Open Access Article
Open Access ArticleNanoslit-concentration-chip integrated microbead-based protein assay system for sensitive and quantitative detection†
Yul Koh‡
 a,
Jin-Kyoung Yang‡b,
Min-Hye Oha,
Homan Kang§
c,
Yoon-Sik Lee*bc and
Yong-Kweon Kim*a
a,
Jin-Kyoung Yang‡b,
Min-Hye Oha,
Homan Kang§
c,
Yoon-Sik Lee*bc and
Yong-Kweon Kim*a
aDepartment of Electrical and Computer Engineering, Seoul National University, Seoul 151-742, Republic of Korea. E-mail: yongkkim@snu.ac.kr; Fax: +82-2-880-9953; Tel: +82-2-880-7440
bSchool of Chemical and Biological Engineering, Seoul National University, Seoul 151-742, Republic of Korea. E-mail: yslee@snu.ac.kr
cNano Systems Institute and Interdisciplinary Program in Nano-Science and Technology, Seoul National University, Seoul 151-742, Republic of Korea
First published on 7th June 2017
Abstract
A nanoslit-concentration-chip (NC-chip) integrated microbead assay system was fabricated to overcome the assay limits of the conventional microbead-based protein assay. The proposed assay system was composed of a microbead-trapping-chip (MT-chip) and a NC-chip as a protein-capturing device and a quantitative and sensitive protein detection device, respectively. Microbeads trapped in the MT-chip formed a three-dimensional microbead packed column leading to efficient protein capturing. The NC-chip was used to trap and concentrate nanoparticle (NP) probes that were specifically captured in the microbead bed of the MT-chip. Furthermore, the nanoslit-patterned membrane of the NC-chip enabled both signal enhancement and quantification of the amount of the trapped NPs. The NC-chip integrated microbead assay system was successfully applied to a protein assay. The sensitivity and reliability of streptavidin detection by HPQ motif containing peptide ligands were compared to those of other reported assay systems. This novel protein assay system could be applied to various microbead-based bioassays, and integration of the NC-chip could further improve their sensitivity and reliability.
Introduction
A microbead-based assay is one of the most powerful methods for detecting biomolecules in lab-on-a-chip systems due to several outstanding merits.1,2 First, the surface of the microbeads can be readily modified by various ligands such as aptamers and antibodies to specifically capture target biomolecules. Second, the microbead-based assay allows much higher biomolecule capturing efficiency compared to the microchannel based assay, since the surface-to-volume ratio of the microbeads is much larger than that of the microfluidic channels.3 Third, the microbead-based microfluidic assay shows efficient mixing of sample solutions by generating torque, whereas the mixing process in conventional microfluidic systems depends on the laminar flow which decreases the mobility of the biomolecules.4 Lastly, the shape and size of the microbeads can be controlled by a batch-type production, leading to uniform loading of ligands to each microbead and subsequent increase in assay reliability. Therefore, microfluidic assays with microbeads have attracted much attention in bio-analytical fields.Recently, various microstructures have been designed to efficiently incorporate microbeads into the microfluidic system; physical isolation,5 magnetic trapping,6,7 and electrical trapping.8 Above all, packing the target-specific microbeads within a specifically designed micro structure for physical isolation is the most frequently used strategy due to its simplicity.9–11 Typically, the microbeads are trapped by a weir or pillars structure in a microfluidic channel, resulting in the formation of a loosely packed 3-dimensional (3-D) microbead-bed. The packed structure of the microbeads reduces the diffusion length of sample solution, which is favorable to capture target biomolecules via specific reaction with ligands on the surface of microbeads. Therefore, microfluidic assay with 3-D microbead-bed has been successfully applied for highly sensitive detection of various biomolecules such as DNA,12 viruses,13 and cancer biomarkers.14
The detection of target–molecule binding event in a microfluidic system mainly relies on the use of fluorescent dye or nanoparticle (NP) probe.6,15 Especially, fluorescent NPs containing target biomolecules have been broadly employed as a secondary probe, and their fluorescence signals were directly measured in the microbead-bed region after their binding to the microbead. However, these direct measurements in the microbead-bed region have been suffered from several critical problems: first, inaccuracy of the fluorescent signal measurement is unavoidable due to the depth of 3-D structured microbead-bead.16 As the optical signals generated from the NP probes are distorted and scattered when the signals are passed through the microbead-bed, they exhibit distance variations when viewed by fluorescent microscopy. Hence, an expensive and complex instrument such as a confocal microscope is necessary to measure the optical signals of NPs accurately in the microbead-bed region. Second, the area of the microbead-bed is usually too large to measure the optical signals. A large amount of the microbeads is required for the sensitive detection of biomolecules, which inevitably give rise to long measuring time due to the increase in the amount of the microbeads and subsequent measuring area.17 Third, the microbeads usually emit its own fluorescent signals which can interfere the sensitive detection of NPs fluorescent signal with decreased signal-to-noise ratio (SNR).18 Therefore, a new measuring method which is not influenced by the structure and background signal of the 3-D microbead-bed is highly required for sensitive and quantitative detection of biomolecules.
Previously, we have developed a nanoslit membrane structure for size-dependent particle trapping,19 which enabled particle separation, concentration, and quantification. Herein, we demonstrate a new assay strategy based on the nanoslit-concentration-chip (NC-chip) integrated microbead assay system for the sensitive and quantitative detection of target biomolecules. Microbeads were trapped in a weir-structured microbead-trapping-chip (MT-chip) to form 3-D packed microbead-bed, and effective binding event occurs between microbeads and NPs based on ligand-target molecules interactions. After the binding, the specifically bound NPs were released from the microbeads, and transported to the NC-chip, where NPs could give highly enhanced fluorescent signals as a result of concentrated NPs within small measuring area. Thus, the NC-chip integrated microbead assay system was successfully applied to streptavidin assay with increased detection sensitivity compared to the conventional microbead based assay systems.
Design and fabrication
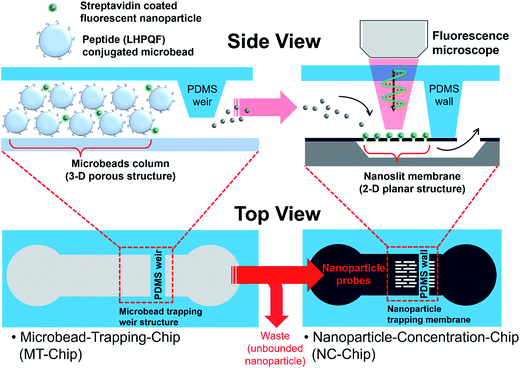
Design of NC-chip integrated microbead assay system
The NC-chip integrated microbead assay system is comprised of two separated devices; the MT-chip for efficient recognition events between target biomolecules adsorbed on NPs and target-specific ligand on microbeads, and the NC-chip for sensitive and quantitative detection of target bound NP probes. As shown in Fig. 1, each part was designed to have a characteristic structure corresponding to each function. In the MT-chip, a weir structure (500 μm width, 80 μm and 20 μm height) was employed to confine 30 μm-sized TentaGel microbeads bearing a peptide ligand for specific reaction with streptavidin coated fluorescent NPs (Stav-FNPs). Stav-FNPs were released from the microbeads and collected in the NC-chip after the binding events in the MT-chip. The NC-chip is equipped with a nanoslit membrane to concentrate Stav-FNPs in a small detection area (500 μm × 300 μm) for sensitive protein detection.Fabrication process and results of NC-chip integrated microbead assay system
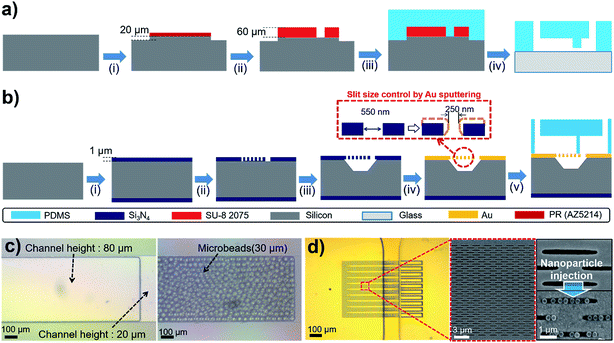
The detailed fabrication process of the MT- and NC-chips are shown in Fig. 2. For fabrication of the MT-chip, the weir-structured microfluidic PDMS mold was prepared by deep reactive ion etching (DRIE) and SU-8 photolithography (Fig. 2a). The first layer of the microfluidic channel was fabricated by etching 20 μm via DRIE. SU-8 photoresist was then patterned on the microfluidic channel at a height of 60 μm to make a weir structure. A PDMS fluid channel was replicated from the mold and bonded to a slide glass using O2 plasma surface treatment. Fig. 2b shows the NC-chip fabrication process. First, a Si3N4 layer (1 μm) was deposited on a silicon wafer by low-pressure chemical vapor deposition (LPCVD) to form a membrane structure. A 500 nm × 4 μm slit pattern array was patterned on the Si3N4 layer by stepper lithography and a dry etching process. The silicon substrate located under the nanoslit-patterned Si3N4 membrane was then removed by 4 μm DRIE and potassium hydroxide (KOH) wet etching for 30 minutes. The wet etching process released the nanoslit-patterned Si3N4 membrane and formed a cavity for bottom fluid channels. Finally, Cr/Au was deposited on the released nanoslit membrane by sputtering to narrow the width of the nanoslit from 500 nm to 250 nm. This additional sputtering process proves to be simple to adjust the width of the nanoslit pattern efficiently, and can be applied to further decrease the width. The PDMS channels of NC-chip was also fabricated using a SU-8 mold. Finally, the PDMS channel and the nanoslit-membrane were clamped by an acrylic plastic housing for sample fluid injection without leakage.The fabrication results of the MT-chip and the NC-chip are shown in Fig. 2c and d. The channel height of MT-chip abruptly decreased from 80 μm to 20 μm at the end of the channel, resulting in the weir-structured channel (Fig. 2c). As the diameter of microbead is 30 μm, this weir structure of MT-chip was sufficient to trap 30 μm-sized microbeads in the 80 μm height zone. Fig. 2d shows the Cr/Au deposited nanoslit membrane and the aligned PDMS channel. The PDMS channel was aligned to guide flow of the sample solution into the membrane. In addition, the enlarged SEM images show that the NC-chip consisted of numerous nanoslit array, in which NPs larger than the slit width could be trapped efficiently as shown in Fig. 4d (more fabrication results and SEM images are shown in Fig. S1 ESI†).
Experimental set up for NC-chip integrated microbead assay system
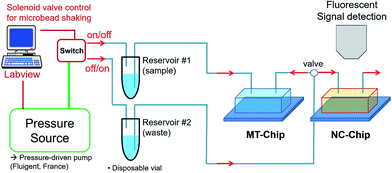
A pressure-based fluidic control system was employed for precise sample transportation, mixing, and washing, instead of using a microsyringe pump. Compared to the microsyringe pump, a pressure-driven flow has advantages of a fast response time and small flow fluctuations. The schematics of the experimental set-up is illustrated in Fig. 3. The sample solutions were loaded in a plastic disposable vial (reservoir #1) and connected to the inlet of the MT-chip for a forward flow. The outlet of the MT-chip was also connected to another plastic disposable vial (reservoir #2) for a backward flow. Forward/backward flows were controlled via solenoid valves with a Labview™ program. The sequential forward/backward flows enable effective mixing of the NPs with the microbeads, leading to effective target binding. After the binding events, only forward flow was allowed for washing, and waste was eluted via an outlet to reservoir #2. For detection, the outlet of the MT-chip was connected to the inlet of the NC-chip with a valve to allow the target bound NPs to flow. All events occurring in the MT- and NC-chips were monitored using a CCD camera-linked microscope (ProgRes C3, JENOPTIK, Germany), and the trapped NPs were also quantified by fluorescence signals using a fluorescence filter set (467–498 nm excitation wavelength, 520 nm emission wavelength).Result and discussion
Experimental design to evaluate the performance of the NC-chip integrated microbead assay system
To demonstrate the efficiency of the proposed system, streptavidin and HPQ containing pentapeptide20 was chosen as a model protein–ligand binding assay. As it has been known that the affinity of the HPQ-containing peptide is depending on the peptide sequence, accuracy of the proposed assay system could be evaluated. We chose three pentapeptides (LHPQF, FHPQG, and IHPQG), which has high, middle, and low affinities to streptavidin. Each pentapeptide was synthesized on TentaGel bead (TG-peptide) by the conventional solid-phase synthesis method. Commercial streptavidin-coated fluorescent NPs (Stav-FNPs, SVFP-0552-5, Spherotech, USA) were used as the target material with a signaling probe. The assay performance of the proposed system was evaluated in two ways. First, the efficiency of NPs trapping in the NC-chip was investigated to find out the detection limit improvements. Next, the accuracy of the assay system was investigated using the LHPQF, FHPQG, and IHPQG conjugated TentaGel beads. The assay results for the three pentapeptides were compared to their known affinities toward streptavidin.Assay results of the NC-chip integrated microbead assay systems
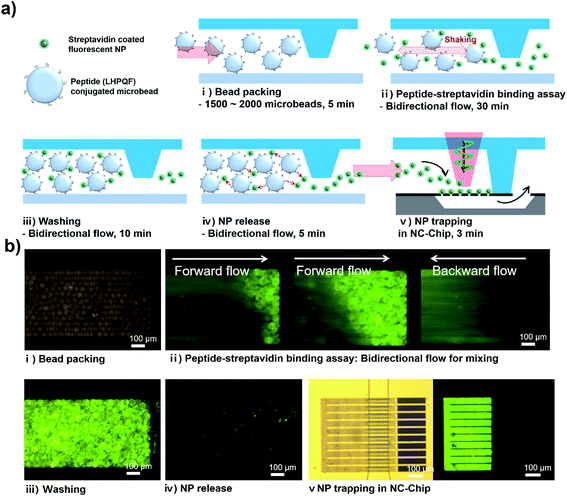
The assay procedure was illustrated in Fig. 4a. (i) First, TG-peptide were trapped in a weir-structure of the MT-chip within 5 min after injection (≈2 × 103). (ii) Stav-FNPs were injected to the TG-peptide packed column, followed by mixing with pressure-based bi-directional flows for 30 min. The forward and backward flows were applied for 2.8 s and 1.2 s at 1.5 kPa for one period and repeated several times. (iii) After the incubation step, the unbounded Stav-FNPs in the MT-chip was washed out by bi-directional flow with a PBS solution (10 mM, pH 7.0) for 10 min. (iv) Next, the captured Stav-FNPs were released from the microbeads by means of chemical and physical treatment. Chemical release of FNPs from TG-peptide was conducted using formamide which can induce dissociation of the streptavidin–peptide binding by conformational change of streptavidin.21 In addition, we found out the additional pressure-based releasing step was essential to release the Stav-FNPs which wasn't released in the first chemical treatment (Fig. S2†). Therefore, a high pressure bi-directional flow was applied to the microbeads to give a vibrational shock to ensure improved releasing of FNPs. The applied pressure was 15 kPa and the duration time of forward/backward flow was 0.55/0.45 s for one cycle period. (v) Finally, a 30 μl of the solution which contained the released FNPs was transported to the NC-chip and injected into the nanoslit membrane for 3 min with 5 kPa. The fluorescence signal of trapped FNPs were detected and quantified using fluorescence microscopy.Each assay steps described above was confirmed by fluorescence measurement. (i) TG-peptide (LHPQF) packed in the MT-chip emitted weak fluorescence signal due to the innate background of the resin beads, which could be considered as a noise leading to decrease in signal sensitivity. (ii) Bi-directional flow depending on the pressure-driven mixing demonstrated that Stav-FNPs (20 μl, 108 ml−1) were reacted well with the pre-packed TG-LHPQF (see Movie S1 ESI†). (iii) After the mixing step, fluorescent signal dramatically increased as a result of specific binding between TG-LHPQF and Stav-FNPs. (iv) Concurrent treatment with formamide and high pressure shaking resulted in successful release of the FNPs, and fluorescence signal level in the region of packed TG-LHPQH substantially decreased to the similar level with the bead background (see Movie S2 ESI†). (v) Finally, the released Stav-FNPs were successfully trapped in the NC-chip (see Movie S3 ESI†). The target FNP recognition in the microbead column, the FNP release from the microbeads, and the FNP trapping and detection in the NC-chip were successfully observed using the fluorescent microscopy. These results clearly demonstrate the basic concept and working principle of the NC-chip integrated microbead assay system.
Enhancement of the detection sensitivity using NC-chip integrated microbead assay system
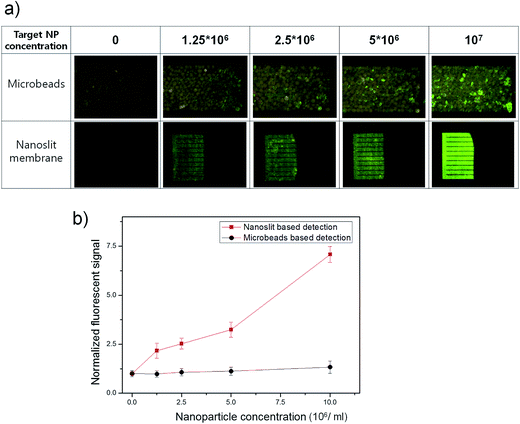
The NC-chip integrated microbead assay system is expected to enhance the detection sensitivity of target NP probes by concentrating the NP probes within the small detection area of NC-chip. Conventional microbead assay usually utilizes large size three-dimensional microbead column to maximize the binding efficiency between the target molecules and the ligand conjugated microbeads. However, this can also decrease the population density of the target NP probes leading to low sensitivity, especially at low target concentrations. We investigated the enrichment effect of the Stav-FNPs via trapping within the NC-chip on the assay sensitivity. For this, Stav-FNPs of various concentrations (1.25 × 106 ml−1, 2.5 × 106 ml−1, 5 × 106 ml−1, and 107 ml−1) were injected and incubated with the packed TG-peptide in the MT-chip. After incubation and washing, the fluorescent signals of the bound Stav-FNPs in the MT-chip were measured for each Stav-FNPs concentrations. Then, the captured Stav-FNPs were released from the TG-peptide in the MT-chip and transported to the NC-chip. Finally, the fluorescent signals from Stav-FNPs trapped on the nanoslit membrane were measured.As shown in Fig. 5a, the fluorescence signals from the FNPs were significantly higher in the NC-chip compared to the signals from the MT-chip. Signals from the NC-chip were homogeneous and strong over the whole detection area, while irregular and overlapped weak fluorescence signals were observed from the microbead-bed in the MT-chip. For more rigorous comparison on the detection sensitivity, fluorescent signals of each image were quantified and normalized using an image analysis software (NI Vision Assistant). As shown in Fig. 5b, the fluorescence signal of FNPs in the NC-chip increased proportionally with Stav-FNPs concentrations ranging from 1.25 × 106 to 107 ml−1. Whereas, the fluorescence signal of FNPs in the MT-chip showed marginal increase up to the concentration of 107 ml−1 Stav-FNPs that showed similar detection limit of conventional peptide-streptavidin microbead assay (Fig. S3†). This increased sensitivity of NC-chip might be attributed to the concentration of Stav-FNPs at the nanoslit membrane. The nanoslit membrane has 500 μm × 300 μm area which is 13.3 times smaller than the area of the microbead-bed in the MT-chip (500 μm × ∼4000 μm). The low background noise of the nanoslit membrane also contributes to an increase in detection sensitivity. As nanoslit membrane (Cr/Au deposited) emit significantly low background fluorescence signal compared to the polymer microbeads, the contrast of the fluorescence of FNPs could be enhanced. The detection limit of FNPs in the NC-chip was expected to be lower than 1.25 × 106 Stav-FNPs ml−1 because the normalized fluorescent signal was substantially high (2.2) at 1.25 × 106 Stav-FNPs ml−1, whereas fluorescence signal in MT-chip showed only small signal difference (1.3) even at higher FNP concentrations (107 ml−1). The detection limit of NC-chip integrated system for the detection of streptavidin was 163 pM, which is lower than the previously reported chip-based biotin-streptavidin assay.22,23 Considering that HPQ peptides are known to exhibit lower affinity to streptavidin than biotin,20,24,25 these results clearly suggest that the NC-chip based detection can significantly enhance the detection sensitivity by concentrating the target Stav-FNPs into a small nanoslit membrane region. The detection sensitivity can be further increased by simply increasing the number of packed microbeads in MT-chip. Furthermore, the NC-chip based detection system is very useful because it can be widely utilized in the microbead-based assay system by simply connecting the NC-chip to the microbead column.
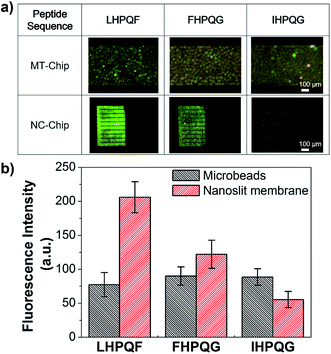
Quantitative analysis of the streptavidin–pentapeptide binding using the NC-chip integrated microbead assay system
Generally, fluorescence signals from the microbead column are hard to quantify with high accuracy because the background fluorescent signals emitted from the microbeads exhibit wide variety due to the size, packing density, and the surface properties of the microbeads. However, the NC-chip integrated microbead assay system can remove the background fluorescent signals emitted from the microbeads, and only measure the signals of the FNPs trapped within the nanoslit membrane which emits low background fluorescent signal. Therefore, compared to the conventional microbead assay system, the proposed assay system could have higher detection accuracy and reliability for the quantitative analysis. To investigate the ability of the quantitative analysis of the NC-chip based detection, streptavidin–peptide binding assay was performed using three different HPQ containing pentapeptides, LHPQF, FHPQG, and IHPQG, which have high, middle, and low affinities to streptavidin. It is expected that fluorescence intensity from the trapped Stav-FNPs in NC-chip reveals the same order of the affinity of HPQ peptides to streptavidin. The experimental procedures were the same as previously described and the concentration of the Stav-FNPs were 107 ml−1. Fig. 6a showed the experimental results of the streptavidin–peptide binding assay using the peptides. The fluorescent signals from the microbead column were very small and irregular, and not matched to the expected assay results. However, the fluorescent signals at the NC-chip were clearly matched to the expected values (LHPQF > FHPQG > IHPQH). The fluorescent signals were also quantified by the image analysis software (NI Vision Assistant) as shown in Fig. 6b, which showed dramatic difference in the detection accuracy and reliability between the MT-chip and the NC-chip. In the MT-chip, the signals of the FNPs were overlapped and buried under the variation of background signals so that affinity tendency cannot be observed. On the other hand, the NC-chip clearly showed the same affinity tendency as expected. From these results, we can conclude that the NC-chip integrated microbead assay system improved the detection accuracy and reliability by reducing the variation of the background fluorescent signals that is an inevitable problem when the microbead is used for bioassay. Moreover, the assay results of the proposed system are highly quantitative, and thus the NC-chip device can be applied to various protein–ligand affinity screening.Conclusion
We demonstrated that the NC-chip integrated microbead assay system could improve the detection sensitivity and signal reliability of the microbead based assay system. The assay system consists of the bio-molecule capturing device (MT-chip) and the signal amplification and detection device (NC-chip) to improve the detection limit with quantification ability. In particular, the nanoslit membrane of the NC-chip can concentrate NPs into a small area, and thus, enhance its fluorescence intensity as well as detection sensitivity. In addition, trapping the FNPs released from the microbead in the NC-chip can contribute to more accurate and reliable bioassay with high SNR of the FNPs as the background signal of nanoslit membrane is substantially low compared to that of microbead. The excellent performance of the NC-chip system was successfully demonstrated via streptavidin–peptide binding assay as a model. The NC-chip integrated microbead assay system showed the improved detection sensitivity over 10 times compared to that of the conventional microbead based assay. This NC-chip integrated microbead assay system has a great potential for the sensitive and quantitative detection of various target molecules in various bio related fields.Acknowledgements
This research was supported by the Pioneer Research Center Program through the National Research Foundation of Korea funded by the Ministry of Science, ICT & Future Planning (NRF-2011-0027888).References
- H. Kawaguchi, Prog. Polym. Sci., 2000, 25, 1171–1210 CrossRef CAS.
- E. Verpoorte, Lab Chip, 2003, 3, 60N–68N RSC.
- J. A. Thompson and H. H. Bau, Microfluid. Nanofluid., 2012, 12, 625–637 CrossRef CAS.
- H. Bruus, Theoretical microfluidics, Oxford University Press, Oxford, New York, 2008 Search PubMed.
- H. Andersson, W. van der Wijngaart, P. Enoksson and G. Stemme, Sens. Actuators, B, 2000, 67, 203–208 CrossRef CAS.
- Y. H. Tennico, D. Hutanu, M. T. Koesdjojo, C. M. Bartel and V. T. Remcho, Anal. Chem., 2010, 82, 5591–5597 CrossRef CAS PubMed.
- H. C. Tekin and M. A. Gijs, Lab Chip, 2013, 13, 4711–4739 RSC.
- Y. S. Huh, A. J. Chung, B. Cordovez and D. Erickson, Lab Chip, 2009, 9, 433–439 RSC.
- K. Sato, M. Tokeshi, H. Kimura and T. Kitamori, Anal. Chem., 2001, 73, 1213–1218 CrossRef CAS PubMed.
- Y. Koh, B. R. Lee, H. J. Yoon, Y. H. Jang, Y. S. Lee, Y. K. Kim and B. G. Kim, Anal. Bioanal. Chem., 2012, 404, 2267–2275 CrossRef CAS PubMed.
- T. Nguyen, R. Pei, M. Stojanovic and Q. Lin, Microfluid. Nanofluid., 2009, 6, 479–487 CrossRef CAS.
- P. B. Monaghan, K. M. McCarney, A. Ricketts, R. E. Littleford, F. Docherty, W. E. Smith, D. Graham and J. M. Cooper, Anal. Chem., 2007, 79, 2844–2849 CrossRef CAS PubMed.
- H. Zhang, T. Xu, C.-W. Li and M. Yang, Biosens. Bioelectron., 2010, 25, 2402–2407 CrossRef CAS PubMed.
- S. W. Han, E. Jang and W.-G. Koh, Sens. Actuators, B, 2015, 209, 242–251 CrossRef CAS.
- H. Zhang, Y. Liu, X. Fu, L. Yuan and Z. Zhu, Microchim. Acta, 2015, 182, 661–669 CrossRef CAS.
- M. Schröder, E. von Lieres and J. Hubbuch, J. Phys. Chem. B, 2006, 110, 1429–1436 CrossRef PubMed.
- X. Zhao and S. A. Shippy, Anal. Chem., 2004, 76, 1871–1876 CrossRef CAS PubMed.
- H. J. Olivos, K. Bachhawat-Sikder and T. Kodadek, ChemBioChem, 2003, 4, 1242–1245 CrossRef CAS PubMed.
- Y. Koh, H. Kang, S. H. Lee, J. K. Yang, J. H. Kim, Y. S. Lee and Y. K. Kim, Lab Chip, 2014, 14, 237–243 RSC.
- K. S. Lam, S. E. Salmon, E. M. Hersh, V. J. Hruby, W. M. Kazmierski and R. J. Knapp, Nature, 1991, 354, 82–84 CrossRef CAS PubMed.
- O. Nord, M. Lukacs, J. Lundeberg and M. Uhlén, Electrophoresis, 2005, 26, 501–510 CrossRef PubMed.
- K.-C. Wang, A. Kumar, S. J. Williams, N. G. Green, K. C. Kim and H.-S. Chuang, Lab Chip, 2014, 14, 3958–3967 RSC.
- C. Benz, H. Retzbach, S. Nagl and D. Belder, Lab Chip, 2013, 13, 2808–2814 RSC.
- L. B. Giebel, R. Cass, D. L. Milligan, D. Young, R. Arze and C. Johnson, Biochemistry, 1995, 34, 15430–15435 CrossRef CAS PubMed.
- P. C. Weber, M. W. Pantoliano and L. D. Thompson, Biochemistry, 1992, 31, 9350–9354 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra02460f |
| ‡ These authors are contributed equally to this work. |
| § Current address: Department of Radiology, Harvard Medical School and Gordon Center for Medical Imaging, Massachusetts General Hospital, Boston, Massachusetts 02129, USA. |
| This journal is © The Royal Society of Chemistry 2017 |