Open Access Article
Open Access ArticleLymphoma cell isolation using multifunctional magnetic nanoparticles: antibody conjugation and characterization†
Soubhagya Laxmi Sahooa,
Chi-Hsien Liu *abcd and
Wei-Chi Wude
*abcd and
Wei-Chi Wude
aGraduate Institute of Biochemical and Biomedical Engineering, Chang Gung University, 259, Wen-Hwa First Road, Kwei-Shan, Tao-Yuan 333, Taiwan
bResearch Center for Chinese Herbal Medicine, Research Center for Food and Cosmetic Safety, College of Human Ecology, Chang Gung University of Science and Technology, 261, Wen-Hwa First Road, Taoyuan, Taiwan
cDepartment of Chemical Engineering, Ming Chi University of Technology, 84, Gung-Juan Road, New Taipei City, Taiwan
dDepartment of Ophthalmology, Chang Gung Memorial Hospital, 5, Fu-Hsing Street, Taoyuan, Taiwan
eCollege of Medicine, Chang Gung University, 259, Wen-Hwa First Road, Taoyuan, Taiwan. E-mail: CHL@mail.cgu.edu.tw
First published on 24th April 2017
Abstract
The early detection of B-cell lymphoma and non-Hodgkin's lymphoma has a wide impact on the diagnosis and therapy of lymphoma patients. Capturing and sorting tumour cells with magnetic nanoparticles (MNPs) has received considerable attention in recent years. Despite these successes, the efficient isolation of circulating tumour cells from complex biological fluids is still under development for the early diagnosis of lymphoma. In this study, MNPs are functionalized with anti-CD20 antibodies using an avidin-biotin linkage, with the aim of achieving specific cancer cell detection and efficient isolation. Anti-CD20 antibody-conjugated MNPs (Ab MNPs) could specifically target CD20-expressing lymphoma cells. The capture efficiency of the Ab MNPs in the lymphoma cell line was >95% with regard to the mixture of two cell lines, as confirmed by flow cytometry and confocal microscopy. Transmission electron microscopy confirmed that the conjugation of an antibody with the MNPs increased the size from 12 to 47 nm. The surface charge of the Ab MNPs was examined by using zeta potential measurements. Furthermore, Prussian blue staining was performed to confirm the interaction of Ab MNPs with the targeted lymphoma cells. Our results indicated that the receptor recognition ability of the antibody was fully retained after conjugation with MNPs. In conclusion, anti-CD20 MNPs can be used for very sensitive detection and quick isolation of CD20-positive lymphoma cells among mixed cells by using only a permanent magnet.
Introduction
Cancers are among the most serious diseases that can ultimately lead to death. Sensitive and rapid isolation of cancer cells from complex bio-fluids is of critical importance for cancer research, prevention and therapy.1 Cell sorting is often used to enrich rare cells for further well-defined culture conditions and to enhance the cell population. The current capture techniques for cancer cells include flow cytometry, magnetic-based sorting devices and microfluidic chips, using for example fluorescence signals, magnetic forces and physical principles.2 Flow cytometry provides precise isolation; however, it involves a sophisticated instrument and needs expensive fluorescent probes for cell labelling. A robust sorting platform is needed to isolate tumour cells for further diagnosis and expansion. Among these techniques, magnetic sorting utilizes targeting magnetic nanoparticles (MNPs), which are biodegradable and have low toxicity.3,4 The MNP-based technology has several advantages, such as a high surface-to-volume ratio, high-binding capacity and specific interactions between nanoparticles.5 Moreover, the diffusion limitation within the micron-sized particles leads to a decrease in their binding efficacy on the targeted cells in biological fluids, such as blood.6 Therefore, the nano-scaled MNPs provide several advantages for capture applications, including a low diffusion barrier, high surface area, stability and specificity.Specific targeting is a key step in realizing the full potential of MNPs in tumour-associated diagnosis and the capture of tumour cells. Researchers have devoted a tremendous amount of time to develop MNPs-based models for the capture and early detection of cancer cells that simultaneously conjugate MNPs to active targeting moieties, such as ligands and monoclonal antibodies. Antibodies are promising moieties for targeting cancer cells, using high affinity and ligand-receptor specificity for the surface antigen on the tumour.7 For example, the anti-CD20 antibody (rituximab) has been applied to the treatment of non-Hodgkin's lymphoma and inflammatory diseases, such as rheumatoid arthritis and myositis.8 The CD20 is a non-glycosylated antigen expressed on B-cell non-Hodgkin's lymphomas,9 and cannot be found on stem cells, pro-B cells, normal plasma cells or other normal tissues.10 In addition, the CD20 on the cell membrane, which is not internalized in response to antibody binding, is a good candidate as a target for cell isolation.
Cancer cells have been successfully detected and isolated by using MNPs based on antibody and antigen interactions.11,12 Cirstoiu-Hapca et al. (2007) and Xu et al. (2014) have used anti-HER2 and anti-GD2 antibodies conjugated with nanoparticles to specifically isolate the tumour cells bearing the surface antigens.6,13 However, only one type of cancer cell was isolated in their targeting studies. In contrast, Song et al. (2011) analyzed the capture efficiencies of tumour cells by using fluorescent MNPs to isolate a small number of spiked tumour cells in a large population of normal cells.14 Magnetic particles labelled with a fluorescent dye for optical detection and conjugated with a monoclonal antibody against the neu receptor have been demonstrated to significantly identify primary and metastatic breast tumours.15 The anti-EGFR antibody-conjugated nanoparticles can be used to capture circulating tumour cells expressing EGFR and in the subsequent diagnostic analysis.16 Wu et al. have successfully developed multi-functional magnetic particles conjugated with anti-EpCAM antibody that could detect endogenous metabolites and isolate rare tumour cell isolation.17 These antibody-conjugated nanoparticles have been proven useful for sensitive detection and rapid isolation of cancer cells in early diagnoses.
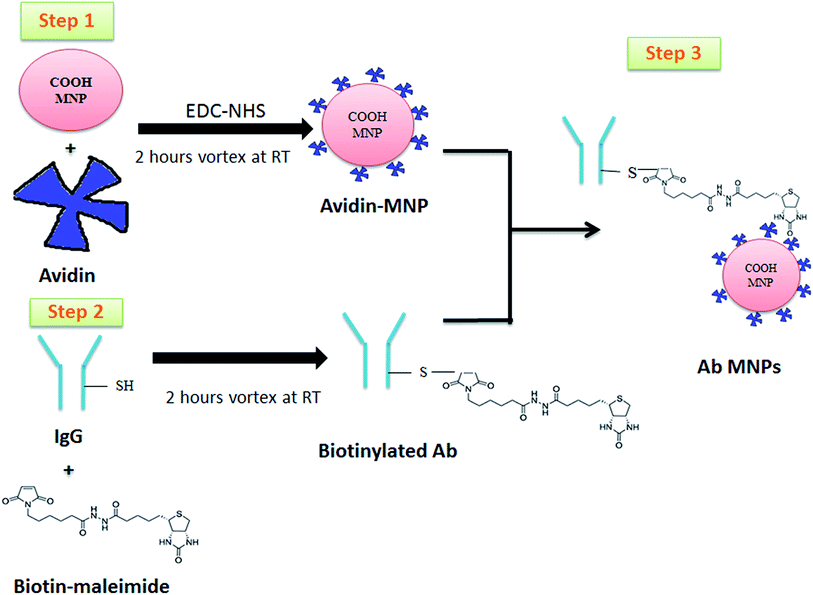
Our objectives are to synthesize and characterize the antibody-conjugated MNPs (Ab MNPs) and to assay their biocompatibility and separation efficacy. The anti-CD20 Ab MNPs were synthesized to detect and isolate lymphoma cells from two kinds of mixed cells via a process based on the high affinity between antigens and antibodies. Specifically, the carboxylic group of MNPs was activated using the EDC/NHS linker, and then avidin was conjugated onto MNPs to form avidin MNPs. Biotin maleimide was conjugated with an anti-CD20 antibody, and in the final step, a biotinylated antibody was added to interact with the avidin MNPs to form Ab MNPs. The morphology and surface charge of the Ab MNPs were examined with a transmission electron microscope and using zeta potential measurements. The use of Ab MNPs to specifically isolate the cancer cells was evaluated by flow cytometry analysis, confocal image and Prussian blue staining.
Experimental section
Materials
All the chemical reagents were commercially purchased and used without further purification. The materials include avidin, biotin maleimide, ethylenediaminetetraacetic acid (EDTA), Hoechst 33342, neutral red, N-(3-dimethylaminopropyl)-ethyl carbodiimide (EDC), hydroxy succinimide (NHS), and Trypan blue solution were purchased from Sigma-Aldrich (MO, USA). Rhodamine B was purchased from Acros Organics (NJ, USA). Potassium thiocyanate and hydrochloric acid were purchased from J.T Baker (PA, USA). Fetal bovine serum (FBS) was purchased from Biological Industries (Haemek, Israel). Paramagnetic iron oxide nanoparticles (COOH-surface modified) were purchased from Taiwan Advance Nanotech (Taoyuan, Taiwan).Functionalization of Ab MNPs
For the targeting approach, the carboxylic group of magnetic nanoparticles (1 mg mL−1) were activated using EDC/NHS linker (0.64 and 1.2 mg mL−1, respectively) at a constant vortex rate for 2 hours at room temperature. Next, avidin (0.2 mg mL−1) was conjugated onto magnetic nanoparticles for 1 hour at room temperature to form avidin MNPs. The unconjugated avidin was removed by washing three times with deionized water using magnetic separation system (Millipore). The anti-CD20 antibodies were purified from the spent media of BCRC 60427 hybridoma cell line by using liquid chromatography and protein A sepharose column (GE). Separately, the biotinylated Ab was prepared as follows. Anti-CD20 antibody (1 mg mL−1) was mixed with biotin maleimide (0.25 mg mL−1) in phosphate buffer (pH 7.2) and reacted for 2 hours at room temperature. The detailed methods are referred to the chemical conjugation textbook.18 In the final step, the biotinylated Ab was mixed with avidin MNPs to form the Ab MNPs for 30 minutes. The Ab MNPs was washed three times to remove unreacted biotinylated Ab and then stored at 4 °C. For the dose effect of MNP isolation, high Ab conjugated MNPs (15.86 μg Ab per mg MNPs) and low Ab conjugated MNPs (8.07 μg Ab per mg MNPs) were prepared by adjusting the amount of biotinylated antibody in the conjugation procedure.Cell culture
The hybridoma cells (BCRC 60427) secreting the immunoglobulin IgG2a, which recognizes the human CD20 antigen, Chinese hamster ovary cell line (CHO, BCRC 60185), and CD20-expressing Ramos lymphoma (BCRC 60252) were obtained from the Bioresource Collection and Research Center (BCRC, Hsinchu, Taiwan). BCRC 60252 was maintained in RPMI-1640 medium supplemented with 10% heat inactivated fetal bovine serum (FBS). BCRC 60427 was maintained in CD hybridoma medium (Thermo Fisher Scientific, Sugar Land, USA). The CD20 free cell line such as CHO BCRC 60185 was maintained in Excel medium. HaCat cells are kindly donated by Prof. Sheu, Hamm-Ming (NCKU, Taiwan) and maintained in DMEM with 10% serum. All cells were incubated at 37 °C under a humidified atmosphere with 5% CO2. The cell morphology and growth were monitored daily using a light microscope. Cell passage was performed every four days to maintain an exponential growth phase. The cell density and viability were determined using a Beckman Coulter counter (MS3 model) and hemocytometer, respectively, prior to all experiments.Characterization
The size and surface property of MNPs were characterized by transmission electron microscopy (TEM). A drop of diluted sample was dispersed onto a 100-mesh copper grid (CF200-Cu, Electron Microscopy Science) and the excess drop was removed with filter paper. The sample containing copper grid was dried for 2 hours at 55 °C prior to TEM analysis. The morphology of the COOH MNPs and Ab MNPs were observed by TEM (JM-1011, JEOL, and Tokyo, Japan). The zeta potential was characterized using a Zetasizer Nano ZS 90 (Malvern, Worcestershire, U.K.) at 32 °C. The tested MNPs under different pH conditions were 1 μg mL−1. The different pH (pH 3, 5, 7, 9, and 11) were obtained by adjusting the amount of HCL (0.1%) and NaOH (0.1%). Fourier transform infrared spectroscopy (Alpha, Bruker, Germany) was used to characterize the presence of specific chemical groups in the modified magnetic nanoparticles. Dried samples (0.8 mg) were mixed with KBr (IR grade, Sigma) powder (80 mg) and compressed into a thin membrane using a desktop pellet press (ICL, Garfield, NJ). The spectra of the samples were then processed by Bruker OPUS software. The loading antibody on the MNPs was determined by Nanodrop spectrophotometer (Thermo, Wilmington, DE).Detection and isolation of CD20 positive lymphocyte cells by Ab MNPs
To demonstrate the abilities of Ab MNPs for detection and isolation of cancer cell, Ramos lymphoma cells were used as target cells. The CD20 free cells including hybridoma cells, HaCaT and CHO cells were used as control. The CD20 positive cells stained with Hoechst 33342 nucleic acid dye with a final concentration of 1 ppm and the control cells were stained with Rhodamine B (with a final concentration of 10 ppm) were mixed with Ab MNPs or COOH MNPs and incubated for 20 minutes at 4 °C to prevent the endocytosis. The target cells bound by their prospective MNPs were washed three times with PBS using a magnet and then imaged with the aid of a confocal microscope and flow cytometry. To demonstrate the Ab MNPs selectivity towards the target cells, we carried out a control experiment with target cancer cells and non-target normal cells. We mixed Ab MNPs with a sample containing 1 × 106 CD20 positive lymphoma cells and CD20 negative cells and incubated for 20 minutes at 4 °C. After magnetic separation, the precipitate was imaged under a confocal microscope and flow cytometry. The number of cells before and after isolation with Ab MNPs was imaged with the confocal microscope to know the efficiency of the Ab MNPs to capture the target cancer cells.Targeting efficiency of Ab MNPs to lymphocyte cells
Magnetic separation was performed by adding Ab MNPs to each 1 mL of sample (1 × 106 cells) as describe in the above procedure. The different concentration (10, 30, 50 μg mL−1) of high Ab conjugated MNPs (15.86 μg Ab per mg MNPs), and low Ab conjugated MNPs (8.07 μg Ab per mg MNPs) were incubated with the cell samples for 20 minutes at 4 °C. Then, a magnet was introduced to the sample tubes, and after 3 minutes the target cells were attached to the tube wall while the supernatant was collected carefully using a pipet. After magnetic separation, the number of isolated cells by Ab MNPs was determined with flow cytometry to know the efficiency of the Ab MNPs to capture the target lymphocyte cells.The specificity of Ab MNPs towards CD20 positive lymphocytes was evaluated by mixing target cells with CD20 free cells in different ratios. The Hoechst 33342 stained lymphocytes were varied from 5 × 104 to 1 × 106 and n CD20 free cells by stained Rhodamine B were fixed at 1 × 106. The mixed cells were incubated with the Ab MNPs (30 μg mL−1) for 20 minutes. The isolation was performed using a magnet and the isolated cells were analyzed by flow cytometry. The isolation efficiency of the Ab MNPs was calculated as follows.
| Isolation efficiency (%) = (isolated CD20 positive cells/initial CD20 positive cells) × 100 |
Biocompatibility studies
The CD20-expressing cells and CHO cells were employed for cytotoxicity evaluation. Cells treated with Ab MNPs at different concentrations were examined with Coulter counter, MTT, and Trypan blue assay to measure cell viability. For MTT assay, cells were seeded into 96-well plates (2 × 104 cells per well) with 100 μL medium and incubated overnight. Subsequently, Ab MNPs at different concentrations (10, 30, 50 μg mL−1) were added to the well and incubated for 24 hours at 37 °C. The control group was incubated with only sterile PBS. Add 200 μL of 0.5 mg mL−1 MTT reagent into each well and incubated for additional 4 hours. After incubation, the supernatants were removed carefully and 200 μL of DMSO was added to each well. Next, plates were shaken on an orbital incubator for 10 minutes in order to dissolve the formazan crystals. Finally, the absorbance of each well was measured by a spectrophotometer. Also, coulter counter and Trypan blue assay was performed to count the cell number and to evaluate the toxicity effect. The cells (2 × 105 cells per well) were cultured in a 6 well plate. The control group were incubated with sterile PBS, whereas different concentrations (10, 30, 50 μg mL−1) of Ab MNPs were added to cells. At each hour, 100 μL of the cell suspension was transferred to an Eppendorf, and the cell number was evaluated by using Beckman Coulter counter (MS3 model) and femocytometer. For all the experiments, measurements were carried out in triplicate. The viability of the cells in the treated groups was calculated according to the following equation.| Viability (%) = (final cell population of treated group/final cell population of control group) × 100 |
Localization of nanoparticles in the lymphoma cell line
Prussian blue staining and potassium thiocyanate method were used to study iron uptake in cells. Equal volumes of 10% potassium ferrocyanide solution and 20% hydrochloric acid solution were freshly mixed to prepare the Prussian blue solution. Ramos lymphoma cells (1 × 106) were incubated with 30 μg mL−1 Ab MNPs or COOH MNPs for one hour at room temperature. Then, washed three times with PBS and incubated 5 minutes with 150 μL ice-cold ethanol (95%). After five minutes, cells were centrifuged to remove ethanol and washed three times with deionized water. Then, the cells were incubated with 150 μL of Prussian blue solution for 20 minutes in the dark. After washing three times in PBS, the cells were counterstained with neutral red (1 ppm) for 2 minutes and imaged under the microscope for Prussian blue staining.For potassium thiocyanate method, 1 × 106 cells from Ramos lymphoma cell line, hybridoma or CHO cells were incubated with 30 μg mL−1 MNPs at 37 °C for 2 hours. The excess iron was removed by washing with PBS, and the iron concentration in 1 × 106 cells was determined using the following thiocyanate-based spectrophotometric assay. The cell samples were mixed with 100 μL of 12 N HCl for 4 hours at room temperature. Then, 400 μL of 5% (w/v) potassium thiocyanate was added to the solution and incubated for 15 minutes. Samples were centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 10 min and 100 μL of the supernatant was added to a 96 well microtiter plate. The absorbance at 480 nm was measured using a microplate reader (Synergy HT, BioTek, Hong Kong). Two calibration curves of MNPs were prepared by using Ab MNPs and COOH MNPs (antibody free). The detailed procedure of potassium thiocyanate method was referred to previous papers.19,20
000 × g for 10 min and 100 μL of the supernatant was added to a 96 well microtiter plate. The absorbance at 480 nm was measured using a microplate reader (Synergy HT, BioTek, Hong Kong). Two calibration curves of MNPs were prepared by using Ab MNPs and COOH MNPs (antibody free). The detailed procedure of potassium thiocyanate method was referred to previous papers.19,20
Results and discussion
Functionalization of antibody magnetic nanoparticles (Ab MNPs)
We firstly chose biotin maleimide to conjugate the biotins to antibodies. The maleimide-containing biotins efficiently react to the thiol groups on the antibody and form a thio-succinimide linkage that can maintain the antibody's targeting ability. The disulfide bonds of antibodies as conjugation sites provide advantages over other reactive groups, such as amines and carboxylates. Avidin is conjugated with COOH MNPs through the EDC/NHS linker, and the biotinylated antibody is eventually bound with COOH MNPs through a biotin–avidin non-covalent interaction to form Ab MNPs. The strong binding affinity of biotin towards avidin is stable during proteolysis and within a wide range of pH and temperature levels and with a variety of denaturing agents.14,21–23 The EDC/NHS linker directly connects carboxylic and amino groups, for conjugating molecules with multiple carboxylic and amino groups. The functionalization procedure of Ab MNPs is shown in the schematic diagram (Fig. 1). The conjugation strategy allows the functional and directional conjugation of the Fc portion of an antibody to MNPs and is directed outward from the surfaces of Ab MNPs, while the Fab (capture site) is largely available for efficient targeting. Conjugating MNPs specifically to the Fc portion decreases the opsonin role of the antibody molecules that are used for targeting the cell surface antigen.24 While conjugating the Fc-directed method, the opsonin activity of the antibody is concealed and reduces the endocytosis of MNPs. Finally, modified MNPs with fewer cytotoxic effects are expected. The target efficiency of the anti-CD20 antibody in relation to the CD20 antigen result in functionalized Ab MNPs. The modified MNPs are characterized and investigated in terms of the isolation of lymphoma cells in the following sections.Characterization of Ab MNPs
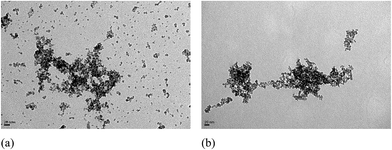
Physical properties were measured using TEM, zeta potential, and Prussian blue staining to confirm the quality of the Ab MNPs. The size and morphology of the Ab MNPs were investigated by TEM (Fig. 2). The particle size was calculated with at least 50 particles chosen at random in both the prepared samples through an image J analysis program. From Fig. 2(a and b), it is clear that the size of the MNPs increased from 12 nm to 47 nm after conjugation with Ab. The properties of the coating materials have a major effect on the diameter, and the diameters of Ab MNPs were shown to be larger than the bare MNPs. Further, the thickness of the coating layer around the surface of the particle increased due to the decrease of water molecules in the magnetic core, which proves the successful conjugation of MNPs with Ab. The smaller size of the MNPs might improve their ability to enter the target site and to avoid endocytosis. Here, we have obtained Ab MNPs of about 47 nm; in a previous study, the sizes of antibody-modified nanoparticles were 340 to 410 nm.25 The TEM images of the antibody-conjugated MNPs revealed a uniform size distribution and no significant change in their morphology, even after the conjugation reaction.Fig. 3 illustrates the zeta-potential of COOH MNPs (unconjugated Ab) and Ab MNPs (conjugated with Ab) as a function of pH, which has been shown to confirm the presence of functional groups on the surface of MNPs. The zeta potential values for COOH MNPs and Ab MNPs were 3 and 5 mV, respectively, at pH 3. The zeta potential of COOH MNPs was observed at around −21 mV. However, the zeta potential of the Ab MNPs shifted from −21 mV to −10 mV at pH 11 (Fig. 3), confirming the successful conjugation of Ab to the surface of the MNPs. The positive zeta potential values started to decrease for both the MNPs as the pH was raised towards 11. The negative zeta values started to increase with the increase in pH, which indicated that the surface charges of both conjugates had increased slightly. The fact that the isoelectric points of COOH MNPs and Ab MNPs were 3.6 and 4.4, respectively, was attributed to the presence of carboxylic groups in the MNPs. The IR spectra of COOH MNPs and Ab MNPs were presented in Fig. S1.† A prominent absorption band at 1717 cm−1 in COOH group (Fig. S1A†) which was due to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretch of carboxylic group. On the other hand, in Ab MNPs (Fig. S1B†), bands related to the presence of protein show up, the amide I and amide II bands at 1646 and 1533 cm−1, respectively. These results confirmed that the MNPs were successfully conjugated with antibodies.
O stretch of carboxylic group. On the other hand, in Ab MNPs (Fig. S1B†), bands related to the presence of protein show up, the amide I and amide II bands at 1646 and 1533 cm−1, respectively. These results confirmed that the MNPs were successfully conjugated with antibodies.
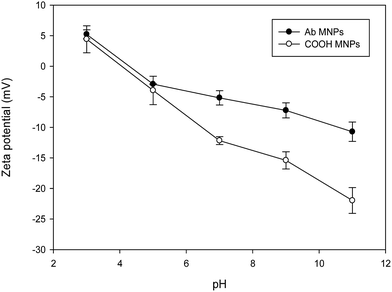 | ||
| Fig. 3 Zeta potentials of COOH MNPs (unconjugated Ab) and Ab MNPs (conjugated with Ab) under different pH conditions. MNPs were dispersed in water ranged from pH 3 to pH 11. | ||
The capture and isolation of CD20-positive lymphocyte cells by Ab MNPs
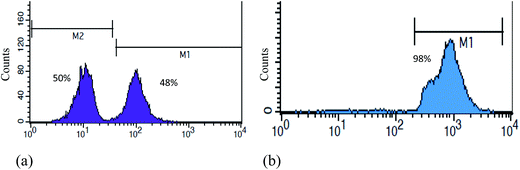
We developed a system in which the constituents are avidin-labelled MNPs, conjugated to a biotin-labelled anti-CD20 antibody (Ab MNPs) (Fig. 1). To demonstrate the specific capturing efficiency of Ab MNPs in targeting CD20-positive lymphocyte cells, we analyzed the MNP-targeted cells using confocal microscopy and flow cytometry. To examine the specific or nonspecific binding effect of MNPs, a magnetic isolation experiment was conducted, as described in the Experimental section. The 106 cells treated by 30 μg of MNPs per mL were used for the cell capture assay. To test the capture efficiency, two kinds of cells (lymphoma and hybridoma cells) were mixed and incubated for 20 minutes with MNPs followed by isolation using a magnet. After magnetic isolation, the flow cytometry analysis indicated that COOH MNPs did not show specificity towards any cell lines. Sequentially, an equal number of mixed cells, that is, 48% of the hybridoma cells (CD20 negative cells) and 50% of the lymphocyte cells (CD20-positive cells) were obtained by COOH MNPs. In contrast, Ab MNPs separated 98% of lymphocyte cells and a negligible percentage (only 2%) of hybridoma cells (Fig. 4b). The fact that COOH MNPs could nonspecifically capture both cell types (Fig. S3†) from mixed cells indicated that COOH MNPs need further modification to enhance the selectivity.26 It also implies that the Ab MNPs display higher binding sensitivity than that of bare MNPs (COOH MNPs). More importantly, our conjugation strategy allows the quick and efficient capture of CD20-positive cells using a low dose of Ab MNPs. In a previous approach, more than an hour of incubation was required for the isolation of the cells.6 The specific binding capacity of Ab MNPs was made possible by targeting the anti-CD20 Ab at the CD20 antigen on lymphocyte cells. These results suggested that the conjugation of anti-CD20 with MNPs can effectively enhance the specific binding of nanoparticles to the cells.The ability of Ab MNPs to recognize the target cells was evaluated in the following experiments. The Ab MNPs were incubated with mixed cells (Fig. 4b), as mentioned in the Experimental section. For the specific isolation, the hybridoma cells (BCRC 60427) and HaCaT cells (skin cells) were used as the negative control and lymphocyte cells expressing CD20 were the target cells. Because the control cells had been stained with Rhodamine B and the target cancer cells with Hoechst dyes, they could be visualized by their different fluorescent qualities when excited under various levels of excitation. If blue fluorescence from the target cell nucleus and red fluorescence on the surface of the cells appeared, we could conclude that the respective cells were detected by the Ab MNPs. The cell mixtures were incubated with Ab MNPs for 20 minutes and isolated by a magnet. Confocal microscopic images of the precipitates were then taken. Fig. S3† shows the experimental results in which the Ab MNPs isolated only CD20-positive lymphoma cells stained with blue fluorescence (from Hoechst 33342), while CD20-free cells stained with red fluorescence (Rhodamine B) were found in the supernatant (as presented in Fig. S4 in the ESI† section). Ab MNPs binding to CD20-negative cells (such as BCRC 60427 cells or HaCaT cells) was negligible, indicating the ability to recognize CD20 antigens with high specificity. The binding of Ab MNPs to CD20-positive cells was confirmed in the confocal images (Fig. S3†). For the recognition and isolation of markers on the cell surface, MNPs are advantageous because the high volume ratio offers a greater surface area for attaching specific ligands and for capturing markers. Most importantly, the size of the MNPs is smaller by orders of magnitude (approximately) than that of a cell, which allows multiple MNPs to be attached to a cell to facilitate magnetic isolation. Hence, the advantages of using the surface-modified MNPs with a target ligand for targeting and isolating cells are that they require only a short incubation time (20 minutes) and a simple washing process with a PBS buffer. The above experiments suggested that the mechanism of interaction between CD20-expressing cells and Ab MNPs is cell surface clustering.27 The high target specificity makes these MNPs ideal as isolation tools for recognizing and sorting cells.
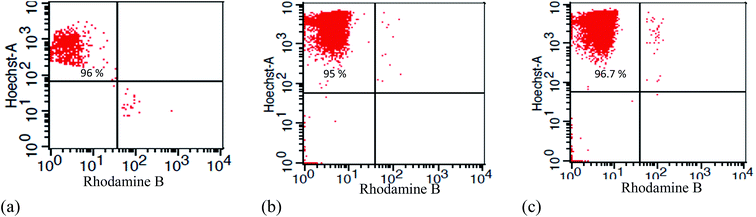
After showing the expected binding, we tested the feasibility of Ab MNPs to isolate target cells from cell mixtures by first preparing mixed samples of CD20-positive cells (lymphoma) and CD20 negative cells, such as hybridoma cells (BCRC 60427), CHO (Chinese hamster ovary cells) and HaCaT cells. The sample was prepared by mixing approximately 1 × 106 target cells and 1 × 106 of other kinds of cells, such as hybridoma, CHO or HaCaT cells, and then incubated with 30 μg mL−1 of Ab MNPs. The lymphoma cells were stained with Hoechst and BCRC 60427, and CHO or HaCaT cells were stained with Rhodamine B to distinguish the target and non-target cells. After incubation and magnetic separation, the magnetically isolated cells were analyzed by flow cytometry. When lymphoma cells mixed with CD20-negative hybridoma cells, the isolated cells contained 96% of CD20-positive cells, as shown in Fig. 5. Similarly, when mixed with either CHO or HaCaT cells, 95% of CD20 positive cells could be isolated by using the Ab MNPs. We also examined the cells that remained in the supernatant, and most of these cells were CD20 negative, as shown in Fig. S4.†
Dose effect of Ab MNPs on the isolation of target lymphoma cells
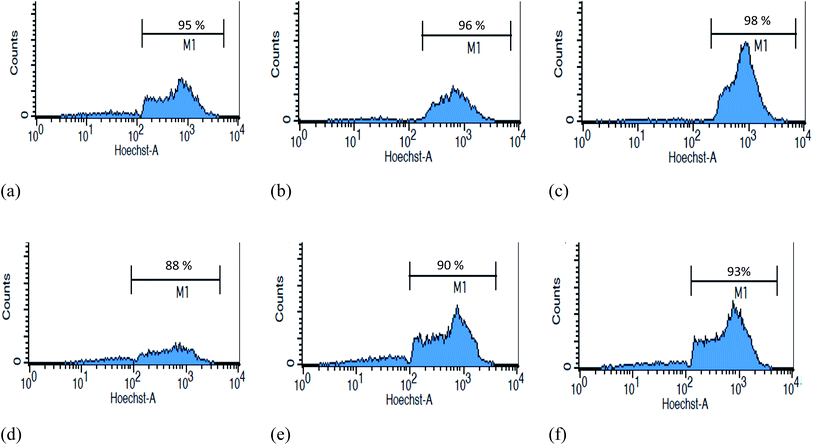
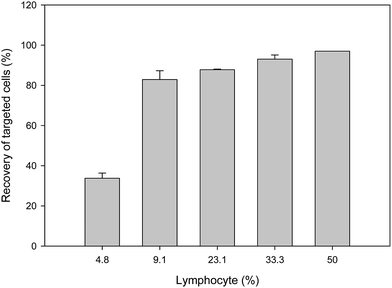
To evaluate the effects of different concentrations, different doses of antibody were conjugated with MNPs to isolate CD20-positive cells and quantified by flow cytometry. A comparison of the high Ab-loaded MNPs (15.86 μg Ab per mg MNPs) and the low Ab-loaded MNPs (8.07 μg Ab per mg MNPs) was evaluated with mixed cells having both CD20-positive cells and CD20-negative cells. Different amounts (10, 30, and 50 μg mL−1) of both high and low Ab-loaded MNPs were incubated with lymphoma (CD20 positive) and CD20-negative cells for 20 minutes and isolated by 3 minutes of magnetic separation. The CD20-positive cell binding increased with increasing concentrations of Ab MNPs. Both the high and low Ab-loaded MNPs showed good separation ability (Fig. 6a–f). The high Ab-loaded MNPs (10 μg mL−1) could efficiently isolate 95% of the CD20-positive cells from CD20-negative hybridoma cells. Furthermore, 30 μg and 50 μg mL−1 of Ab MNPs separated 96% and 98% of CD20-positive cells, respectively. In contrast, the low Ab-loaded MNPs showed that 10 μg mL−1 of MNPs separated 88% of CD20-positive cells (Fig. 6d). The cell-capture rate of high concentrations of Ab MNPs had higher separation efficacy even at a low concentration (10 μg mL−1) of Ab MNP treatment of cells. Whereas, with a low concentration of Ab MNPs, at 10 μg, 30 μg and 50 μg mL−1, separated 88%, 90% and 95% of CD20-positive cells, respectively, and resulted in a few more CD20 negative cells compared with the high Ab MNPs (Fig. 6a–f). This proves the successful Ab conjugation on MNPs since lower concentrations of Ab MNPs can specifically bind to CD20-positive cells. The isolation of the CD20 cells demonstrated some differences in binding, further indicating the Ab effect on MNPs. However, in the studied concentration range, Ab MNPs with high Ab conjugation exhibited more specific isolation of lymphocyte cells than those with low Ab modification on MNPs. A similar result was obtained when CD20-positive cells were mixed with CD20-negative CHO cells, as indicated in Fig. S7.† In a previous report, it was suggested that an optimal concentration of Ab would be necessary to achieve efficient cellular targeting of MNPs.25 The above studies showed that both Ab concentrations (high and low) showed specificity towards the target cells.Generally, in the early stage of cancer, very few cancer cells are present within a large number of normal cells in the blood. To determine the specificity of Ab MNPs towards less CD20-positive lymphocyte cells, the target cells were mixed with non-target cells in different ratios (Fig. 7); stained (Hoechst 33342) target cells ratio were varied (5 × 104–1 × 106), and stained (Rhodamine B) non-target cells ratio were fixed (1 × 106). The mixed cells were incubated with fixed concentrations (30 μg mL−1) of Ab MNPs for 20 minutes and then isolated using a magnet and analyzed by flow cytometry. The results showed that even if we used low concentrations of target cells, the Ab MNPs could recognize the specific target cells against the large population of normal cells. When the target cells were very rare (i.e. only 5 × 104 target lymphocyte cells mixed with 106 non-target cells), only 31% of target cells were isolated.
Biocompatibility studies
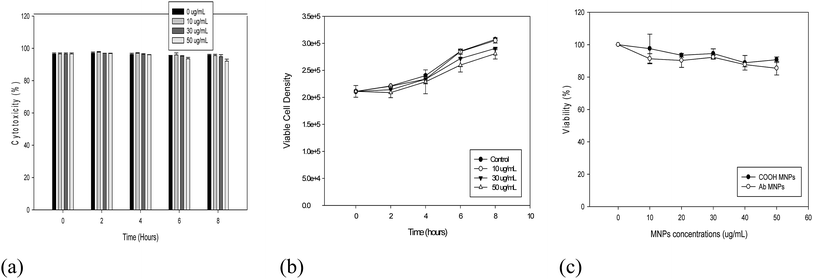
Factors such as surface conjugation, charges and sizes of MNPS are key aspects in understanding the biocompatibility of the functionalized MNPs. A high concentration of MNPs has been reported to be toxic to in vitro cells due to the generation of reactive oxygen species.28 On the contrary, trivial effects on the viability of various cell lines have been reported for these MNPs by other authors.29,30 Herein, we evaluated the biocompatibility of modified Ab MNPs using several complementary approaches, such as Trypan blue, cell counting and MTT assay for CD20-positive lymphocyte cells. The results of Trypan blue assay and cell counting (Fig. 8a and b) confirmed the biocompatibility of Ab MNPs. The cell viability was above 90% after 8 hours of incubation. Trypan blue test has been proposed as the standard method to validate cell viability after MNP incubation. The results of cell counting showed that cell density remained the same after 8 hours of incubation with Ab MNPs. The MTT assay (Fig. 8) showed that cell activity was not significantly affected by the presence of Ab MNPs at 24 hours of treatment (>80% viability in relation to the control sample), even at the highest concentration (50 μg mL−1). Moreover, Ab MNPs toxicity effect was also evaluated with normal cell (CHO cells) and presented in Fig. S8.† Higher viability has been observed for the two cell types when compared to the control conditions. MTT assay shows that Ab MNPs do not exert acute adverse effect on CHO cells and lymphoma cells even at high dosage, suggesting that it can be applicable in vivo applications. The MTT assay is based on the reduction of soluble yellow tetrazolium into insoluble purple formazan crystals by mitochondrial succinate dehydrogenase of the viable cell. Thus, the rate of formazan crystal formation is directly proportional to the total number of viable cells, which is measured in terms of absorbance.31 A modified MTT assay has been adopted here to avoid the influence of MNPs on the colorimetric assay.32 Fig. 8c, shows that the viability of cells exhibited a slightly decreasing trend from 1 hour to 24 hours of co-culture for both bare and Ab MNPs in a similar manner to that of the control sample (without MNPs), and the viability remained more than 80%. All the Trypan blue dye, MTT assay, and cell counting results confirmed the compatibility of the Ab MNPs developed in this study. Cell viability could be maintained within the 24 h incubation using 50 μg mL−1 of MNPs.Localization of nanoparticles in the lymphoma cell line
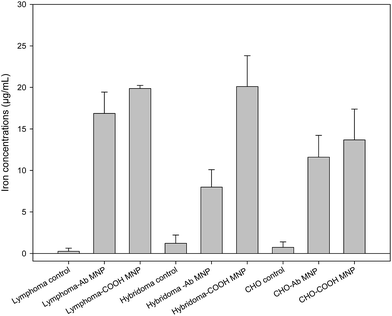
The Prussian blue staining test was employed to detect the localization of the nanoparticles with antibody-conjugated and unconjugated MNPs on CD20-positive lymphocyte cells (Fig. S9†). This method detects iron within the treated cells through the reduction of ferric ions into the ferrous state, which is shown as a blue colour.33 The cells treated with COOH MNPs (antibody free) showed occasional blue spots outside of the cells (Fig. S9a†), whereas the cells treated with antibody-conjugated nanoparticles showed clear blue shells around the cells, confirming the presence of Ab MNPs around the cell membranes (Fig. S9b†). There is abundant literature on MNPs conjugation efficiency; however, the results are difficult to compare due to the different experimental protocols, such as size, incubation time, surface coating, longer incubation time and higher concentration. Moreover, longer incubation time and higher concentrations of MNPs enable high interactions with the cell surface and increase the detection efficiency.34 High concentrations of MNPs for an extended incubation may reduce viability and enhance cell aggregation.35 Therefore, the required quantity of MNPs for cellular uptake must be balanced with their biocompatibility for efficient diagnosis or treatment.29 To evaluate the selectivity of Ab MNPs, the iron concentration was also obtained by the potassium thiocyanate method (Fig. 9). In brief, the iron content of different cells (1 × 106) was measured after 2 hours of incubation with 30 μg of Ab MNPs or COOH MNPs at 37 °C. The results showed that a small difference in the amount of accumulated iron in the two cell lines used as controls could be due to either different MNP sizes or the type of cell line. Our potassium thiocyanate data indicated that Ab MNPs bound effectively at the level of 16.8 μg per 10−6 cells on the cell surface of lymphocytes. In contrast, half the quantity of Ab MNPs was deposited on CD20-free hybridoma and CHO cells at the level of 7.9 and 8.8 μg per 106 cells, respectively. Nonspecific iron binding (13–20 μg per 106 cells) was observed in the three cell lines when isolation was performed by using COOH MNPs. This result confirmed the nonspecific binding of COOH MNPs to cells. Recently, the location of functionalized MNPs in MCF-7 cells after 24 h incubation has been observed by optical microscopy and TEM. Most of the MNPs were distributed in the periplasmic area.30 In addition, their results for Prussian blue staining and ferrozine-based assay also indicated that MNPs (21 μg per 106 cells) can be internalized effectively (21 μg per 106 cells) by MCF-7 cells. In contrast, the major location of our Ab MNPs or COOH MNPs was on the surface of cellular membranes in the lymphoma cells, as indicated in Fig. S9b.† It is important to point out the dose of our MNPs was 13-fold lower than in Calero's paper, which might have resulted in the difference.30Conclusion
In this study, we demonstrated a simple and robust conjugation method by using antibodies and MNPs. The specificity for targeting the thiols of the Fc fragment in an antibody can be improved by using maleimide conjugation. The synthetic platform described herein was efficient. It has the potential for further optimization for antibody conjugation on different nanoparticles. In addition, the lymphocytes cells were successfully detected and isolated from mixed samples containing other cells and target lymphocytes cells. The Ab MNPs fully retained their antibody binding capacity and could detect and isolate the CD20-positive cells from the mixed cells. The capture efficiency of Ab MNPs for lymphoma cells was above 95% under our experimental conditions. The capture efficiency was confirmed by complementary techniques, such as flow cytometry and confocal microscopy. Based on this simple antibody conjugation on MNPs, this platform enables robust cell sorting by using only a permanent magnet. The present approach demonstrated a facile, time saving and economical synthesis of Ab MNPs that can be used as an efficient capture platform for tumour cells.Conflict of interest
The authors would like to declare that no potential conflicts of interest exist.Acknowledgements
We express gratitude to Ministry of Science and Technology (MOST 105-2221-E-182-070), Chang Gung University (BMRP 758) and Chang Gung Memorial Hospital (CMRPD 1E0352) for funding and supporting this research.References
- M. Yu, S. Stott, M. Toner, S. Maheswaran and D. A. Haber, Circulating tumor cells: approaches to isolation and characterization, J. Cell Biol., 2011, 192, 373–382 CrossRef CAS PubMed.
- C. Wyatt Shields Iv, C. D. Reyes and G. P. López, Microfluidic cell sorting: a review of the advances in the separation of cells from debulking to rare cell isolation, Lab Chip, 2015, 15, 1230–1249 RSC.
- Z. Bakhtiary, A. A. Saei, M. J. Hajipour, M. Raoufi, O. Vermesh and M. Mahmoudi, Targeted superparamagnetic iron oxide nanoparticles for early detection of cancer: possibilities and challenges, Nanomedicine: Nanotechnology, Biology and Medicine, 2016, 12, 287–307 CrossRef CAS PubMed.
- D. Guldner, J. K. Hwang, M. C. D. Cardieri, M. Eren, P. Ziaei and M. G. Norton, et al., In vitro evaluation of the biological responses of canine macrophages challenged with PLGA nanoparticles containing monophosphoryl lipid A, PLoS One, 2016, 11(11), e0165477 Search PubMed.
- S. H. Huang and R. S. Juang, Biochemical and biomedical applications of multifunctional magnetic nanoparticles: a review, J. Nanopart. Res., 2011, 13, 4411–4430 CrossRef.
- H. Xu, Z. P. Aguilar, L. Yang, M. Kuang, H. Duan and Y. Xiong, et al., Antibody conjugated magnetic iron oxide nanoparticles for cancer cell separation in fresh whole blood, Biomaterials, 2011, 32, 9758–9765 CrossRef CAS PubMed.
- M. A. Firer and G. Gellerman, Targeted drug delivery for cancer therapy: the other side of antibodies, J. Hematol. Oncol., 2012, 5, 1–16 CrossRef PubMed.
- S. H. Lim, S. A. Beers, R. R. French, P. W. M. Johnson, M. J. Glennie and M. S. Cragg, Anti-CD20 monoclonal antibodies: historical and future perspectives, Haematologica, 2010, 95, 135–143 CrossRef CAS PubMed.
- L. Al-Zoobi, S. Salti, A. Colavecchio, M. Jundi, A. Nadiri and G. S. Hassan, et al., Enhancement of rituximab-induced cell death by the physical association of CD20 with CD40 molecules on the cell surface, Int. Immunol., 2014, 26, 451–465 CrossRef CAS PubMed.
- P. Jain and S. O'Brien, Anti-CD20 monoclonal antibodies in chronic lymphocytic leukemia, Expert Opin. Biol. Ther., 2013, 13, 169–182 CrossRef CAS PubMed.
- F. M. Kievit, Z. R. Stephen, O. Veiseh, H. Arami, T. Wang and V. P. Lai, et al., Targeting of primary breast cancers and metastases in a transgenic mouse model using rationally designed multifunctional SPIONs, ACS Nano, 2012, 6, 2591–2601 CrossRef CAS PubMed.
- H. J. Hathaway, K. S. Butler, N. L. Adolphi, D. M. Lovato, R. Belfon and D. Fegan, et al., Detection of breast cancer cells using targeted magnetic nanoparticles and ultra-sensitive magnetic field sensors, Breast Cancer Res., 2011, 13, R108 CrossRef CAS PubMed.
- A. Cirstoiu-Hapca, L. Bossy-Nobs, F. Buchegger, R. Gurny and F. Delie, Differential tumor cell targeting of anti-HER2 (Herceptin®) and anti-CD20 (Mabthera®) coupled nanoparticles, Int. J. Pharm., 2007, 331, 190–196 CrossRef CAS PubMed.
- E. Q. Song, J. Hu, C. Y. Wen, Z. Q. Tian, X. Yu and Z. L. Zhang, et al., Fluorescent–magnetic-biotargeting multifunctional nanobioprobes for detecting and isolating multiple types of tumor cells, ACS Nano, 2011, 5, 761–770 CrossRef CAS PubMed.
- F. M. Kievit, Z. R. Stephen, O. Veiseh, H. Arami, T. Wang and V. P. Lai, et al., Targeting of primary breast cancers and metastases in a transgenic mouse model using rationally designed multifunctional SPIONs, ACS Nano, 2012, 6, 2591–2601 CrossRef CAS PubMed.
- S.-M. Jo, S.-H. Noh, Z. Jin, Y. Lim, J. Cheon and H.-S. Kim, Simple and efficient capture of EGFR-expressing tumor cells using magnetic nanoparticles, Sens. Actuators, B, 2014, 201, 144–152 CrossRef CAS.
- J. Wu, X. Wei, J. Gan, L. Huang, T. Shen and J. Lou, et al., Multifunctional Magnetic Particles for Combined Circulating Tumor Cells Isolation and Cellular Metabolism Detection, Adv. Funct. Mater., 2016, 26, 4016–4025 CrossRef CAS PubMed.
- G. T. Hermanson, 8-Tags and Probes. Bioconjugate Techniques, Academic Press, San Diego, 1996, pp. 297–416 Search PubMed.
- S. H. Tseng, M. Y. Chou and I. M. Chu, Cetuximab-conjugated iron oxide nanoparticles for cancer imaging and therapy, Int. J. Nanomed., 2015, 10, 3663–3685 CAS.
- Y.-C. Lu, P.-C. Luo, C.-W. Huang, Y.-L. Leu, T.-H. Wang and K.-C. Wei, et al., Augmented cellular uptake of nanoparticles using tea catechins: effect of surface modification on nanoparticle-cell interaction, Nanoscale, 2014, 6, 10297–10306 RSC.
- F. Rusmini, Z. Zhong and J. Feijen, Protein immobilization strategies for protein biochips, Biomacromolecules, 2007, 8, 1775–1789 CrossRef CAS PubMed.
- M. Wilchek, E. A. Bayer and O. Livnah, Essentials of biorecognition: the (strept)avidin–biotin system as a model for protein–protein and protein–ligand interaction, Immunol. Lett., 2006, 103, 27–32 CrossRef CAS PubMed.
- S. Sun, M. Ma, N. Qiu, X. Huang, Z. Cai and Q. Huang, et al., Affinity adsorption and separation behaviors of avidin on biofunctional magnetic nanoparticles binding to iminobiotin, Colloids Surf., B, 2011, 88, 246–253 CrossRef CAS PubMed.
- R. Rezaeipoor, R. John, S. G. Adie, E. J. Chaney, M. Marjanovic and A. L. Oldenburg, et al., Fc-directed antibody conjugation of magnetic nanoparticles for enhanced molecular targeting, J. Innovative Opt. Health Sci., 2009, 2, 387–396 CrossRef PubMed.
- L. Nobs, F. Buchegger, R. Gurny and E. Allémann, Biodegradable nanoparticles for direct or two-step tumor immunotargeting, Bioconjugate Chem., 2006, 17, 139–145 CrossRef CAS PubMed.
- K. Yan, P. Li, H. Zhu, Y. Zhou, J. Ding and J. Shen, et al., Recent advances in multifunctional magnetic nanoparticles and applications to biomedical diagnosis and treatment, RSC Adv., 2013, 3, 10598–10618 RSC.
- A. Cirstoiu-Hapca, L. Bossy-Nobs, F. Buchegger, R. Gurny and F. Delie, Differential tumor cell targeting of anti-HER2 (Herceptin) and anti-CD20 (Mabthera) coupled nanoparticles, Int. J. Pharm., 2007, 331, 190–196 CrossRef CAS PubMed.
- A. Hanini, A. Schmitt, K. Kacem, F. Chau, S. Ammar and J. Gavard, Evaluation of iron oxide nanoparticle biocompatibility, Int. J. Nanomed., 2011, 6, 787–794 CAS.
- M. Calero, L. Gutiérrez, G. Salas, Y. Luengo, A. Lázaro and P. Acedo, et al., Efficient and safe internalization of magnetic iron oxide nanoparticles: two fundamental requirements for biomedical applications, Nanomedicine: Nanotechnology, Biology and Medicine, 2014, 10, 733–743 CrossRef CAS PubMed.
- M. Calero, M. Chiappi, A. Lazaro-Carrillo, M. J. Rodriguez, F. J. Chichon and K. Crosbie-Staunton, et al., Characterization of interaction of magnetic nanoparticles with breast cancer cells, J. Nanobiotechnol., 2015, 13, 16 CrossRef PubMed.
- S. Shukla, A. Jadaun, V. Arora, R. K. Sinha, N. Biyani and V. K. Jain, In vitro toxicity assessment of chitosan oligosaccharide coated iron oxide nanoparticles, Toxicol. Rep., 2015, 2, 27–39 CrossRef CAS.
- S. Laurent, C. Burtea, C. Thirifays, U. O. Häfeli and M. Mahmoudi, Crucial ignored parameters on nanotoxicology: the importance of toxicity assay modifications and “cell vision”, PLoS One, 2012, 7, e29997 CAS.
- M. Neri, C. Maderna, C. Cavazzin, V. Deidda-Vigoriti, L. S. Politi and G. Scotti, et al., Efficient in vitro labeling of human neural precursor cells with superparamagnetic iron oxide particles: relevance for in vivo cell tracking, Stem Cells, 2008, 26, 505–516 CrossRef CAS PubMed.
- T. Schlorf, M. Meincke, E. Kossel, C. C. Gluer, O. Jansen and R. Mentlein, Biological properties of iron oxide nanoparticles for cellular and molecular magnetic resonance imaging, Int. J. Mol. Sci., 2010, 12, 12–23 CrossRef PubMed.
- S. Naqvi, M. Samim, M. Abdin, F. J. Ahmed, A. Maitra and C. Prashant, et al., Concentration-dependent toxicity of iron oxide nanoparticles mediated by increased oxidative stress, Int. J. Nanomed., 2010, 5, 983–989 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra02084h |
| This journal is © The Royal Society of Chemistry 2017 |