It was then lyophilized, the eluted mixture was re-dissolved in methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (5
water (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 95, v/v) and subjected to LC-UV analysis.
95, v/v) and subjected to LC-UV analysis.
The adsorption capacity of functionalized magnetic nanoparticles was also evaluated using the standard nucleosides. Five milligram adsorbent was incubated with 1 mL aqueous solution of adenosine (rA) for 20 min at different concentrations (0.5 μg mL−1, 1 μg mL−1, 5 μg mL−1, 10 μg mL−1, 20 μg mL−1, 50 μg mL−1 and 100 μg mL−1). pH of the solutions was adjusted to 9 by ammonia solution and vortexed. After 30 min, the magnetic sorbent was separated by an external magnet and adenosine was desorbed by the formic acid solution. Similar procedure was adopted to calculate the adsorption capacity of guanidine (rG), (rC) and (rU). The adsorption capacity was calculated by eqn (1):
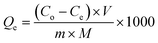 | (1) |
Selectivity of the material was assessed by four standard cis-diol compounds in the presence of four non cis-diol compounds. Five mixtures with increasing ratio of non cis-diol compounds were prepared (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1
1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, 1
10, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, 1
50, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 and 1
100 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500) and extractions were made under optimized conditions. Recovery of material was evaluated by spiking 6 standard cis-diol compounds (rA, rC, rU, rG, m1A and m5U) in urine samples and extractions were made in three replicates, followed by LC-MS analysis.
500) and extractions were made under optimized conditions. Recovery of material was evaluated by spiking 6 standard cis-diol compounds (rA, rC, rU, rG, m1A and m5U) in urine samples and extractions were made in three replicates, followed by LC-MS analysis.
Urine samples of 10 healthy and 10 lung cancer patients were collected and treated according to the Ethical guidelines (Table S1†). Briefly, urine samples were collected early morning and centrifuged for 10 min at 4 °C with 5000 g to remove the cell debris. The supernatant was filtered by membrane filter (13 mm × 0.22 μM) and centrifuged again. The supernatant was collected and stored at −80 °C. Pooled urine samples were diluted and pH of the samples was adjusted to 9. Five milligrams of material was added to 100 μL of alkylated urine and mixture was incubated for 30 min. Material was separated via an external magnet, washed twice with methanol and water to remove the non-specific attachments. Finally the bound cis-diol compounds were eluted with 2% formic acid and LC-MS analysis was carried out.
2.5 LC-MS analysis and database search
Optimization experiments were carried out on Agilent 1200 series HPLC system equipped with UV detector (Agilent Technologies, Singapore). Zorbax SB-C18 column (150 mm × 2.1 mm i.d., 5 μm, Agilent Technologies) was used for HPLC analysis and all the experiments were performed at column temperature of 30 °C. Mobile phase consisted of 5 mM ammonium formate as solvent A and methanol as solvent B. Flow rate was set as 1 mL min−1 with linear gradient of 10 min 5–30% B, 0.01 min 30–80% B, 15 min 80% B, 0.01 min 80–5% B and 8 min 5% B. Creatinine was used as an internal standard for the quantification of urinary nucleosides. Initially the concentration of creatinine was determined in 50 times diluted urine. Water and acetonitrile were used as mobile phase A and mobile phase B with flow rate of 0.8 mL min−1 at 30 °C and wavelength set at 235 nm.LC-MS experiments were performed on Agilent 1260 series HPLC system (Agilent Technologies, Singapore), consisting of an auto sampler, vacuum degasser, binary pump and combined to 6460 triple quadrupole mass spectrometer, equipped with electrospray ionization interface. Separation experiments were performed on an Agilent Zorbax SB-C18-T column (150 mm × 2.1 mm i.d., 5 μm, Agilent Technologies). Column temperature was fixed at 30 °C. Five mM ammonium formate as mobile phase A and methanol as mobile phase B with flow rate of 0.8 mL min−1 were used for separation. Gradient of 0–40 min from 5% B to 90% B, 40–43 min 90% B, 43–45 min from 90% B to 5% B, and 45–60 min 5% B were used. MRM (multiple reaction monitoring) with positive ion mode was applied for the analysis of standard ribosylated metabolites and the parameters of MRM mode are given in Table S2.† MS scans were acquired in positive ion mode and mass range was selected as m/z 100–1000. Quantification of urinary nucleosides was done by MRM in the positive mode. The optimal conditions for ESI source were as follows: interface temperature 300 °C, heat block temperature 400 °C, DL temperature 250 °C, heating gas 10 L min−1, nebulizing gas 2 L min−1 and drying gas 10 L min−1. Compound-specific MS parameters for the MRM transitions were optimized by direct infusion. The data processing and acquisition were performed on Analyst 1.5 Software (AB Sciex, Applied Biosystems). The cis-diol compounds were identified based on the accurate molecular mass (<3 ppm), METLIN database (http://www.metlin.scripps.edu) and RNA modification database (http://www.mods.rna.albany.edu/mods/modifications). MS/MS fragment information was used for the identification.
3 Results and discussion
3.1 Synthesis and characterization of magnetic sorbent
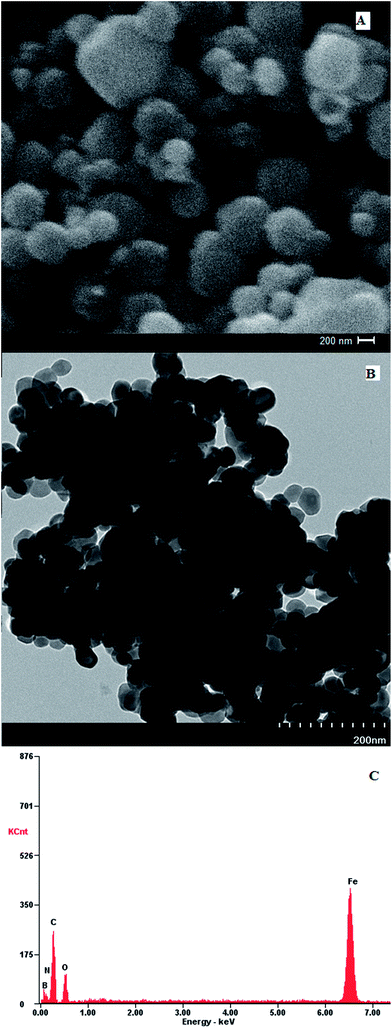
Polydopamine assisted 4-formylphenylboronic acid Fe3O4 nanoparticles (Fe3O4@PDA-FPBA) have been prepared by a simple and cost effective method. The support material has better dispersibility, easier modification ability and inherent magnetic property. Dopamine is decorated onto the surface to enhance the dispersion of magnetic particles. Dopamine self-polymerizes to polydopamine, increasing the reactive sites for the attachment of boronate affinity around magnetic nanoparticles. The surface area of the particles increases which is beneficial for the nucleoside extraction. The polydopamine coating also reduces the cost of analysis by eliminating the need of dendrimers; which are considered less effective for the extraction of small molecule cis-diol compounds such as nucleosides.38 Magnetic SPE operation contributes towards the rapid extraction of nucleosides by using an external magnet during magnetic solid phase extraction (MSPE). Tedious procedure of centrifugation after each step is therefore avoided, resulting in shorter analysis time. The attachment of boronic acid with cis-diol compounds is reversible covalent interaction, resulting in the formation of five or six membered cyclic esters. The pKa of boronic acid is 8.8 which is suitable for the covalent bond formation at basic pH and at acidic pH the covalent bond is broken, eluting the bound cis-diol molecules.Coating of Fe3O4@PDA-FPBA is confirmed by Fourier transform infrared spectroscopy (FTIR), morphology by scanning electron microscopy (SEM) and transmission electron microscopy (TEM), elemental composition by energy dispersive X-ray spectroscopy (EDX) and BET surface area by nitrogen adsorption porosimetry. SEM image shows the morphology of particles (Fig. 1A) and approximate particle size distribution in the range of 80–120 nm. No agglomeration is visible which may be attributed to the dispersibility of iron oxide particles as well as the coating by dopamine. Morphology and size is further determined by TEM (Fig. 1B) and results are complementary to SEM.
EDX spectrum of Fe3O4@PDA-FPBA shows that C, N, B, O and Fe are present in appropriate ratios 24.7%, 8.1%, 5.9%, 16.2% and 43.7% respectively (Fig. 1C) which confirms the coating of boronic acid on magnetic nanoparticles. Nitrogen adsorption porosimetry exhibits the specific surface area for functionalized material as 64 m2 g−1 in comparison to 41 m2 g−1 for iron oxide particles.
Fig. S1† shows the IR spectrum of Fe3O4 nanoparticles and Fe3O4@PDA-FPBA. The absorption band at 600 cm−1 represents Fe–O (Fig. S1A†). The stretch band at 3400 cm−1 belongs to –OH groups at the surface of synthesized material whereas O–H bend is present at 1400 cm−1. Peak at 2940 cm−1 corresponds to C–H stretch of benzene ring. Characteristic peak of B–O is present at 1369 cm−1 (Fig. S1B†).
3.2 Protocol optimization
Initially LC-UV analysis of four standard cis-diol compounds, i.e. adenosine (rA), cytidine (rC), guanidine (rG) and uridine (rU), is carried out to optimize the LC conditions and finding the retention times of four standards via UV detection at 245 nm (Fig. S2†). Fe3O4@PDA-FPBA nanoparticles are then applied to the standard mixture in the presence of four non cis-diol compounds; 2′-deoxyadenosine (dA), 2′-deoxycytidine (dC), 2′-deoxyguanidine (dG) and Thymine (T) as interfering species. Chromatogram A in Fig. 2 represents LC-UV chromatogram of nucleoside mixture prior to extraction where all eight components of sample mixture are detected. dG and T are simultaneously co-eluted under these chromatographic conditions. Four cis-diol compounds are detected after the enrichment by boronic acid functionalized magnetic nanoparticles (chromatogram B). Non cis-diols are completely eliminated during the extraction process. There is the formation of five or six membered ester under basic pH and only cis-diol compounds are attached to the magnetic sorbent. They are eluted in the acidic condition.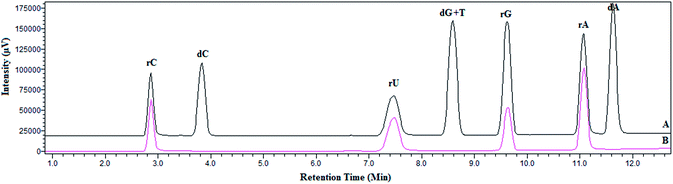 | ||
| Fig. 2 LC chromatogram of standard nucleosides (A) mixture of cis-diol and non cis-diol compounds and (B) after extraction with Fe3O4@PDA-FPBA nanoparticles. Detection wavelength is set at 254 nm. | ||
The other extraction parameters include amount of material, extraction pH, extraction time and formic acid content for elution process. They are optimized using standard nucleosides at 20 ng mL−1 concentration of each analyte in the sampling solution. Fig. 3A shows increase in extraction efficiency for all the standard nucleosides by increasing the adsorbent amount. After 5 mg adsorbent amount, no considerable increase in peak intensity is observed; attaining the extraction equilibrium with no sites available for interaction of nucleosides. Thus 5 mg of material is used in further experiments.
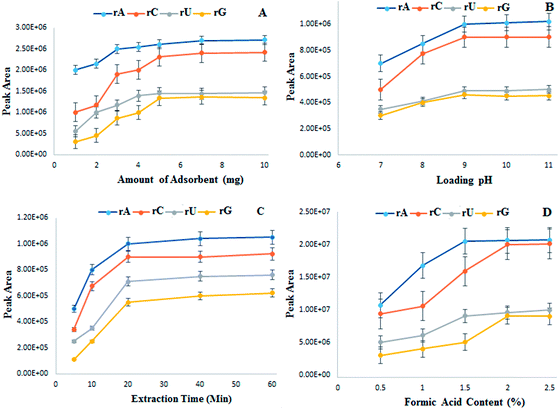 | ||
| Fig. 3 Optimization of extraction parameters by Fe3O4@PDA-FPBA (A) amount of sorbent (B) loading pH (C) extraction time and (D) formic acid content for elution. | ||
pH of loading solution is selected from 7 to 11 considering that nucleosides and glycosylated moieties bind under basic conditions. Fig. 3B exhibits the peak intensity increasing with the pH increase and becoming stable at pH 9. Extraction time is optimized from 5 to 60 minutes, however after 30 minutes there is no significant increase in peak intensities (Fig. 3C). An appropriate pH of eluting solvent is also necessary as the adsorption and elution processes are pH dependent. Different concentrations of formic acid are added to elute the bound nucleosides from magnetic sorbent. The elution is incomplete at lower formic acid content, however all the nucleosides are eluted with 2% formic acid (Fig. 3D).
3.3 Selectivity, recovery and adsorption capacity
The selectivity of magnetic sorbent is tested by using different ratios of four cis-diol compounds and four non cis-diol compounds. Five mixtures are prepared with increasing ratios of non cis-diol compounds (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1
1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, 1
10, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, 1
50, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 and 1
100 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500). LC-MS results prior to enrichment show the presence of all eight compounds with varying peak intensity, however after the extraction with boronic acid functionalized magnetic nanoparticles, four cis-diol compounds are detected (Fig. 4). No apparent change in the extraction pattern is observed with the increasing concentration of non-cis-diol compounds in the mixture. Improved enrichment performance can be attributed to the surface area of material and the strong affinity for nucleosides. Furthermore, PDA coating provides multiple reactive sites for the attachment of boronic acid moieties.
500). LC-MS results prior to enrichment show the presence of all eight compounds with varying peak intensity, however after the extraction with boronic acid functionalized magnetic nanoparticles, four cis-diol compounds are detected (Fig. 4). No apparent change in the extraction pattern is observed with the increasing concentration of non-cis-diol compounds in the mixture. Improved enrichment performance can be attributed to the surface area of material and the strong affinity for nucleosides. Furthermore, PDA coating provides multiple reactive sites for the attachment of boronic acid moieties.
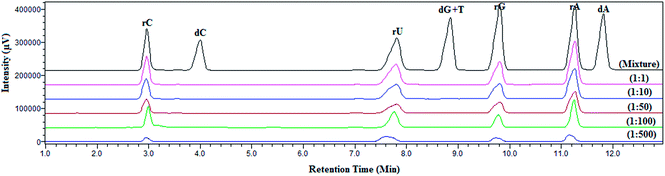 | ||
Fig. 4 Selectivity assessment of Fe3O4@PDA-FPBA at different ratios of cis-diol to non cis-diol compounds from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 1 1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500. Detection wavelength is set at 254 nm. 500. Detection wavelength is set at 254 nm. | ||
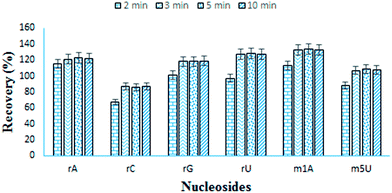
The recovery of nucleosides is calculated by spiking six standard nucleosides into the urine samples. Six nucleosides are selected on the basis of their difference in hydrophilicity. Recovery is evaluated in terms of the eluting time of nucleosides, i.e. 1 min, 2 min, 3 min, 5 min and 10 min. Recovery improves till 3 min and after that no significant improvement is observed. Material shows recoveries from 87% to 133% at optimized conditions (Fig. 5). The nucleoside, m1A shows the highest recovery (133%) while rC, the lowest recovery (87%). Although 87% recovery of rC is relatively lower but still it is better than some previously reported materials. Lower recovery of rC is because of the more hydrophilicity which results in lower interaction towards boronic acid.
The adsorption capacity of Fe3O4@PDA-FPBA is determined by using four standard nucleosides, rA, rC, rU and rG. Material shows excellent adsorption capacity towards all four standard cis-diol compounds (adenosine 197.3 μg g−1, cytidine 183.9 μg g−1, guanidine 163.1 μg g−1 and uridine 186.5 μg g−1). Adsorption capacity of material is also compared to the previously reported materials (Table S3†) and this material exhibits better capacity than most of the boronic acid functionalized materials as well as some metal oxides. Lower capacity as compared to zirconium based materials may be due to the covalent bonding in boronic acid functionality, which is much stronger than Lewis acid base mechanism in metal oxides. Still this material shows better efficiency in terms of recovery, sensitivity and adsorption capacity.
3.4 Extraction of nucleosides from urine
Urine is a rich source of metabolite biomarkers because metabolic by-products are released directly or in-directly into urine. The complexity of urine is relatively less as compared to plasma or serum, so it is easier to extract trace level moieties from urine. Nucleosides have been chosen for this study as they are potential cancers biomarkers. They are extracted as urinary metabolites from lung cancer patients. In comparison the healthy controls are also tested. Furthermore, endogenous ribosylated metabolites from human urine samples of healthy and lung cancer patients are extracted through Fe3O4@PDA-FPBA nanoparticles. LC-MS/MS analysis identifies total of 47 modified nucleosides and ribosylated metabolites from healthy urine samples after extraction with Fe3O4@PDA-FPBA (Fig. S3†). Before the quantification of urinary nucleosides, creatinine as an internal standard is quantified because creatinine concentration is related to the urinary nucleosides. This is standard method to normalize the ribosylated metabolites present in urine as creatinine excretion is constant for longer period of time. The obtained concentration of creatinine is 8.1 ± 0.45 and 7.4 ± 0.3 in healthy controls and lung cancer samples respectively. The identified metabolites with their formula, retention times, mass and content ratio of lung cancer patients with healthy controls are given in Table 1. It is observed that the content ratio of nucleosides has increased in the lung cancer patients. Some of ribosylated metabolites and modified nucleosides have increased more than 1.5 folds as compared to the healthy controls. Apart from that, total of 64 ion pairs with neutral losses are detected and 17 of them are still unidentified (Table S4†). The large number of identified nucleosides highlights the performance of magnetic sorbent and the sample preparation protocol. In case of lung cancer patients, increased levels of modified nucleosides and ribosylated metabolites are observed. This phenomenon can be due to the increased turnover of RNAs which result in increased concentration of nucleoside metabolites. Concentration decreases for some metabolites, which reflects the irregularity in the metabolism during cancer.| Sr. No. | Name | Formula | Retention time (min) | Precursor ions | Product ions | Content ratios of lung cancer (vs. healthy controls) |
|---|---|---|---|---|---|---|
| 1 | n-Ribosylhistidine | C11H17N3O6 | 2.3 | 288.1985 | 156.0097 | 0.69 ± 0.09 |
| 2 | Nicotinamide riboside | C11H15N2O5+ | 2.4 | 255.1029 | 123.0665 | 0.61 ± 0.13 |
| 3 | Orotidine | C10H12N2O8 | 2.5 | 288.9656 | 157.0401 | 1.13 ± 0.07 |
| 4 | Urate-3-ribonucleoside | C10H12N4O7 | 2.7 | 301.0751 | 169.0332 | 1.09 ± 0.04 |
| 5 | Cytidine | C9H13N3O5 | 2.9 | 244.1115 | 112.1203 | 1.10 ± 0.21 |
| 6 | Pseudouridine | C9H12N2O6 | 3.9 | 245.1906 | 113.1380 | 1.29 ± 0.31 |
| 7 | 3-Methylcytidine | C10H15N3O5 | 4.5 | 257.9985 | 126.0670 | 1.09 ± 0.08 |
| 8 | 1-Ribosyl-N-acetylhistamine | C12H19N3O5 | 4.7 | 286.1385 | 154.0980 | 6.91 ± 0.69 |
| 9 | Uridine | C9H12N2O6 | 5.1 | 245.5703 | 113.1705 | 1.32 ± 0.39 |
| 10 | 3-(3-Amino-3-caroxypropyl)-uridine | C10H13N3O6 | 5.6 | 345.9935 | 214.0861 | 2.73 ± 0.49 |
| 11 | 5-Carbamoylmethyluridine | C11H15N3O7 | 6.3 | 302.5920 | 170.0829 | 0.83 ± 0.09 |
| 12 | Isoguanosine | C10H13N5O5 | 6.9 | 284.1020 | 152.0812 | 1.95 ± 0.30 |
| 13 | Adenosine | C10H13N5O4 | 7.1 | 267.9041 | 135.9810 | 1.07 ± 0.13 |
| 14 | 5,6-Dihydrouridine | C9H14N2O6 | 7.9 | 247.0909 | 115.1501 | 0.41 ± 0.03 |
| 15 | 1-Methyl-N2-ethylguanosine | C13H19N5O5 | 8.5 | 326.0937 | 194.0287 | 0.95 ± 0.23 |
| 16 | Guanosine | C10H13N5O5 | 9.1 | 284.1006 | 152.1931 | 2.29 ± 0.15 |
| 17 | Inosine | C10H12N4O5 | 10.0 | 269.0881 | 137.2984 | 0.65 ± 0.18 |
| 18 | 1-Methyladenosine | C11H15N5O4 | 10.2 | 282.0991 | 150.1073 | 0.51 ± 0.08 |
| 19 | 6-Hydroxyl-1,6-dihydropurine ribonucleosides | C10H14N4O5 | 10.9 | 271.1009 | 139.1569 | 0.63 ± 0.05 |
| 20 | Ribosylpyridinonecarboxamide | C11H12N2O6 | 11.7 | 271.1032 | 139.0527 | 1.13 ± 0.20 |
| 21 | 1-Methylinosine | C11H14N4O5 | 12.1 | 283.1037 | 159.9908 | 1.51 ± 0.11 |
| 22 | 1-Methylguanosine | C11H15N5O5 | 12.7 | 298.2099 | 166.1091 | 1.10 ± 0.07 |
| 23 | N6,N6-Dimethyladenosine | C12H17N5O4 | 12.9 | 296.1331 | 164.0911 | 1.05 ± 0.09 |
| 24 | 5-Methyluridine | C10H14N2O6 | 13.1 | 258.9901 | 127.1281 | 0.60 ± 0.05 |
| 25 | 3-Methyluridine | C10H14N2O6 | 13.3 | 259.1052 | 127.0538 | 0.99 ± 0.13 |
| 26 | N2-Methylguanosine | C11H15N5O5 | 13.5 | 297.9817 | 166.1082 | 1.89 ± 0.27 |
| 27 | Xanthosine | C10H12N4O6 | 13.5 | 285.1025 | 153.1190 | 1.38 ± 0.29 |
| 28 | N2,N2-Dimethylguanosine | C12H17N5O5 | 13.9 | 312.0979 | 180.0884 | 1.24 ± 0.13 |
| 29 | 5′-Deoxyadenosine | C10H13N5O3 | 14.0 | 251.9986 | 136.1228 | 0.89 ± 0.11 |
| 30 | 5-Methoxycarbonylmethyluridine | C12H16N2O8 | 14.3 | 317.1012 | 185.0753 | 2.46 ± 0.22 |
| 31 | N4-Acetylcytidine | C11H15N3O6 | 14.4 | 286.1034 | 154.0771 | 1.47 ± 0.31 |
| 32 | 5′-Deoxy-5′-methylthioadensine | C11H15N5O3S | 14.8 | 298.0952 | 136.0603 | 1.51 ± 0.09 |
| 33 | 5-Methylaminomethyl-2-thiouridine | C11H17N3O5S | 15.0 | 304.1801 | 172.0319 | 1.88 ± 0.20 |
| 34 | 5-Carbamoylmethyl-2-thiouridine | C11H15N3O6S | 15.5 | 318.1039 | 172.0792 | 0.47 ± 0.02 |
| 35 | 2-Hydroxyluridine | C9H14N2O6 | 15.6 | 247.0901 | 115.0487 | 0.90 ± 0.17 |
| 36 | 8-Oxo-guanosine | C10H13N5O6 | 15.9 | 300.1007 | 168.1294 | 0.66 ± 0.05 |
| 37 | Succinyladenosine | C14H17N5O8 | 16.1 | 384.1142 | 252.1081 | 0.81 ± 0.07 |
| 38 | 1-Ribosyl-N-ω-valerylhistamine | C15H25N3O5 | 16.6 | 328.0099 | 196.2276 | 2.17 ± 0.16 |
| 39 | N6-Threonylcarbamoyladenosine | C15H20N6O8 | 17.2 | 413.1721 | 281.1013 | 0.60 ± 0.05 |
| 40 | 5-Methylaminomethyluridine | C11H17N3O6 | 17.6 | 288.3212 | 126.6546 | 1.40 ± 0.17 |
| 41 | N6-Methyladenosine | C11H15N5O4 | 17.9 | 282.3101 | 150.1009 | 1.23 ± 0.09 |
| 42 | 3-Methylcytidine | C10H15N3O5 | 18.1 | 258.1310 | 126.0417 | 1.11 ± 0.08 |
| 43 | 7-Methylguanosine | C11H17N5O5 | 21.7 | 299.8727 | 168.0931 | 2.50 ± 0.21 |
| 44 | N2,N2,7-Trimethylguanosine | C13H20N5O5+ | 23.5 | 326.1455 | 194.0995 | 1.49 ± 0.22 |
| 45 | 1-Ribosyl-N-propionylhistamine | C13H21N3O5 | 24.3 | 300.0992 | 168.1360 | 7.49 ± 0.88 |
| 46 | N2,7-Dimethylguanosine | C12H19N5O5 | 25.5 | 314.1532 | 182.1290 | 0.59 ± 0.09 |
| 47 | 4-((1H-Imidazol-2-yl)methyl)phenol-1-glucoside | C16H20N2O6 | 25.8 | 337.2192 | 175.1176 | 6.91 ± 2.09 |
4 Conclusion
A magnetic material coated with polydopamine for the functionalization of 4-formylphenylboronic acid (Fe3O4@PDA-FPBA) is developed by simple and convenient method. SEM, TEM, EDX, BET and FTIR characterization results reveal the morphology, size, surface area and coating of boronic acid on magnetic material. Polydopamine coating provides reactive sites for boronic acid attachment. The designed material is applied for the extraction of standard nucleosides under optimized extraction parameters. Material possesses higher sensitivity and selectivity towards cis-diol biomolecules due to the reversible covalent bond formation with boronic acid. Material is also applied to extract the endogenous ribosylated metabolites and modified nucleosides from urine samples of healthy as well as lung cancer patients. Forty seven ribosylated metabolites are identified which is so far the highest number of nucleosides identified by any adsorbent. Their content ratios are studied by LC-MS/MS. This rapid, selective and cost effective method has the potential to extract nucleosides based cancer biomarkers.Ethical consent
Human urine samples were collected from the healthy volunteers after taking their written consent and the approval by the ethical committee of the Institute.Acknowledgements
This work is supported by the Research Project funded by Bahauddin Zakariya University, Multan (60800), Pakistan and Higher Education Commission, Pakistan. Furthermore, the authors declare that they have no conflict of interest.References
- C. Y. Jao and A. Salic, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 15779–15784 CrossRef CAS PubMed.
- W. Struck-Lewicka, R. Kaliszan and M. J. Markuszewski, J. Pharm. Biomed. Anal., 2014, 101, 50–57 CrossRef CAS PubMed.
- W.-Y. Hsu, C.-J. Chen, Y.-C. Huang, F.-J. Tsai, L.-B. Jeng and C.-C. Lai, PLoS One, 2013, 8, e81701 Search PubMed.
- W.-Y. Hsu, W. T.-L. Chen, W.-D. Lin, F.-J. Tsai, Y. Tsai, C.-T. Lin, W.-Y. Lo, L.-B. Jeng and C.-C. Lai, Clin. Chim. Acta, 2009, 402, 31–37 CrossRef CAS PubMed.
- B. O. Feng, M.-H. Zheng, Y.-F. Zheng, A.-G. Lu, J.-W. Li, M.-L. Wang, J.-J. Ma, G.-W. Xu, B.-Y. Liu and Z.-G. Zhu, J. Gastroenterol. Hepatol., 2005, 20, 1913–1919 CrossRef PubMed.
- S. La, J. Cho, J.-H. Kim and K.-R. Kim, Anal. Chim. Acta, 2003, 486, 171–182 CrossRef CAS.
- Y. Mao, X. Zhao, S. Wang and Y. Cheng, Anal. Chim. Acta, 2007, 598, 34–40 CrossRef CAS PubMed.
- K.-R. Kim, S. La, A. Kim, J.-H. Kim and H. M. Liebich, J. Chromatogr. B: Biomed. Sci. Appl., 2001, 754, 97–106 CrossRef CAS.
- S.-H. Cho, M. H. Choi, W.-Y. Lee and B. C. Chung, Clin. Biochem., 2009, 42, 540–543 CrossRef CAS PubMed.
- K. H. Schram, Mass Spectrom. Rev., 1998, 17, 131–251 CrossRef CAS PubMed.
- P. Nehls, J. Adamkiewicz and M. F. Rajewsky, J. Cancer Res. Clin. Oncol., 1984, 108, 23–29 CrossRef CAS PubMed.
- C. Reynaud, C. Bruno, P. Boullanger, J. Grange, S. Barbesti and A. Niveleau, Cancer Lett., 1992, 61, 255–262 CrossRef CAS PubMed.
- E. Rodríguez-Gonzalo, R. Hernández-Prieto, D. García-Gómez and R. Carabias-Martínez, J. Pharm. Biomed. Anal., 2014, 88, 489–496 CrossRef PubMed.
- W. Struck, M. Waszczuk-Jankowska, R. Kaliszan and M. J. Markuszewski, Anal. Bioanal. Chem., 2011, 401, 2039–2050 CrossRef CAS PubMed.
- A. Floegel, N. Stefan, Z. Yu, K. Mühlenbruch, D. Drogan, H.-G. Joost, A. Fritsche, H.-U. Häring, M. Hrabě de Angelis, A. Peters, M. Roden, C. Prehn, R. Wang-Sattler, T. Illig, M. B. Schulze, J. Adamski, H. Boeing and T. Pischon, Diabetes, 2013, 62, 639–648 CrossRef CAS PubMed.
- A. Zhang, H. Sun, P. Wang, Y. Han and X. Wang, Analyst, 2012, 137, 293–300 RSC.
- G. A. Theodoridis, H. G. Gika, E. J. Want and I. D. Wilson, Anal. Chim. Acta, 2012, 711, 7–16 CrossRef CAS PubMed.
- P. Liu, C.-B. Qi, Q.-F. Zhu, B.-F. Yuan and Y.-Q. Feng, Sci. Rep., 2016, 6, 21433 CrossRef CAS PubMed.
- H. Li, Q. Qin, L. Qiao, X. Shi and G. Xu, Chem. Commun., 2015, 51, 11321–11324 RSC.
- J. M. Hoffman, Q. A. Soltow, S. Li, A. Sidik, D. P. Jones and D. E. Promislow, Aging Cell, 2014, 13, 596–604 CrossRef CAS PubMed.
- L. Ren, Z. Liu, Y. Liu, P. Dou and H.-Y. Chen, Angew. Chem., Int. Ed., 2009, 48, 6704–6707 CrossRef CAS PubMed.
- T. Cheng, H. Li, Y. Ma, X. Liu and H. Zhang, Anal. Bioanal. Chem., 2015, 407, 3525–3529 CrossRef CAS PubMed.
- C. Wang, H. Xu and Y. Wei, Anal. Chim. Acta, 2016, 902, 115–122 CrossRef CAS PubMed.
- S.-T. Wang, W. Huang, W. Lu, B.-F. Yuan and Y.-Q. Feng, Anal. Chem., 2013, 85, 10512–10518 CrossRef CAS PubMed.
- J.-M. Chu, C.-B. Qi, Y.-Q. Huang, H.-P. Jiang, Y.-H. Hao, B.-F. Yuan and Y.-Q. Feng, Anal. Chem., 2015, 87, 7364–7372 CrossRef CAS PubMed.
- S.-T. Wang, W. Huang, Y.-F. Deng, Q. Gao, B.-F. Yuan and Y.-Q. Feng, J. Chromatogr. A, 2014, 1361, 100–107 CrossRef CAS PubMed.
- H. Fan, P. Chen, C. Wang and Y. Wei, J. Chromatogr.A, 2016, 1448, 20–31 CrossRef CAS PubMed.
- Q. Wu, D. Wu and Y. Guan, Anal. Chem., 2014, 86, 10122–10130 CrossRef CAS PubMed.
- X. Zhu, J. Gu, J. Zhu, Y. Li, L. Zhao and J. Shi, Adv. Funct. Mater., 2015, 25, 3847–3854 CrossRef CAS.
- H. P. Jiang, C. B. Qi, J. M. Chu, B. F. Yuan and Y. Q. Feng, Sci. Rep., 2015, 5, 7785 CrossRef CAS PubMed.
- S.-T. Wang, D. Chen, J. Ding, B.-F. Yuan and Y.-Q. Feng, Chem.–Eur. J., 2013, 19, 606–612 CrossRef CAS PubMed.
- M. Wierucka and M. Biziuk, TrAC, Trends Anal. Chem., 2014, 59, 50–58 CrossRef CAS.
- S.-H. Huo and X.-P. Yan, Analyst, 2012, 137, 3445–3451 RSC.
- G.-T. Zhu, X.-M. He, X. Chen, D. Hussain, J. Ding and Y.-Q. Feng, J. Chromatogr. A, 2016, 1437, 137–144 CrossRef CAS PubMed.
- Y. Huang, Y. Wang, Q. Pan, Y. Wang, X. Ding, K. Xu, N. Li and Q. Wen, Anal. Chim. Acta, 2015, 877, 90–99 CrossRef CAS PubMed.
- X.-S. Li, G.-T. Zhu, Y.-B. Luo, B.-F. Yuan and Y.-Q. Feng, TrAC, Trends Anal. Chem., 2013, 45, 233–247 CrossRef CAS.
- H. Li, Y. Shan, L. Qiao, A. Dou, X. Shi and G. Xu, Anal. Chem., 2013, 85, 11585–11592 CrossRef CAS PubMed.
- D. Li, Y. Chen and Z. Liu, Chem. Soc. Rev., 2015, 44, 8097–8123 RSC.
- X.-T. Peng, L. Jiang, Y. Gong, X.-Z. Hu, L.-J. Peng and Y.-Q. Feng, Talanta, 2015, 132, 118–125 CrossRef CAS PubMed.
- W.-H. Zhou, C.-H. Lu, X.-C. Guo, F.-R. Chen, H.-H. Yang and X.-R. Wang, J. Mater. Chem., 2010, 20, 880–883 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Additional information includes FT-IR spectra for the characterization of material, LC-UV chromatogram of standard nucleosides, table regarding MRM parameters, table of comparison with previously reported materials and table summarizing the masses of ions detected in LC-MS/MS analysis. See DOI: 10.1039/c6ra28369a |
| This journal is © The Royal Society of Chemistry 2017 |