Open Access Article
Open Access ArticleFunction analysis and application of IAP1/2 of Autographa californica multiple nucleopolyhedrovirus†
Yuejun Fu *a,
Leixi Caoa,
Shuju Wub and
Aihua Lianga
*a,
Leixi Caoa,
Shuju Wub and
Aihua Lianga
aKey Laboratory of Chemical Biology and Molecular Engineering of Ministry of Education, Institute of Biotechnology, Shanxi University, Taiyuan 030006, PR China. E-mail: yjfu@sxu.edu.cn; Fax: +86 351 7011499; Tel: +86 351 7016125
bSchool of Life Science and Technology, Harbin Normal University, Harbin, 150025, PR China
First published on 24th April 2017
Abstract
The baculovirus Autographa californica multiple nucleopolyhedrovirus (AcMNPV) possesses two genes, iap1 and iap2, which encode the inhibitors of apoptosis proteins (IAP). One previous work showed that transient expression of Ac-IAP1 was capable of inducing apoptosis of Sf9 cells, but others demonstrated that Ac-IAP1 had anti-apoptosis activity against apoptosis induced by HearNPV in Tn-Hi5 cells. So the function of Ac-IAP1 remained unclear. To further define the function of Ac-IAPs, we used a Bac-to-Bac baculovirus expression system to generate two recombinant baculoviruses, AcMNPV-iap1-egfp and AcMNPV-iap2-egfp. Function analysis showed that Ac-IAP1 could induce the apoptosis process of Sf9 cells in a dose-dependent manner, even in the medium starvation mode. However, Ac-IAP2 could promote cell proliferation and the BIR2 domain in Ac-IAP2 played an important role in anti-apoptotic ability. Moreover, an insecticidal potency test showed that the larvae of Helicoverpa armigera in the AcMNPV-iap2-egfp group had a higher mortality rate (61.11%) and a lower pupation rate, although the time of death and pupation were delayed, compared with those in AcMNPV-egfp and AcMNPV-iap1-egfp groups, which confirmed that the recombinant virus AcMNPV-iap2-egfp had better insecticidal efficiency. This study confirmed the function of Ac-IAPs and developed a useful AcMNPV-iap2-egfp, which provided the experimental basis for the mechanistic analysis of Ac-IAPs and the theoretical foundation for using and modifying AcMNPV.
1. Introduction
Apoptosis is an important defense against virus infection, and replication of most insect viruses can induce apoptosis. To inhibit this highly conserved self-destruct process and expedite the related virus multiplication and dissemination, baculoviruses acquire auxiliary genes from the ancestral host insect to control host physiology, and the inhibitor of the apoptosis (iap) gene is one of these host homologs.1–3 The iap genes were identified firstly in baculoviruses, Cydia pomonella granulovirus (CpGV) and Orgyia pseudotsugata multiple nucleopolyhedrovirus (OpMNPV), as these genes encoded an anti-apoptotic protein that suppressed virus-induced apoptosis of Spodoptera frugiperda 21 (Sf21) cells upon infection with a recombinant Autographa californica multiple nucleopolyhedrovirus (AcMNPV) harboring a defective p35 gene.4,5 Homologues of the baculovirus iap genes were subsequently detected in yeast, nematodes, insects and mammals.6–8Baculoviral IAPs are classified into six groups, IAP1–IAP6, according to their sequence homology.1 However, only a subset of these IAPs function as apoptosis suppressors and the role of many other IAPs are unknown in cellular survival and differentiation. In most of the baculoviruses, IAP3 has an anti-apoptosis function.9–13 IAPs have 1–3 baculovirus IAP repeats (BIRs) in the N-terminal region and a RING finger domain in the C-terminal region. BIRs are unique to IAPs and consist of ∼70 amino acids with the signature sequence CX2CX16HX6C, which coordinate a zinc ion and mediate protein–protein interaction. An individual BIR domain is required for anti-apoptotic activity of IAPs. The RING finger domain functions as an E3 ubiquitin ligase, and consists of approximately 40 amino acids, including eight cysteine and histidine residues that form a binding site for two zinc ions, which promotes the transfer of ubiquitin onto selected protein targets, whereby the IAPs are involved in the proteasome-mediated degradation of target proteins.14
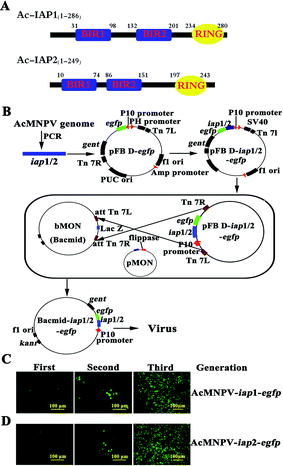
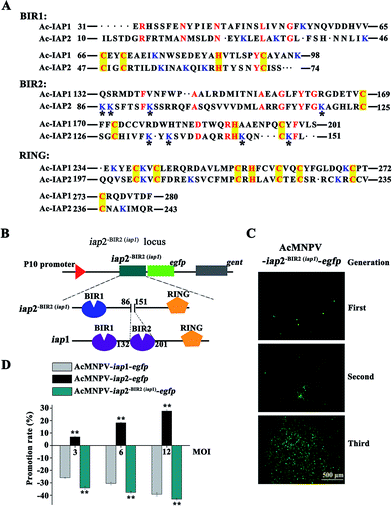
The genome of AcMNPV contains two iap genes, iap1 and iap2. Ac-IAP1 and Ac-IAP2 have two well-conserved BIRs and a RING domain. BIR1 domain is more conserved than BIR2 domain across species or one species different IAPs (Fig. 1A).1 To disclose the role of the AcMNPV iap genes, the over-expressing vectors, pIZ/V5-iap1, pIZ/V5-iap2, and recombinant HearNPV expressing AcMNPV iap1 and iap2 were constructed and the results indicated that both AcMNPV IAP1 and IAP2 functioned independently as apoptosis inhibitors.5 However, transient expression of Ac-IAP1 was capable of inducing apoptosis and stimulating caspase-3 protease activity in various lepidopteran and dipteran cell lines.15 So, the function of Ac-IAPs remained unknown. In this study, Bac-to-Bac baculovirus expression system was used to generate two recombinant baculoviruses, AcMNPV-iap1-egfp and AcMNPV-iap2-egfp, the effects of AcMNPV-iap1/2-egfp on Sf9 (Spodoptera frugiperda 9) cells and Helicoverpa armigera larvae were examined, and the function of BIR2 domain in Ac-IAP2 was analyzed preliminarily, which further confirmed the role of Ac-IAP1/2 during viral infection and offered experimental basis for using AcMNPV-iap2-egfp as a stronger insecticide and a more effective insect cell expression system.
2. Materials and methods
2.1 Materials
Sf9 cells, AcMNPV, AcMNPV-egfp, E. coli DH10B and pFastBac dual-egfp were maintained in our laboratory. Helicoverpa armigera eggs were purchased from Jiyuan Baiyun biotechnology Corp Ltd (Jiyuan, China). Restriction enzyme Sam I, Xho I and T4 DNA ligase enzyme were purchased from Takara Biotechnology Corp Ltd (Dalian, China). The anti-β-actin and anti-GFP antibody were purchased from Sangon Biotechnology Corp (Shanghai, China). Cell lysis buffer and BCA Protein Assay kit were purchased from Beyotime Biotechnology Corp (Jiangsu, China). Roxithromycin, vitamin C and compound vitamin B tablets were purchased from Tianjin yabao pharmaceutical technology Corp Ltd (Tianjin, China). Other chemical reagents were reagent grade and purchased from Sangon Biotech (Shanghai, China).The composition of artificial diet for Helicoverpa armigera larvae was as followed: 50 g soybean flour, 75 g maize flour, 20 g yeast extract powder, 2 g calcium benzoate, 10 g agar powder, 7.5 g vitamin C, 0.375 g compound vitamin B, 75 mg roxithromycin, 15 ml 36% (v/v) glacial acetic acid, 2 ml 10% (v/v) formaldehyde and 500 ml water.
2.2 Cell culture
The Sf9 cells were cultured at 27 °C in serum-free insect cell culture medium (HyClone, USA). Cell cultures were prepared and maintained according to standard cell culture procedures.162.3 Construction of recombinant bacmid-iap1/2-egfp
As shown in Fig. 1B, to generate bacmid-iap1-egfp, we first amplified the iap1 gene fragment from the AcMNPV genome and primers iap1-Sam I–F (5′-TACCCGGGTATGAACGAGG-ACACG-3′, Sam I site was marked in bold) and iap1-Xho I–R (5′-GCCTCGAGGCACC-ACAAATATTTTTAT-3′, Xho I site was marked in bold) were used in polymerase chain reaction (PCR). The PCR product was cloned between the Sam I site and Xho I site in the pFastBac dual-egfp (pFB D-egfp) vector. DNA sequences were determined using an ABI Prism 3100 DNA sequencer (Applied Biosystems). The resulting plasmid (pFB D-iap1-egfp), namely, intermediate vector was transformed into the Escherichia coli strain DH10 Bac and transposed into the AcMNPV genome, followed by PCR verification with primer M13-F (5′-CGCCAGGGTTTTCCCAGTCACGAC-3′) and M13-R (5′-CACACAGGAAACA GCTATGAC-3′). On the other hand, the existence of gentamicin and kanamycin resistant genes in the recombinant bacmid could act as selection markers. Tandem gene iap1-egfp was inserted in the place of the polyhedron gene (located in the wide type baculovirus genome) via Tn7 transpositional recombination. The resulting bacmid was named as bacmid-iap1-egfp (Fig. 1B).Analogously, the construction process of bacmid-iap2-egfp was primarily to generate an intermediate vector, iap2 gene was amplified from the AcMNPV genome using the primers iap2-Sam I–F (5′-ATCCCGGGTATGAATTTGAT GCAATTT-3′, Sam I site was marked in bold) and iap2-Xho I–R (5′-GGCTCGAGACTGAGGTAATG TTTCGAT-3′, Xho I site was marked in bold). The PCR product was inserted into pFastBac dual-egfp vector and pFB D-iap2-egfp was generated. pFB D-iap2-egfp was transposed into the AcMNPV genome and bacmid-iap2-egfp was obtained (Fig. 1B). Ac-IAP1 and Ac-IAP2 were analyzed using software of the EBI server (http://www.ebi.ac.uk/) and ExPASy sever (http://www.expasy.ch/tool/) for the prediction of domains and motifs (Fig. 1A).17
2.4 Generation of recombinant viruses AcMNPV-iap1/2-egfp
Sf9 cells were washed with PBS and seeded into 96 well plates (5 × 104 cells per well). Then, cells were transfected with 0.2 μg bacmid and 0.6 μl liposome TransFast™ Transfection Reagent (USA), and subjected to fluorescence microscopic observation at 24 h post transfection. The fluorescence intensity of EGFP was monitored by Nikon eclipse Ti microscope. At 4 d post transfection, the supernatant, which contained the first generation recombinant virus, was collected. The first generation recombinant virus was added to freshly seeded Sf9 cells, the culture was continued and the fluorescence intensity of these cells was measured daily as above. When there had enough intense fluorescence, the cells supernate and the medium, which contained the second generation recombinant virus, were collected. In turn, we obtained the third generation recombinant virus. In this step, we got the recombinant viruses AcMNPV-iap1/2-egfp (Fig. 1C and D) which had only BV form in the history of viral life. The foreign genes, iap1/2-egfp, were inserted in the place of the polyhedron gene in the construction process, so the ODV virus particle couldn't form in this method.2.5 Measurement of viral titers
The optical density method was used to measure titers of virus. The action system was listed in Table 1. The mixing reagents were incubated at 56 °C for 1 h. Then the mixture was centrifuged at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 871 g for 5 min to separate the immunoprecipitates, and the supernatants were collected. OD260 values were measured by Synergy Mx (BioTek, USA). The following formula was used to calculate the virus titer: virus titer = OD260 × dilution factor × 1012.
871 g for 5 min to separate the immunoprecipitates, and the supernatants were collected. OD260 values were measured by Synergy Mx (BioTek, USA). The following formula was used to calculate the virus titer: virus titer = OD260 × dilution factor × 1012.
| System | Control (μl) | Test (μl) |
|---|---|---|
| PBS | 960 | 860 |
| 10% SDS | 40 | 40 |
| Virus | — | 100 |
2.6 Plaque assay
Sf9 cells were seeded in 6-well plates at 80% confluence, and the medium was replaced with 1 ml serum-free insect cell culture medium after 1 h. After 30 min incubation at room temperature, 1 ml serially diluted AcMNPV-iap1/2-egfp, AcMNPV-egfp or AcMNPV was added to each well and incubated for 2 h at room temperature, respectively. The cells were washed once with fresh serum-free insect cell culture medium before 1.8% nutrient agarose overlay was added. After 4–5 d, plaques were counted and detected by fluorescence microscopy in AcMNPV-iap1/2-egfp and AcMNPV-egfp treatment groups. For AcMNPV groups, 0.01% neutral red staining reagent was added in 1.8% nutrient agarose to identify plaques. Plaques were counted from 12 to 24 h post infection (h p.i.).2.7 Western blot analysis of Ac-IAP1/2
Monolayer cultures of ∼6 × 106 Sf9 cells in 6-well microtiter plates were infected with AcMNPV-iap1/2-egfp at 3 × 1010 vp ml−1 (1 MOI), respectively. Total protein of cells infected with virus was resolved on SDS-PAGE gels, transferred onto PVDF membranes (Millipore, Billerica, MA, USA) for western blot analysis, and then blocked for 1 h in blocking buffer with slow rotation on an orbital shaker. PVDF membranes were incubated with primary anti-β-actin (diluted at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000) or anti-GFP (diluted at 1
2000) or anti-GFP (diluted at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000) antibody overnight at 4 °C, and were washed four times with PBST for 15 min. Blots were incubated with secondary antibody (IRDye 680LT, diluted at 1
2000) antibody overnight at 4 °C, and were washed four times with PBST for 15 min. Blots were incubated with secondary antibody (IRDye 680LT, diluted at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, licor, USA) for 1 h with slow rotation on an orbital shaker, washed four times with PBST for 15 min and then detected by ODYSSY CLx (Gene Company Limited, USA).
000, licor, USA) for 1 h with slow rotation on an orbital shaker, washed four times with PBST for 15 min and then detected by ODYSSY CLx (Gene Company Limited, USA).
2.8 MTT assay
The effects of AcMNPV-iap1-egfp, AcMNPV-iap2-egfp on Sf9 cells proliferation were examined by MTT assay. This assay was based on cellular conversion of a tetrazolium salt into a formazan product that was easily detected using 96-well plate reader. Therefore, MTT assay was a simple and sensitive method to study cellular survival and cellular growth, cytotoxicity, cell attachment and apoptosis. Equivalent number of Sf9 cells was added to a 96-well microtiter plates, followed by adding the medium to give the final volume of 100 μl. Then, cells were infected with AcMNPV, AcMNPV-iap1-egfp and AcMNPV-iap2-egfp for 48 h at the concentration of 0.75, 1.5, 3 and 6 × 1010 vp ml−1 (1.5, 3, 6, 12 MOI), respectively. Then tetrazolium salt was added to each well and further incubated for 4 h, the medium were discarded, and DMSO (100 μl) was added to each well. The absorbance at 490 nm of each well was recorded using a microplate reader (BioTek, USA). Inhibition rate was calculated according to the following equation: inhibition rate (%) = (C − T)/C × 100%, promoting rate (%) = (T − C)/C × 100%, where T and C represented the mean optical density of the treated sample and the control, respectively. Experiments were performed six times and the average values ± SD were recorded.2.9 Budded virus (BV) production in Sf9 cells
To determine the effect of Ac-IAP1/2 overexpression on progeny virus production in Sf9 cells, monolayer cultures of 3 × 106 Sf9 cells in 6-well microtiter plates were infected with AcMNPV, AcMNPV-iap1-egfp and AcMNPV-iap2-egfp for 1–4 d p.i. at a multiplicity of infection (MOI) of 0.001 (3 × 103 pfu ml−1), respectively. Following incubation for 2 h, the virus-containing culture medium was removed and fresh medium was added. Culture medium and cells was harvested at specific time point. Cells were resuspended in PBS buffer and released the viruses by repeated freeze–thaw cycles. BV production was determined by the plaque assay.2.10 Function analysis of BIR2 domain in Ac-IAP2
The construction process of bacmid-iap2-BIR2(iap1)-egfp was identical to the construction of bacmid-iap2-egfp, except that the Ac-iap2 gene was replaced with its mutant iap2-BIR2(iap1). iap2-BIR2(iap1) gene was obtained by over lap extension PCR, and its BIR2 domain in iap2 was replaced by the BIR2 domain of iap1. We first amplified the iap1 and iap2 gene fragments from the AcMNPV genome by using their respective primers, and amplified the BIR2 domain of iap1 (named B fragment) with the primers B–F (5′-AATGAATCGATGAGACAGTCGCGTATGGAT-3′) and B–R (5′-GTCTTCT ATTGCGTTCGACAATACAAAATA-3′), and amplified the upstream fragment of iap2 gene BIR2 domain (named A fragment) with the primers iap2-Sam I–F and A–R (5′-ATCCATACGCGACTGTCTCATCGATTCATT-3′), and amplified the downstream fragment of iap2 gene BIR2 domain (named C fragment) with the primers C–F (5′-TATTTTGTATTGTCGAACGCAATAGAAGAC-3′) and iap2-Xho I–R. Then we amplified A–B gene fragment with primers iap2-Sam I–F and B–R by using A and B fragment as templates. Then we amplified iap2-BIR2(iap1) gene fragment with the primers iap2-Sam I–F and iap2-Xho I–R by using A–B and C fragment as templates. Finally, the bacmid-iap2-BIR2(iap1)-egfp and AcMNPV-iap2-BIR2(iap1)-egfp viruses were constructed according to the previous process.The effects of AcMNPV-iap1-egfp, AcMNPV-iap2-egfp and AcMNPV-iap2-BIR2(iap1)-egfp on Sf9 cells proliferation were examined by MTT assay. Sf9 cells were infected with AcMNPV-iap1-egfp, AcMNPV-iap2-egfp and AcMNPV-iap2-BIR2(iap1)-egfp for 48 h at the concentration of 1.5, 3 and 6 × 1010 vp ml−1 (3, 6, 12 MOI), respectively.
2.11 Prediction of three dimensional structure of Ac-IAP1/2 and analysis of the distribution of hydrogen bonds and electrostatic surfaces of their BIR2 domain
We obtained the amino acid sequence of Ac-IAP1/2 and their BIR2 domain in the GenBank database and SMART (Simple Modular Architecture Research Tool) database. Then, these sequences were submitted to the Swiss-Model server,18–21 which was a fully automated protein structure homology-modelling server, to predict the three dimensional structure of Ac-IAP1/2, Ac-IAP2-BIR2(iap1) and their BIR2 domain and we got the corresponding .pdb files. Finally, Swiss-PdbViewer 3.7 software was used to compute hydrogen bond (H-bond) and electrostatic surfaces of the BIR2 domain in Ac-IAPs.2.12 Anti-insect activity analysis
The larvae of Helicoverpa armigera, as the permissive host for AcMNPV, were used to compare the virulence of AcMNPV, AcMNPV-iap1-egfp and AcMNPV-iap2-egfp. The larvae were reared in 24-well microtiter plates with artificial diet, under the condition of 28 ± 2 °C, 70–80% humidity, and natural photoperiod. The biological activity against third instar larvae of the recombinant virus AcMNPV-iap1/2-egfp was evaluated. The mortality and pupation rate for AcMNPV-iap1/2-egfp and AcMNPV-egfp treatment groups were determined by feeding of 3rd instar larvae of Helicoverpa armigera on a piece of artificial diet (27 mm3) with 20 μl, 1 × 107 pfu ml−1 of each viruses daily for five days, respectively. There were 36 larvae in each treatment group. The average length, mortality and pupation of larvae were recorded daily.2.13 Statistical analysis
For each experiment, six independent experiments were performed. Statistical significance was assessed using t-test by using excel. Results were considered significant for P < 0.05 and very significant for P < 0.01.3. Results and discussion
3.1 Ac-IAP1/2 and Ac-IAP2-BIR2(iap1) were overexpressed during the infection process of AcMNPV-iap1/2-egfp and AcMNPV-iap2-BIR2(iap1)-egfp, respectively
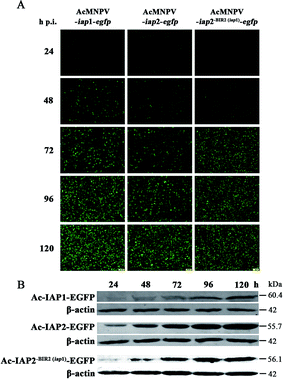
Since IAP1, IAP2 and Ac-IAP2-BIR2(iap1) were fused respectively to EGFP in its N-terminal end and were transcribed driven by the late strong P10 promoter, we could monitor the expression level of Ac-IAP1, Ac-IAP2 and its mutant Ac-IAP2-BIR2(iap1) by detecting the expression of EGFP. Green fluorescence detection and western blot were used to analyze the expression level of Ac-IAP1, Ac-IAP2 or Ac-IAP2-BIR2(iap1) in Sf9 cells infected with AcMNPV-iap1/2-egfp and AcMNPV-iap2-BIR2(iap1)-egfp, respectively, during 24–120 h p.i. As shown in Fig. 2, green fluorescence intensity in infected cells and the expression of Ac-IAP1/2-EGFP and Ac-IAP2-BIR2(iap1)-EGFP were significantly enhanced with the infection time, and both of Ac-IAP1/2-EGFP and Ac-IAP2-BIR2(iap1)-EGFP reached the top expression level at 120 h p.i.3.2 Ac-IAP1 induced apoptosis of Sf9 cells, while Ac-IAP2 promoted cell proliferation
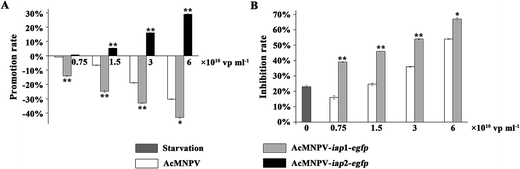
Monolayer cultures of ∼5 × 104 Sf9 cells in 96-well microtiter plates were infected with AcMNPV, AcMNPV-iap1-egfp and AcMNPV-iap2-egfp for 48 h, respectively. As shown in Fig. 3A, in the AcMNPV and AcMNPV-iap1-egfp treatment groups, with the increase of concentration of viruses, the inhibition rates were increased markedly. Moreover, compared with AcMNPV treatment group, the inhibition rate was dramatically improved in a dose dependent manner in the AcMNPV-iap1-egfp treatment group, suggesting that Ac-IAP1 could induce apoptosis of host cells. In addition, to make the nutrition starvation model, Sf9 cells were treated with half quantity of medium for 24 h, and then infected with the AcMNPV and AcMNPV-iap1-egfp at the same concentrations. We found that AcMNPV-iap1-egfp could also inhibit Sf9 cells proliferation (Fig. 3B). So, we confirmed that whether under normal growth or nutrition stress, Ac-IAP1 could induce cell apoptosis, which was consistent with the previous findings. Ikeda et al. reported that the transient expression of Ac-IAP1 stimulated caspase-3-like protease activity and induced apoptosis in Sf9 cells.15 However, Zeng et al. reported that the transient expression of Ac-IAP1 suppressed apoptosis of Tn-Hi5 cells during HearNPV infection.5 It is very likely that different iaps may be functional in different specific cells and hosts. And Huang et al. reported that Ac-IAP1 was unable to inhibit apoptosis induced by Bax and Fas in mammalian 293 cells.22 It was speculated that Ac-IAP1 was likely to be a key factor limiting the host domain of AcMNPV.Conversely, AcMNPV-iap2-egfp could promote the cell proliferation under the test concentrations compared with the AcMNPV treatment, which demonstrated that Ac-IAP2 could stimulate Sf9 cell proliferation (Fig. 3A).
3.3 Ac-IAP2 promoted progeny virus production in Sf9 cells
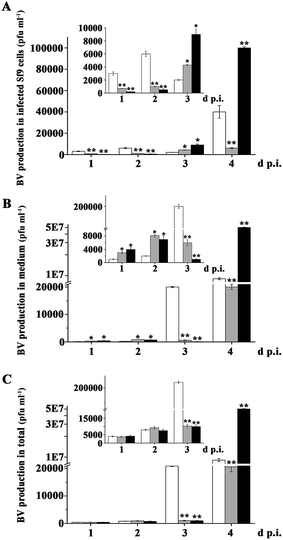
In order to expedite multiplication, baculoviruses inhibited host cells apoptosis, which was induced by viruses DNA replication. So, we examined BV production in virus-infected cells. As shown in Fig. 4A and Table S1,† there had more BV production in the AcMNPV-infected cells compared with that in AcMNPV-iap1-egfp and AcMNPV-iap2-egfp treatment groups at 1–2 d p.i. During 3–4 d p.i., BV production in AcMNPV-iap2-egfp-infected cells was strikingly high compared with that in AcMNPV and in AcMNPV-iap1-egfp-infected cells. However, BV production in AcMNPV-iap1-egfp-infected cells was lower than that in AcMNPV-infected cells at 4 d p.i.On the other hand, there was a relative low BV production in the medium of the AcMNPV treated group at 1–2 d p.i. In total, at 1–2 d p.i., BV production in three treatment groups was comparable. Compared to that in AcMNPV group, there was an extremely high BV production in AcMNPV-iap2-egfp overall at 4 d p.i. and a lower BV production in AcMNPV-iap1-egfp overall at 3–4 d p.i. (Fig. 4B and C). That is, Ac-IAP2 could accelerate multiplication of progeny virus via inhibiting cell apoptosis.
AcMNPV is similar to BmNPV which possesses two inhibitors of apoptosis (IAP) proteins, Bm-IAP1 and Bm-IAP2.23 The amino acid sequence analysis showed that Ac-IAP1 and Ac-IAP2 were extremely similar to Bm-IAP1 and Bm-IAP2, and the identity rates were up to 94.1% and 96%, respectively. Previous studies indicated that the Bm-IAP2 possessed three putative functional domains: BIR domain, BIR-like (BIRL) domain, and RING finger domain. Bm-iap2 is required for efficient viral propagation, and BIR or BIRL domain plays an important role in BmNPV infection, while the RING domain does not contribute to BmNPV propagation.24,25 So, we speculated there was a key domain in Ac-IAP2 that determined the function of the anti-apoptotic and pro-multiplication (of progeny virus) of Ac-IAP2.
3.4 BIR2 domain in Ac-IAP2 was essential for its anti-apoptotic function
We have previously defined the Ac-IAP1 and Ac-IAP2 had similar domains (Fig. 1A), but had the opposite function (Fig. 3). Thus, it is imperative to clarify the function of main domain in Ac-IAP1 and Ac-IAP2. Homologous sequence alignment showed that although the BIR domains of Ac-IAP1 and Ac-IAP2 had typical C2HC motifs, we found more lysine residues in the BIR2 domain of Ac-IAP2, while the BIR2 domain of Ac-IAP1 don't (Fig. 5A). Notably, the lysine residues were the potential ubiquitination sites, which might related to the stability and function of Ac-IAPs. Therefore, we hypothesized that BIR2 domain was essential for the anti-apoptotic ability of Ac-IAP2. To test this hypothesis, the baculovirus AcMNPV-iap2-BIR2(iap1)-egfp was generated (Fig. 5B and C). Green fluorescence detection and western blot analysis showed that Ac-IAP2-BIR2(iap1) was expressed stably and increased during 24–120 h p.i. in Sf9 cells infected with AcMNPV-iap2-BIR2(iap1)-egfp (Fig. 2B). MTT assay showed that the inhibition rates were 25.99%, 30.53%, 39.23% and 34.23%, 37.81%, 43.2% respectively when Sf9 cells were infected with AcMNPV-iap1-egfp and AcMNPV-iap2-BIR2(iap1)-egfp at 3, 6 and 12 MOI for 48 h (Fig. 5D). Contrary to AcMNPV-iap2-egfp, AcMNPV-iap2-BIR2(iap1)-egfp could inhibit Sf9 cells proliferation in a dose-dependent manner. And compared with AcMNPV-iap1-egfp treatment group, the inhibition rate was dramatically improved in the AcMNPV-iap2-BIR2(iap1)-egfp treatment group. Only the BIR2 domain was different between AcMNPV-iap2-egfp and AcMNPV-iap2-BIR2(iap1)-egfp, so we concluded that BIR2 domain was essential for the anti-apoptotic ability of Ac-IAP2.Recently reports suggested that insect IAPs had the N-terminal leader, which carried the unique signal-responsive degrons initating apoptosis. The baculovirus IAPs (especially Op-IAP3) stability and potent antiapoptotic activity was because of the elimination of the N-terminal leader. Op-IAP3 preserved its anti-apoptosis by binding to its targeted cellular IAP.26,27 In this study, the amino acid sequence before the BIR1 domain in Ac-IAP2 was extremely short, that is, Ac-IAP2 didn't have the N-terminal “leader”, which suggested that Ac-IAP2 was likely to have the same anti-apoptotic mechanism as Op-IAP3. Of course, its RING domain also needed to have E3 ubiquitin ligase activity. Ac-IAP2 and Sf-IAP could dimerize.
Previous studies showed that RING domain of OP-IAP3 had E3 ubiquitin ligase activity, which was closely related to the proteasome degradation pathway and associated with the protein own half-life.1 Some reports speculated OP-IAP3's half-life was extended because of the absence of its own 17 lysine residues, which might be the ubiquitination sites, so RING domain could not perform itself degradation function, and therefore OP-IAP3 was stable and had the anti-apoptotic ability.27 According to this presumption, if the RING domain of Ac-IAP2 also had E3 ubiquitin ligase activity, the 8 lysine residues in its BIR2 domain (Fig. 5A), which were not existed in the BIR2 domain of Ac-IAP1, were completely unable to enhance its anti-apoptotic function. Therefore, it was possible that BIR2 domain of Ac-IAP2 performed its function in another pathway that didn't depend on E3 ubiquitin ligase activity. BIR2 domain might determine the function of Ac-IAPs by changing their advanced structure. Of course, the other domains might be important for Ac-IAP2's anti-apoptotic ability. The mechanism of Ac-IAPs function was very complex, the function of the other domains in Ac-IAPs should be analyzed in future work.
3.5 BIR2 domain affected the function of Ac-IAP1/2 by affecting their three-dimensional conformation
Structure determined function. To analyze the relationship between the BIR2 domains and function of Ac-IAP1/2 and Ac-IAP2-BIR2(iap1) protein, we used Swiss-Model server and Swiss-PdbViewer 3.7 software to predict the three dimensional structure of Ac-IAP1/2 and Ac-IAP2-BIR2(iap1) and indicate the position of their BIR2 domains (Fig. 6A). We found that the three-dimensional structure simulation of the mutant Ac-IAP2-BIR2(iap1) formed by the BIR2 functional domain substitution was more loose than Ac-IAP1/2 structure. So, we speculated that the BIR2 domains might affect IAPs function by influencing the formation of their advanced structure. We further analyzed the H-bond and electrostatic surfaces of their BIR2 domain. As shown in Fig. 6B and C, we couldn't find any roles of H-bond in the structure of BIR2 in Ac-IAP1/2. However, the BIR2 domain of Ac-IAP1 and Ac-IAP2 had exactly the opposite surface potential, which showed that BIR2 domain was most likely to affect their function by affecting the three-dimensional conformation of Ac-IAP1/2.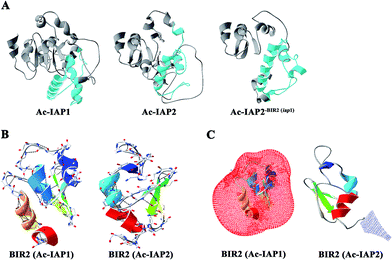 | ||
| Fig. 6 Three dimensional structure simulation of Ac-IAPs and their BIR2 domain. (A) Three dimensional structure of Ac-IAP1/2 and Ac-IAP2-BIR2(iap1). Swiss-Model server was used to predict the three dimensional structure of Ac-IAP1/2 and Ac-IAP2-BIR2(iap1). Ac-IAP1/2 and Ac-IAP2-BIR2(iap1) had the high homology with 3t6p.1.A (described as baculoviral IAP repeat-containing protein 2) template in PDB database,28 so their structures were constructed by using 3t6p.1.A model as the template, and the BIR2 domains were marked with blue. (B) Distribution analysis of H-bond in the BIR2 domain of Ac-IAP1/2. The BIR2 domain of Ac-IAP1/2 was constructed by using the 1q4q.4.A (described as apoptosis 1 inhibitor) and 4mtz.1.A (described as E3 ubiquitin-protein ligase XIAP) model as the template, respectively.29 The BIR2 domain was colored by secondary structure succession. And H-bonds were marked by the program Swiss-PdbViewer 3.7 and drawn by a black dotted line. (C) The electrostatic surface analysis of BIR2 domain in Ac-IAP1/2. Electrostatic surface is calculated by the program Swiss-PdbViewer 3.7. Different color codes are −1.8kT/e (red) and 1.8kT/e (blue) where k is the Boltzmann constant, T is the temperature in kelvin, and e is the charge of the electron. The BIR2 domain was colored by secondary structure succession. Positively charged residues were shown in blue and negatively charged in red. | ||
3.6 Recombinant AcMNPV-iap2-egfp had high anti-insect activity
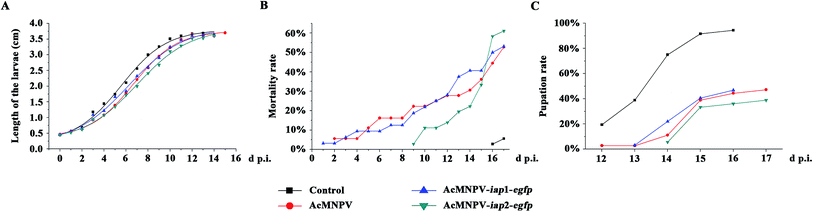
In theory, a higher cytotoxic virus would result in higher insecticidal rates, and AcMNPV-iap1-egfp might have a higher insecticidal efficiency. As shown in Fig. 7 and Tables 2–4, compared with control treatment, the larvae in the viruses' treatment groups were growing slowly, and had higher mortality rates and lower pupation rates. Three treatments (AcMNPV-egfp, AcMNPV-iap1-egfp and AcMNPV-iap2-egfp) were compared with each other: (1) the larva average growth rate in the AcMNPV-iap1-egfp treatment was faster than that in the other two groups at the early infection stage (1–6 d p.i.); (2) at the late infection stage (7–15 d p.i.), compared with AcMNPV-egfp and AcMNPV-iap1-egfp groups, the larva in AcMNPV-iap2-egfp group were slow growing, while the average growth rate in the other two groups was almost the same on the whole.| d.p.i. | Length (cm) | |||
|---|---|---|---|---|
| Mock | AcMNPV | AcMNPV-iap1-egfp | AcMNPV-iap2-egfp | |
| a Data shown are mean values of triplicate assays ± SD, ** means the difference is significant at the 0.01 level between the AcMNPV or AcMNPV-iap1/2-egfp and mock treatments in the t-test. * means the difference is significant at the 0.05 level between the AcMNPV or AcMNPV-iap1/2-egfp and mock treatments in the t-test. | ||||
| 0 | 0.45 ± 0.0654 | 0.45 ± 0.056 | 0.46 ± 0.07 | 0.45 ± 0.05 |
| 1 | 0.55 ± 0.062 | 0.56 ± 0.05 | 0.56 ± 0.082 | 0.52 ± 0.063 (*) |
| 2 | 0.69 ± 0.046 | 0.63 ± 0.07 (**) | 0.65 ± 0.082 (*) | 0.62 ± 0.082 (**) |
| 3 | 1.18 ± 0.142 | 0.92 ± 0.20 (**) | 1.09 ± 0.180 (*) | 0.94 ± 0.175 (**) |
| 4 | 1.44 ± 0.136 | 1.08 ± 0.23 (**) | 1.22 ± 0.187 (**) | 1.08 ± 0.181 (**) |
| 5 | 1.74 ± 0.254 | 1.39 ± 0.33 (**) | 1.66 ± 0.226 (*) | 1.35 ± 0.307 (**) |
| 6 | 2.11 ± 0.352 | 1.76 ± 0.33 (**) | 1.84 ± 0.314 (**) | 1.72 ± 0.246 (**) |
| 7 | 2.56 ± 0.4 | 2.21 ± 0.305 (**) | 2.32 ± 0.434 (**) | 2 ± 0.385 (**) |
| 8 | 3 ± 0.37 | 2.59 ± 0.402 (**) | 2.6 ± 0.435 (**) | 2.41 ± 0.319 (**) |
| 9 | 3.26 ± 0.297 | 2.95 ± 0.388 (**) | 2.9 ± 0.407 (**) | 2.68 ± 0.342 (**) |
| 10 | 3.51 ± 0.225 | 3.23 ± 0.303 (**) | 3.25 ± 0.288 (**) | 3.11 ± 0.407 (**) |
| 11 | 3.6 ± 0.105 | 3.49 ± 0.277 (**) | 3.47 ± 0.225 (*) | 3.29 ± 0.386 (**) |
| 12 | 3.67 ± 0.066 | 3.65 ± 0.1775 (*) | 3.61 ± 0.086 (*) | 3.52 ± 0.167 (**) |
| 13 | 3.69 ± 0.099 | 3.61 ± 0.103 (*) | 3.63 ± 0.048 | 3.53 ± 0.045 (**) |
| 14 | 3.65 ± 0.07 | 3.6 ± 0.033 | 3.63 ± 0.05 | 3.6 ± 0 |
| d p.i. | Mortality rate (%) | |||
|---|---|---|---|---|
| Mock | AcMNPV | AcMNPV-iap1-egfp | AcMNPV-iap2-egfp | |
| a The half-lethal time of AcMNPV-iap1-egfp was marked in bold. | ||||
| 1 | 3.13 | |||
| 2 | 5.56 | 3.13 | ||
| 3 | 5.56 | 6.25 | ||
| 4 | 5.56 | 9.38 | ||
| 5 | 11.11 | 9.38 | ||
| 6 | 16.17 | 9.38 | ||
| 7 | 16.17 | 12.5 | ||
| 8 | 16.17 | 12.5 | ||
| 9 | 22.22 | 18.75 | 2.78 | |
| 10 | 22.22 | 21.88 | 11.11 | |
| 11 | 25 | 25 | 11.11 | |
| 12 | 27.78 | 28.13 | 13.89 | |
| 13 | 27.78 | 37.5 | 19.44 | |
| 14 | 30.56 | 40.63 | 22.22 | |
| 15 | 36.11 | 40.63 | 33.33 | |
| 16 | 2.78 | 44.44 | 50 | 58.33 |
| 17 | 5.56 | 52.78 | 53.13 | 61.11 |
| d p.i. | Pupation rate (%) | |||
|---|---|---|---|---|
| Mock | AcMNPV | AcMNPV-iap1-egfp | AcMNPV-iap2-egfp | |
| 1–11 | — | — | — | — |
| 12 | 19.44 | 2.78 | — | — |
| 13 | 38.89 | 2.78 | 3.13 | — |
| 14 | 75 | 11.11 | 22 | 5.56 |
| 15 | 91.67 | 38.89 | 40.63 | 33.33 |
| 16 | 94.44 | 44.44 | 46.88 | 36.11 |
| 17 | — | 47.22 | — | 38.89 |
As shown in Fig. 7B, the average mortality rate of the larvae in the three treatment groups (AcMNPV-egfp, AcMNPV-iap1-egfp and AcMNPV-iap2-egfp) were 52.78%, 53.13% and 61.11% respectively. Pupation rate is also one of the important indices for detecting pesticide effect, it was 47.22%, 46.88% and 38.89% in AcMNPV-egfp, AcMNPV-iap1-egfp and AcMNPV-iap2-egfp group, respectively (Fig. 7C). Although there was a delay in the pupation and the death, the average mortality rate of AcMNPV-iap2-egfp group was higher than the others; the average pupation rate was also lower than the others; and the half-lethal time of the three virus-treated groups was almost the same (Table 3), which suggested that AcMNPV-iap2-egfp had higher anti-insect activity. We hypothesized that this was due to the large proliferation of progeny virus caused by the inhibition of apoptosis by Ac-IAP2.
From the above results, we concluded that although AcMNPV-iap1-egfp had a stronger cytotoxicity, AcMNPV-iap2-egfp could be used as an effective insecticide.
4. Conclusions
The function of Ac-IAP1/2 was confirmed in vitro and in vivo. Ac-IAP1 induced Sf9 cells apoptosis in a dose-depend manner, however, Ac-IAP2 promoted Sf9 cell proliferation and recombinant virus AcMNPV-iap2-egfp had a better insecticidal efficiency. In theory, AcMNPV-iap2-egfp has multiple functions: (1) in the field of agricultural plant protection, as the microbial pesticides, it is advantageous to the virus proliferation and the insect-resistant activity; (2) in terms of insect cell expression system, it can prolong the cell survival time and improve the expression level of foreign protein; (3) the EGFP harbored in this virus could provide a detection tag to analyze the distribution state of recombinant virus during the field application or the infection process in the host cells; (4) more importantly, AcMNPV is widely used as a viral pesticide and insect expression system, we used its own iap2 gene to improve its resistance and expression efficiency and the biological safety is reliable.Conflict of interest
The authors declare no competing financial interest.Acknowledgements
This project was supported by grants from ‘National Natural Science Foundation of China (No. 31272100 and 31372199)’, ‘Natural Science Foundation of Shanxi Province (No. 2014011038-1)’.Notes and references
- R. J. Clem, Semin. Cell Dev. Biol., 2015, 39, 72–79 CrossRef CAS PubMed.
- M. B. Sonja, Annu. Rev. Microbiol., 2008, 62, 171–192 CrossRef PubMed.
- S. Katsuma, S. Kawaoka, K. Mita and T. Shimada, Insect Biochem. Mol. Biol., 2008, 38, 1080–1086 CrossRef CAS PubMed.
- M. J. Birnbaum, R. J. Clem and L. K. Miller, J. Virol., 1994, 68, 2521–2528 CAS.
- X. Zeng, F. Nan, C. Liang, J. Song, Q. Wang, J. M. Vlak and X. Chen, Sci. China, Ser. C: Life Sci., 2009, 52, 761–770 CrossRef PubMed.
- J. Silke and D. L. Vaux, J. Cell Sci., 2001, 114, 1821–1827 CAS.
- R. Budhidarmo and C. L. Day, Semin. Cell Dev. Biol., 2015, 39, 80–90 CrossRef CAS PubMed.
- J. Silke and D. Vucic, Methods Enzymol., 2014, 545, 35–65 CAS.
- M. Ikeda, K. Yanagimoto and M. Kobayashi, Virology, 2004, 321, 359–371 CrossRef CAS PubMed.
- M. P. Carpes, M. E. de Castro, E. F. Soares, A. G. Villela, F. J. Pinedo and B. M. Ribeiro, Arch. Virol., 2005, 150, 1549–1562 CrossRef PubMed.
- C. Liang, J. de Lange, X. Chen, M. M. van Oers, J. M. Vlak and M. Westenberg, Virus Res., 2012, 165, 107–111 CrossRef CAS PubMed.
- M. C. Green, K. P. Monser and R. J. Clem, Virus Res., 2004, 105, 89–96 CrossRef CAS PubMed.
- D. P. Miller, T. Luque, N. E. Crook, D. Winstanley and D. R. O'Reilly, Arch. Virol., 2002, 147, 1221–1236 CrossRef CAS PubMed.
- C. W. Wright and R. J. Clem, J. Biol. Chem., 2002, 227, 2454–2462 CrossRef PubMed.
- M. Ikeda, H. Yamada, H. Ito and M. Kobayashi, J. Gen. Virol., 2011, 92, 2654–2663 CrossRef CAS PubMed.
- M. Mohammad, H. Mazhar, M. Leila and A. Sassan, J. Virol., 2015, 89, 8182–8192 CrossRef PubMed.
- Q. Yu, Virus Genes, 2010, 41, 298–304 CrossRef CAS PubMed.
- M. C. Peitsch, Bio/Technology, 1995, 13, 658–660 CrossRef CAS.
- N. Guex and M. C. Peitsch, Electrophoresis, 1997, 18, 2714–2723 CrossRef CAS PubMed.
- T. Schwede, J. Kopp, N. Guex and M. C. Peitsch, Nucleic Acids Res., 2003, 31, 3381–3385 CrossRef CAS PubMed.
- J. Kopp and T. Schwede, Nucleic Acids Res., 2004, 32, 230–234 CrossRef PubMed.
- Q. Huang, Q. L. Deveraux, S. Maeda, G. S. Salvesen, H. R. Stennicke, B. D. Hammock and J. C. Reed, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 1427–1432 CrossRef CAS.
- R. J. Clem, Curr. Top. Microbiol. Immunol., 2005, 289, 113–129 CAS.
- H. Ito, H. Bando, T. Shimada and S. Katsuma, Biochem. Biophys. Res. Commun., 2014, 454, 581–587 CrossRef CAS PubMed.
- C. Ono, T. Kamagata, H. Taka, K. Sahara, S. Asano and H. Bando, Virus Res., 2012, 165, 197–206 CrossRef CAS PubMed.
- R. Vandergaast, J. K. Mitchell, N. M. Byers and P. D. Friesen, J. Virol., 2015, 89, 4481–4493 CrossRef CAS PubMed.
- N. M. Byers, R. L. Vandergaast and P. D. Friesen, J. Virol., 2016, 90, 533–544 CrossRef CAS PubMed.
- E. C. Dueber, A. J. Schoeffler, A. Lingel, J. M. Elliott, A. V. Fedorova, A. M. Giannetti, K. Zobel, B. Maurer, E. Varfolomeev, P. Wu, H. J. Wallweber, S. G. Hymowitz, K. Deshayes, D. Vucic and W. J. Fairbrother, Science, 2011, 334, 376–380 CrossRef CAS PubMed.
- J. Chai, N. Yan, J. R. Huh, J. W. Wu, W. Li, B. A. Hay and Y. Shi, Nat. Struct. Biol., 2003, 10, 892–898 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra03711b |
| This journal is © The Royal Society of Chemistry 2017 |