Open Access Article
Open Access ArticleProduction of exopolysaccharide by Bifidobacterium longum isolated from elderly and infant feces and analysis of priming glycosyltransferase genes
Shuang Yana,
Guozhong Zhao *ac,
Xiaoming Liuac,
Jianxin Zhaoac,
Hao Zhangac and
Wei Chen
*ac,
Xiaoming Liuac,
Jianxin Zhaoac,
Hao Zhangac and
Wei Chen *abc
*abc
aState Key Laboratory of Food Science and Technology, School of Food Science and Technology, Jiangnan University, Wuxi 214122, P. R. China. E-mail: zhaoguozhongsun@gmail.com; chenwei66@jiangnan.edu.cn; Fax: +86 0510 85329081; Tel: +86 0510 85912087
bBeijing Innovation Centre of Food Nutrition and Human Health, Beijing Technology & Business University, Beijing 100048, P. R. China
cInternational Joint Research Center for Probiotics & Gut Health, Jiangnan University, Wuxi 214122, P. R. China
First published on 20th June 2017
Abstract
The exopolysaccharide (EPS) production of several Bifidobacterium longum strains isolated from infant and elder feces was determined. The relationship between EPS production and tolerance to artificial gastric and intestinal juices was analyzed. Moreover, priming glycosyltransferase (pGT) gene fragments of these strains were amplified and sequenced. The results indicate that their tolerance correlated well with EPS production, especially the production of cell-surface-bound exopolysaccharide (EPS-b). The EPS-b production by strains isolated from elderly volunteers was found to be significantly higher than that produced by strains isolated from infants. Lastly, the phylogenetic tree of the pGT gene sequence fragments showed that the pGT genes of infant-originated B. longum strains had greater homology than those of elder-originated strains.
1 Introduction
Human gut microbiota is an extremely complex ecosystem that consists of 10 times more cells than the entire human body. Research data have indicated that gut microbes affect various human functions, such as the development of the immune system and intestinal cell proliferation and differentiation, and they are related to many human diseases.1–5 Numerous commensal bacteria appear to have co-evolved and interact with their human hosts.6 Studies have been conducted to map the bacterial members of the human gut microbiota. However, the accurate molecular mechanisms involved in the interactions between commensal bacteria and their hosts remain largely unknown. The cell envelopes of gut commensal bacteria, which contain proteins and carbohydrates, are important interfaces between the organisms and their hosts.6Bacteria of the genus Bifidobacterium were first discovered in 1899 by Tissier, which are one of the bacterial groups that dominate the human gut microbiota, especially in breast fed infants.7 Certain Bifidobacterium strains interact with their host and are considered to be potential probiotics. For example, Bifidobacteria have been reported to be associated with antimicrobial activity against enteropathogens, to inhibit of colon cancer and ulcerative colitis, and to improve the symptoms of constipation.8–11
The production of exopolysaccharides (EPSs) by Bifidobacteria is one of the proposed mechanisms for their probiotic activities. EPSs are carbohydrate polymers synthesized by certain bacteria and either form an extracellular layer closely attached to the cell surface or are free in the surrounding.12 Because the composition of monosaccharides and the chemical bonds to connect these monosaccharide are diverse, EPSs synthesized by different bacteria have variable structures. Bacterial EPSs consist of repeating subunits of either mono- or oligosaccharides, generating homo- or heteropolysaccharides, respectively. Experimental data have demonstrated the importance of capsular polysaccharides produced by pathogen in pathogenesis.13 In contrast, EPSs produced by probiotics, such as Bifidobacteria, are important in maintaining commensalism between host and bacteria by modifying the physical properties of cell surfaces. For example, EPSs may increase the tolerance of the bacteria to the gastrointestinal environment and protect the bacteria against the host immune system.14,15 Surface EPSs produced by Bifidobacterium breve UCC2003 were reported to increase its stress tolerance against low pH and bile, and affected the gut persistence of the strain; however, they did not affect initial colonization. Furthermore, compared with the isogenic mutants that did not produce surface EPS (EPS− strain), the surface EPS-producing strain UCC2003 (EPS+ strain) induced a weak adaptive immune response after oral gavage of these strains.15 EPSs produced by B. longum 35624 were demonstrated to reduce the proinflammatory host responses to the strain.16 Therefore, the production of EPSs could be regarded as a beneficial property for the screening of probiotic strains. In addition, EPS-producing lactic acid bacteria (LAB) are used in the food industry; for example, during the fermentation of yogurts, the texture and viscosity of the products can be improved by the EPSs produced by LAB.17
In this study, we isolated several B. longum strains from human feces, and determined the production of EPSs (either bound to cell surfaces or released into their surroundings, i.e. EPS-b or EPS-r) and their relationship with the tolerance of these strains to simulated gastrointestinal juices. Furthermore, we used PCR techniques to determine the gene fragments that code for priming glycosyltransferase (pGT), which catalyzes the first step in the synthesis of EPS.18 Unlike other glycosyltransferase (GT) genes that are often either unique or dissimilar to each other, pGT genes found in different bacterial strains are fairly similar to each other, especially at the carboxy terminus.19 Therefore, we designed primers based on the conserved areas of pGT genes and amplified the pGT genes of our strains to analyze the homology of these genes and their relationship to EPS production.
2 Materials and methods
2.1 Sample collection and isolation of Bifidobacteria
We recruited 30 volunteers for collection of fecal samples: 15 elderly people (older than 90 years of age) and 15 infants (less than 1 year of age). Fecal samples were collected in sterile tubes (50 mL) with 25 mL of sterilized 30% (v/v) glycerol containing 0.05% (w/v) L-cysteine hydrochloride and kept at 4 °C. All of the samples were transported to the laboratory within 48 h and stored at −80 °C.The samples were homogenized and the fecal suspensions were then serially diluted with sterilized normal saline solution containing 0.05% (w/v) L-cysteine hydrochloride (pH 7.0). Appropriate dilutions of fecal samples were spread on MRS plates containing 0.05% (w/v) L-cysteine hydrochloride and 50 μg mL−1 mupirocin (MRS-M). The plates were incubated in a Whitley DG250 anaerobic workstation (Don Whitley Scientific Limited, Shipley, UK) at 37 °C for 48 h. Colonies that exhibited different morphologic features were isolated and identified. Two Bifidobacterium longum strains, JCM 7052 and JCM 7053 (here renumbered as 642 and 643, respectively), obtained from the Japan Collection of Microorganisms were used as reference strains.
2.2 Bacterial genomic DNA purification
All of the isolated strains were inoculated into MRS broth and cultured anaerobically at 37 °C for 48 h. 1 mL of the culture were centrifuged (6000g for 5 min, 4 °C) and bacterial cell pellets were collected, DNA was then extracted from the pellets using a Rapid Bacterial Genomic DNA Isolation Kit (Sangon Biotech, Shanghai). Finally, the purified DNA was dissolved in 100 μL of TE buffer (pH 8.0).2.3 Identification of isolated strains by 16S rRNA gene sequencing and carbohydrate fermentation tests
16S rRNA gene from all of the isolated strains was amplified by PCR with the universal primers 27F (5′-AGA GTT TGA TCC TGG CTC AG-3′) and 1492R (5′-ACG GCT ACC TTG TTA CGA CTT-3′). PCR was carried out in 30 μL final volume samples containing 15 μL 2× Taq MasterMix (Cwbiotech, Changping, Beijing), 0.4 μM of each primer, and 50 ng of DNA templates. The PCR program consisted of 30 cycles after initial incubation at 95 °C for 5 min. Each cycle consisted of a denaturation step at 95 °C for 30 s, an annealing step at 55 °C for 30 s, an extension step at 72 °C for 60 s, and a final extension step at 72 °C for 5 min. The PCR products were detected using agarose gel electrophoresis and stored at 4 °C until sequencing.Carbohydrate fermentation tests were carried out in MRS broth supplemented with different carbohydrates (Table 3) and 0.04 g L−1 bromocresol purple as a pH indicator. Strains that caused a yellow-to-purple color change were considered to be able to ferment the carbohydrate present in the medium.
2.4 Determination of growth curve
Strains were cultured for three generations, inoculated into 100 mL of MRS broth, and cultured anaerobically at 37 °C for 40 h. During the culture procedure, the optical density of the culture broth at 600 nm was determined with a spectrophotometer.2.5 Isolation and determination of exopolysaccharides
After anaerobic culturing at 37 °C for 10 to 25 h (at the end of the exponential phase of each strain), 10 mL of the culture was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 min at 4 °C. The suspension and cell pellets were used to extract cell-surface-bound exopolysaccharide (EPS-b) and released exopolysaccharide (EPS-r), respectively. The extraction of EPS-b and EPS-r was conducted using a method described previously.12 The crude EPS-b and EPS-r were dissolved into a final volume of 2.5 mL, respectively. The amounts of EPS-b and EPS-r were determined using the phenol/sulfuric acid method.20 The results were presented as mg per g biomass (dry weight).
000g for 10 min at 4 °C. The suspension and cell pellets were used to extract cell-surface-bound exopolysaccharide (EPS-b) and released exopolysaccharide (EPS-r), respectively. The extraction of EPS-b and EPS-r was conducted using a method described previously.12 The crude EPS-b and EPS-r were dissolved into a final volume of 2.5 mL, respectively. The amounts of EPS-b and EPS-r were determined using the phenol/sulfuric acid method.20 The results were presented as mg per g biomass (dry weight).
2.6 Tolerance to simulated gastrointestinal environment
Simulated gastric and intestinal juices were prepared using the previous method.21 Simulated gastric juice (pH 3.0) contained 3 g L−1 pepsin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15
15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000; BBI Life Sciences, CA); simulated intestinal juice (pH 8.0) contains 1 g L−1 trypsin (1
000; BBI Life Sciences, CA); simulated intestinal juice (pH 8.0) contains 1 g L−1 trypsin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 250; Sinopharm Chemical Reagent Co., Ltd, China). Both simulated gastric and intestinal juices were sterilized using 0.22 μm membrane.
250; Sinopharm Chemical Reagent Co., Ltd, China). Both simulated gastric and intestinal juices were sterilized using 0.22 μm membrane.
Strains were cultured to the end of the exponential phase and then centrifuged at 6000g for 5 min at 4 °C, and the cell pellets were washed with sterile normal saline solution and resuspended in simulated gastric juice. After the suspensions were incubated anaerobically at 37 °C for 3 h, the viable counts of the tested strains were determined by the pour plate method. The suspensions were centrifugated, and each cell pellet was resuspended in the same volume of simulated intestinal juice. After anaerobic incubation at 37 °C for 2 h, the viable counts of the tested strains were determined. The tolerances to simulated gastric and intestinal juices were represented by the survival rates after treatment with the above juices.
2.7 Amplification and sequence analysis of pGT genes
Gene sequences coding for pGT were found and downloaded from the GenBank database (Table 1). Sequence alignment was conducted using DNAMAN software (vision 6.0, Lynnon Biosoft, San Ramon, CA), and specific primers for pGT genes were designed based on the conserved regions of pGT genes. The PCR conditions for pGT genes with specific primers were the same as those for 16S rRNA gene, except for the annealing temperature (Table 2). The PCR products were detected by agarose gel electrophoresis and stored at 4 °C until sequencing. Multiple sequences from the 3′-terminal region of the pGT genes of each of the B. longum strain were aligned using Clustal X2 software (http://www.clustal.org/). The phylogenetic tree was created with MEGA5.1 software (http://www.megasoftware.net/) with the neighbor-joining method.| Strains | Genome accession no. | pGT gene | Location in the genome |
|---|---|---|---|
| B. longum NCC2705 | AE014295.3 | cpsD/rfbP | BL0237/BL0249 |
| B. longum 105-A | AP014658.1 | cpsD | BL105A_0405 |
| B. longum 35624 | CP013673.1 | cpsD | B624_0342 |
| B. longum AH1206 | CP016019.1 | cpsD | BL1206_0418 |
| B. longum BBMN68 | CP002286.1 | cpsD | BBMN68_1012 |
| B. longum JCM 1217 | AP010888.1 | cpsD | BLLJ_0364 |
| B. longum CECT 7347 | CALH01000027.1 | cpsD | BN57_533 |
| B. longum KACC 91563 | CP002794.1 | rfbP | BLNIAS_02272 |
| B. longum NCIMB8809 | CP011964.1 | rfbP | B8809_0330 |
| B. longum BG7 | CP010453.1 | rfbP | BBG7_0429 |
| B. longum CCUG30698 | CP011965.1 | rfbP | BBL306_0360 |
| B. longum F8 | FP929034.1 | rfbP | BIL_15040 |
| Primer | Sequence (5′–3′) | Annealing temperature (°C) |
|---|---|---|
| cpsD-F | TTCTCYGTGCGCATGGAATC | 55 |
| cpsD-R | CCCATAATSGACCAGTTCTGCAC | |
| rfbP-F | GATTCYGAGACCATGCGTAC | 55 |
| rfbP-R | GCATARTCCGACTGTTCCTGAG |
2.8 Statistical analysis
Results were expressed as the average of three independent trials. Statistical analysis was conducted using SPSS 20.0 software (SPSS Inc., Chicago, IL). Pearson's correlation was used to determine the relationship between the tolerance to the gastrointestinal environment and EPS production. Independent-samples t-tests were used to determine the significant differences between two groups.3 Results
3.1 Isolation and identification of Bifidobacteria
Three hundred and twenty bacteria strains were isolated from the 30 fecal samples. Ninety-three of these strains were identified as B. longum by 16S rRNA gene sequencing. We chose 21 strains from different fecal samples and two reference strains for subsequent analyses and carbohydrate fermentation tests (Table 3). Different B. longum strains exhibited different fermentation properties. This may indicate that they possessed different genotypes or phenotypes. It was reported previously that two biovars can be distinguished within species of B. longum. Biovar a ferments mannose and is more frequently found in human adults; biovar b is unable to ferment mannose and is more frequently found in neonates.22 Thus, the tested strains were divided into two groups; 11 strains were mannose-positive, and the remaining strains were mannose-negative.| Strain | Glucose | D-Fructose | Melezitose | D-Mannose | D-Glucosamine | D-Xylose | Arabic gum | Cellobiose | L-Arabinose | Salicin | Gluconate | Rafinose |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a +/−, the ability to metabolize this carbohydrate is very weak. | ||||||||||||
| B. longum 642 | + | + | + | − | − | + | − | − | + | − | − | + |
| B. longum 643 | + | + | − | − | − | − | − | − | + | − | − | + |
| B. longum 685 | + | + | − | − | − | + | − | − | + | − | − | + |
| B. longum 686 | + | + | + | + | − | + | − | − | + | − | − | + |
| B. longum 687 | + | + | + | + | − | + | − | − | + | − | − | + |
| B. longum 688 | + | + | + | + | − | + | − | − | + | − | − | + |
| B. longum 689 | + | + | − | + | − | + | − | − | + | − | − | + |
| B. longum 752 | + | + | + | + | − | + | − | − | + | − | − | + |
| B. longum 756 | + | + | + | + | − | + | − | − | + | − | − | + |
| B. longum 760 | + | + | − | + | − | + | − | − | + | − | − | + |
| B. longum 762 | + | + | + | + | − | + | − | − | + | − | − | + |
| B. longum GX-17-A9 | + | + | + | − | − | + | + | − | + | − | − | + |
| B. longum HUB-37-A12 | + | + | + | + | − | + | − | − | + | − | − | + |
| B. longum M2-3-F02 27 | + | + | − | − | +/− | − | − | − | − | − | − | + |
| B. longum M1-20-R01-3 | + | + | + | − | − | + | − | − | + | − | − | + |
| B. longum M1-20-R01-B15 | + | + | + | − | − | + | − | − | + | − | − | + |
| B. longum M2-C-F01-14 | + | + | − | − | − | + | − | + | − | − | − | + |
| B. longum C-1-A13 | + | + | + | + | − | + | − | − | + | − | − | + |
| B. longum C-6-A3 | + | + | + | − | − | + | − | − | + | − | − | + |
| B. longum C-11-A10 | + | + | − | − | − | + | − | − | + | − | − | + |
| B. longum HUB-6-1 | + | + | − | − | − | + | − | − | + | − | − | + |
| B. longum HUB-2-25 | + | + | + | − | − | + | − | − | + | − | − | + |
| B. longum HAN-4-25 | + | + | + | + | − | + | − | − | + | − | − | + |
3.2 Growth characteristics of Bifidobacterial strains
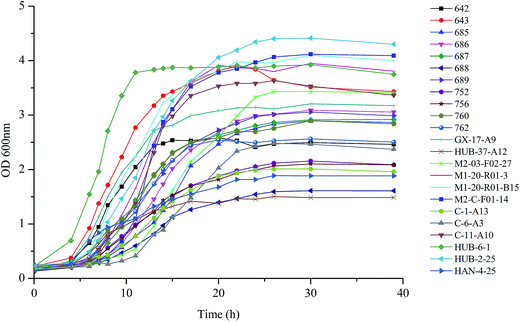
As shown in Fig. 1, each of the B. longum strains exhibited different growth rates in MRS broth. B. longum HUB-6-1 proliferated rapidly after inoculation and reached the stationary phase after 10 h. B. longum HUB-37-A12 exhibited a relatively flat growth curve and its optical density during the stationary phase was lower than other strains. Most of the strains reached the stationary phase 20 to 25 h after inoculation. Previous studies showed that the EPS production started to decline at the beginning of the stationary phase.12,23 Therefore, we collected the cultures of each strain at the end of their exponential phases and determined the production of EPS and biomass.3.3 EPS production and bacterial tolerance to simulated gastrointestinal environment
We determined the EPS production of the 23 selected strains (Table 4). B. longum strains 762, HUB-37-A12, and HAN-4-25 produced negligible amounts of EPS-r and EPS-b; strains M2-03-F02-27, 760, and HUB-2-25 produced only EPS-b; the remaining strains produced both EPS-r and EPS-b. In addition, we found that the EPS-b production levels of strains isolated from elderly volunteers were significantly higher than those of strains isolated from infants (p < 0.05). However, there was no significant difference in EPS-r production between strains from elder and infant samples (Fig. 2A). As shown in Table 4, strains M1-20-R01-3, M1-20-R01-B15, and C-11-A10 exhibited the highest tolerances to simulated gastric and intestinal juices, which corresponded with maximum EPS production. However, strains M2-03-F02-27 and HUB-2-25, which produced only EPS-b, also had relatively high survival rates after treatment with simulated gastric and intestinal juices. The correlations between EPS production and tolerance to the gastrointestinal environment were significant (p < 0.05); especially for EPS-b production, with Pearson correlation coefficients of 0.781 and 0.831 for tolerance to simulated gastric and intestinal juices, respectively, and apparent linear relations (Fig. 3). These results indicate that EPS bound to the cell surface is important for the survival of Bifidobacteria in adverse environments, such as the gastrointestinal tract. Furthermore, we found that the amount of EPS-b produced by mannose (+) strains was higher than that produced by mannose (−) strains (p < 0.05; Fig. 2B). These results suggest that the synthesis of cell-surface-bound exopolysaccharide may associated with the metabolism of mannose, however, the corresponding biological mechanisms are unclear.| Strain | Source | EPS-r mg per g biomass | EPS-b mg per g biomass | Amplification of pGT gene | Survival rate (%) | |
|---|---|---|---|---|---|---|
| Gastric juice | Intestinal juice | |||||
| a cpsD or rfbP represented that pGT gene in this strain was successfully amplified with primer cpsD-F/R or rfbP-F/R; N.D., pGT gene was not detected. | ||||||
| B. longum 642 | Reference | 27.40 ± 1.07 | 27.06 ± 1.60 | cpsD | 49.72 ± 1.44 | 48.96 ± 3.40 |
| B. longum 643 | Reference | 23.23 ± 0.55 | 21.67 ± 0.63 | rfbP | 61.08 ± 2.76 | 32.07 ± 2.56 |
| B. longum 685 | Infant fecal | 46.33 ± 0.80 | 23.43 ± 0.89 | rfbP | 52.91 ± 2.69 | 55.49 ± 3.32 |
| B. longum 686 | Infant fecal | 26.48 ± 1.02 | 37.96 ± 1.31 | rfbP | 65.03 ± 0.67 | 55.57 ± 2.13 |
| B. longum 687 | Infant fecal | 25.49 ± 0.63 | 33.29 ± 1.19 | rfbP | 49.41 ± 2.81 | 49.75 ± 2.54 |
| B. longum 688 | Infant fecal | 31.02 ± 1.17 | 6.06 ± 0.70 | — | 33.09 ± 2.81 | 46.47 ± 0.98 |
| B. longum 689 | Infant fecal | 29.23 ± 1.30 | 10.97 ± 1.35 | — | 30.04 ± 0.89 | 30.56 ± 2.39 |
| B. longum 752 | Infant fecal | 21.50 ± 0.84 | 6.56 ± 0.69 | rfbP | 38.32 ± 0.70 | 21.80 ± 1.22 |
| B. longum 756 | Infant fecal | 29.42 ± 0.87 | 5.56 ± 0.35 | rfbP | 58.41 ± 1.79 | 30.91 ± 3.06 |
| B. longum 760 | Infant fecal | 0.65 ± 0.48 | 1.19 ± 0.49 | rfbP | 32.63 ± 0.56 | 26.27 ± 1.40 |
| B. longum 762 | Infant fecal | 0.32 ± 0.54 | 0.03 ± 0.35 | cpsD | 47.96 ± 2.14 | 32.19 ± 2.79 |
| B. longum GX-17-A9 | Infant fecal | 8.41 ± 0.71 | 1.27 ± 0.66 | cpsD | 36.40 ± 1.70 | 16.63 ± 1.30 |
| B. longum HUB-37-A12 | Infant fecal | 0.68 ± 0.52 | 0.21 ± 0.32 | rfbP | 34.48 ± 1.43 | 20.29 ± 1.82 |
| B. longum M2-03-F02-27 | Infant fecal | 0.64 ± 0.46 | 15.26 ± 0.91 | rfbP | 57.34 ± 2.53 | 40.96 ± 3.26 |
| B. longum M1-20-R01-3 | Elder fecal | 55.31 ± 1.15 | 32.78 ± 1.11 | cpsD | 94.54 ± 2.34 | 75.57 ± 2.07 |
| B. longum M1-20-R01-B15 | Elder fecal | 43.91 ± 0.73 | 48.70 ± 0.79 | cpsD | 89.32 ± 3.72 | 65.25 ± 1.96 |
| B. longum M2-C-F01-14 | Elder fecal | 11.30 ± 1.01 | 31.47 ± 0.98 | — | 55.38 ± 1.62 | 46.15 ± 1.94 |
| B. longum C-1-A13 | Elder fecal | 19.55 ± 0.92 | 30.82 ± 1.06 | rfbP | 70.15 ± 1.85 | 64.30 ± 0.57 |
| B. longum C-6-A3 | Elder fecal | 7.91 ± 0.41 | 34.58 ± 1.31 | cpsD | 76.28 ± 1.23 | 58.04 ± 3.02 |
| B. longum C-11-A10 | Elder fecal | 69.27 ± 1.80 | 32.45 ± 0.74 | rfbP | 86.33 ± 0.62 | 79.38 ± 2.09 |
| B. longum HUB-6-1 | Elder fecal | 1.82 ± 0.80 | 11.58 ± 0.80 | rfbP | 36.11 ± 1.94 | 21.92 ± 1.44 |
| B. longum HUB-2-25 | Elder fecal | 0.39 ± 0.61 | 37.14 ± 0.72 | cpsD | 76.11 ± 2.21 | 68.24 ± 1.71 |
| B. longum HAN-4-25 | Elder fecal | 0.45 ± 0.45 | 0.12 ± 0.35 | cpsD | 52.22 ± 1.83 | 35.29 ± 2.35 |
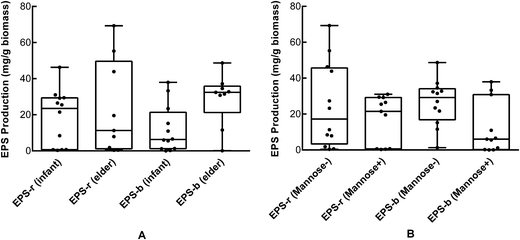 | ||
| Fig. 2 (A) Productions of EPS-r and EPS-b of B. longum strains isolated from elder and infant feces. (B) Productions of EPS-r and EPS-b of B. longum strains that fermented or did not ferment mannose. | ||
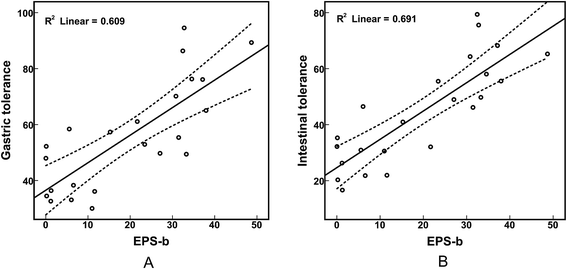 | ||
| Fig. 3 Correlations between production of EPS-b and tolerance to simulated gastric juice (A) and intestinal juice (B). | ||
3.4 Determination of pGT genes and sequence analysis
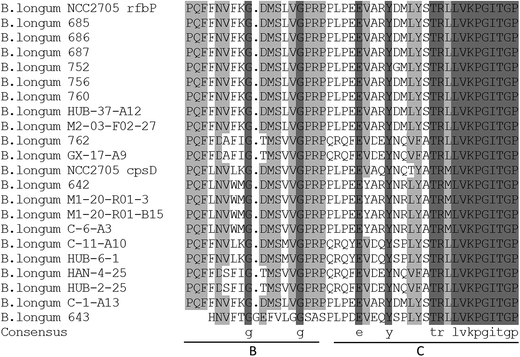
The gene presumed to code for pGT in the chosen B. longum strains was amplified (Table 4). There were three strains in which the pGT gene was not detected (strains 688, 689, and M2-C-F01-14); all three of these strains produced both EPS-r and EPS-b.The amino acid sequences predicted from the PCR products of the pGT gene exhibited high homology in the partial C-terminal regions (Fig. 4). Within these sequences, blocks B and C were involved in interaction with the lipid carrier and in sugar-specific recognition, respectively.18 Phylogenetic trees (Fig. 5) of these DNA and amino acid sequences were constructed using a neighbor-joining method, including two pGT gene fragments of B. longum NCC2705 from GenBank. The sequences of strains isolated from infant feces were largely clustered together (except for strains 762 and GX-17-A9), and were more similar to the sequence of rfbP gene from strain B. longum NCC2705. Most of the remaining sequences had higher homology with the sequence of cpsD gene from strain B. longum NCC2705. The sequences of strains HAN-4-25, HUB-2-25, GX-17-A9 and 762 were separate from those of other strains.
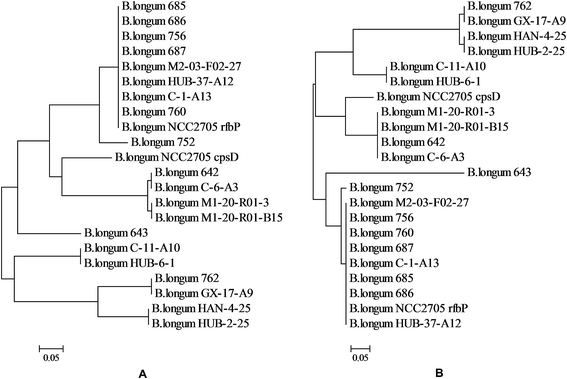 | ||
| Fig. 5 Phylogenetic trees of the internal fragments of priming glycosyltransferases genes obtained by PCR amplifications (A) and the corresponding amino acid sequences (B). | ||
4 Discussion
Bifidobacteria have been recognized as the predominance of intestinal microbiota in vaginally delivered infants; their numbers decline with the increasing age of their hosts.24–26 Analysis of the diversity of Bifidobacteria within infant and maternal gut microbiota showed that B. longum was the dominant Bifidobacterium species in both groups, constituting 56.2% and 38.2% of the total Bifidobacteria, respectively.27 Similarly, B. longum comprised the majority of Bifidobacterial strains isolated in the present study.EPSs produced by Bifidobacteria are considered to be significant in interaction mechanisms between these bacteria and their hosts. Some commensal bacteria producing capsular polysaccharides have been predicted to involved in the decoration of hosts' intestinal mucosa.28 The determination of EPS productions by different Bifidobacterium species showed no significant differences between the species, and EPS production was significantly correlated with the tolerance ability against low pH and bile salts.14 In this study, two types of EPSs produced by different B. longum strains were determined. Most of the tested strains produced both types of EPSs. However, several strains produced only EPS-b, and others produced neither of the two types of EPS. No strain was found to produce only EPS-r; this was attributed to the synthesis process of EPS, whereby it is assembled on the extracellular surface of cytomembranes from repeating units.29 EPS-b production of strains isolated from elder volunteers was significantly higher than those of strains isolated from infants, which may indicate that commensal Bifidobacteria co-evolved with their hosts.6
EPS synthesis is a complex process that is catalyzed by several enzymes coded by eps clusters. In previous studies, PCR techniques were used to detect fragments of genes coding glucansucrases, fructansucrases and glycosyltransferase synthesizing β-D-glucan, that are involved in the biosynthesis of homopolysaccharides in EPS-producing Bifidobacteria. However, these attempts were unsuccessful. This may indicate that most Bifidobacteria carry genes associated with the synthesis of heteropolysaccharides.30–32 The initial step in EPS synthesis is the pGT-catalysed addition of a sugar-1-phosphate to a lipid carrier molecule anchored to the cell membrane. Additional sugar moieties are linked to the initial monosaccharide by forming glucosidic bonds, catalyzed by glycosyltransferases (GTs), to form repeating units.33 The sugars used to form repeating units are in an activated form (nucleoside di- or monophosphorylated sugars), and the structures of different repeating units are dependent upon the GTs involved in their syntheses. Previous studied have demonstrated that the production of EPS was altered or interrupted after inactivation of the pGT gene, indicating that pGTs are necessary for the biosynthesis of EPS.34,35 Most genes coding for GTs are dissimilar to each other; however, the pGT genes in various bacteria are relatively similar and define conserved amino acid sequences in a specific group of pGTs, particularly in the carboxy terminus.19 The relatively high homology may be related to the presence of domains that interact with lipid carrier.18 The genome of B. longum NCC2705 contains two pGT genes coding for galactosyltransferase (cpsD) and undecaprenyl-phosphate sugar phosphotransferase (rfbP). However, in other B. longum strains, whose genomes have been completely sequenced, only one or neither of the two genes was found. Similarly, 20 of the 23 strains analyzed in this study contained either rfbP or cpsD gene fragments; no pGT genes were detected in the remaining strains. Similarly, Hidalgo-Cantabrana et al.36 selected four primer pairs from 24 degenerated primers and detected at least one pGT gene (cpsD or rfbP) in 63 out of 99 Bifidobacterial strains.
Analyses of EPS production by our tested strains showed there were three strains were unable to produce both types of EPS. However, these strains contained pGT genes. This discrepancy may result from the complex synthesis of EPS, which contains many steps and is catalyzed by a series of enzymes coded by genes contained in a complex gene cluster. This include genes coding for enzymes involved in the transportation of the repeated EPS subunits to the extracellular surface of the cytomembrane and in the assembly of the repeating units.18 The expression of these genes is regulated by regulatory genes and affected by environmental and nutritional conditions. Furthermore, three B. longum strains were positive for EPS production but contained no pGT genes. These results suggest that the sequences of the pGT genes in these strains may have less homology with those of other strains or that there may be other types of pGTs coded by genes that could not amplified using our primers.
The phylogenetic trees constructed using the pGT gene fragments and their corresponding amino acid sequences showed that the sequences amplified with the rfbP-F/R primer pair were mostly clustered together (except for strains 643, C-11-A10, and HUB-6-1) and that these clustered sequences were mostly from strains isolated from infant feces (except for strain C-1-A13). Furthermore, the two sequences amplified with the cpsD-F/R primer pair from infant sample strains were also clustered together. These results suggest that the pGT genes of infant-originated B. longum strains had greater homology than those of elder-originated pGT genes.
5 Conclusion
We determined the EPS productions by B. longum strains isolated from elder and infant feces. In addition, we determined the tolerances of B. longum strains to simulated gastric and intestinal juices. These tolerances were found to correlate with EPS production; particularly with the production of EPS-b, which is often regarded as a capsular polysaccharide. Moreover, we determined that EPS-b production in strains isolated from elderly volunteers was significantly higher than that in strains isolated from infants, and this was the first time to report this result. Furthermore, mannose (−) strains produced more EPS-b than mannose (+) strains. Lastly, PCR was used to detect gene fragments coding for pGT, which catalyzes the first step of EPS production. These gene fragments were present in most of the B. longum strains tested. The phylogenetic tree of these sequences indicated that the pGT genes of infant-originated B. longum strains had greater homology than those of the elder-originated strains.Acknowledgements
This work was supported by the Young Scientists Fund in National Natural Science Foundation of China (No. 31401682), the Young Scientists Fund in Science and Nature Foundation of Jiangsu Province (No. BK 20140146), the 111 Project B07029, the Program for Changjiang Scholars and Innovative Research Team in University (IRT1249), and the Program of Collaborative innovation center of food safety and quality control in Jiangsu Province.References
- N. Qin, F. Yang, A. Li, E. Prifti, Y. Chen, L. Shao, J. Guo, E. Le Chatelier, J. Yao, L. Wu, J. Zhou, S. Ni, L. Liu, N. Pons, J. M. Batto, S. P. Kennedy, P. Leonard, C. Yuan, W. Ding, Y. Chen, X. Hu, B. Zheng, G. Qian, W. Xu, S. D. Ehrlich, S. Zheng and L. Li, Nature, 2014, 513, 59–64 CrossRef CAS PubMed.
- S. L. Russell, M. J. Gold, M. Hartmann, B. P. Willing, L. Thorson, M. Wlodarska, N. Gill, M. R. Blanchet, W. W. Mohn, K. M. McNagny and B. B. Finlay, EMBO Rep., 2012, 13, 440–447 CrossRef CAS PubMed.
- M. P. Conte, S. Schippa, I. Zamboni, M. Penta, F. Chiarini, L. Seganti, J. Osborn, P. Falconieri, O. Borrelli and S. Cucchiara, Gut, 2006, 55, 1760–1767 CrossRef CAS PubMed.
- Z. Guo, J. Zhang, Z. Wang, K. Y. Ang, S. Huang, Q. Hou, X. Su, J. Qiao, Y. Zheng, L. Wang, E. Koh, H. Danliang, J. Xu, Y. K. Lee and H. Zhang, Sci. Rep., 2016, 6, 20602 CrossRef CAS PubMed.
- M. E. Dumas, R. H. Barton, A. Toye, O. Cloarec, C. Blancher, A. Rothwell, J. Fearnside, R. Tatoud, V. Blanc, J. C. Lindon, S. C. Mitchell, E. Holmes, M. I. McCarthy, J. Scott, D. Gauguier and J. K. Nicholson, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 12511–12516 CrossRef CAS PubMed.
- M. Ventura, F. Turroni, M. O. Motherway, J. MacSharry and D. van Sinderen, Trends Microbiol., 2012, 20, 467–476 CrossRef CAS PubMed.
- M. Gueimonde, K. Laitinen, S. Salminen and E. Isolauri, Neonatology, 2007, 92, 64–66 CrossRef PubMed.
- S. Fukuda, H. Toh, K. Hase, K. Oshima, Y. Nakanishi, K. Yoshimura, T. Tobe, J. M. Clarke, D. L. Topping, T. Suzuki, T. D. Taylor, K. Itoh, J. Kikuchi, H. Morita, M. Hattori and H. Ohno, Nature, 2011, 469, 543–547 CrossRef CAS PubMed.
- J. Singh, A. Rivenson, M. Tomita, S. Shimamura, N. Ishibashi and B. S. Reddy, Carcinogenesis, 1997, 18, 833–841 CrossRef CAS PubMed.
- H. Ishikawa, I. Akedo, Y. Umesaki, R. Tanaka, A. Imaoka and T. Otani, J. Am. Coll. Nutr., 2003, 22, 56–63 CrossRef PubMed.
- P. V. P. Guerra, L. N. Lima, T. C. Souza, V. Mazochi, F. J. Penna, A. M. Silva, J. R. Nicoli and E. V. Guimaraes, World J. Gastroenterol., 2011, 17, 3916–3921 CrossRef PubMed.
- R. Tallon, P. Bressollier and M. C. Urdaci, Res. Microbiol., 2003, 154, 705–712 CrossRef CAS PubMed.
- S. K. Mazmanian and D. L. Kasper, Nat. Rev. Immunol., 2006, 6, 849–858 CrossRef CAS PubMed.
- G. Alp and B. Aslim, Anaerobe, 2010, 16, 101–105 CrossRef CAS PubMed.
- S. Fanning, L. J. Hall, M. Cronin, A. Zomer, J. MacSharry, D. Goulding, M. O. Motherway, F. Shanahan, K. Nally, G. Dougan and D. van Sinderen, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 2108–2113 CrossRef CAS PubMed.
- E. Schiavi, M. Gleinser, E. Molloy, D. Groeger, R. Frei, R. Ferstl, N. Rodriguez-Perez, M. Ziegler, R. Grant, T. F. Moriarty, S. Plattner, S. Healy, M. O'Connell Motherway, C. A. Akdis, J. Roper, F. Altmann, D. van Sinderen and L. O'Mahony, Applied and Environmental Microbiology, 2016, 82, 7185–7196 CrossRef CAS PubMed.
- P. Ruas-Madiedo, J. Hugenholtz and P. Zoon, Int. Dairy J., 2002, 12, 163–171 CrossRef CAS.
- C. Provencher, G. LaPointe, S. Sirois, M. R. Van Calsteren and D. Roy, Appl. Environ. Microbiol., 2003, 69, 3299–3307 CrossRef CAS PubMed.
- L. Wang, D. Liu and P. R. Reeves, J. Bacteriol., 1996, 178, 2598–2604 CrossRef CAS PubMed.
- M. DuBois, K. A. Gilles, J. K. Hamilton, P. t. Rebers and F. Smith, Anal. Chem., 1956, 28, 350–356 CrossRef CAS.
- P. Chen, Q. X. Zhang, H. Dang, X. M. Liu, F. W. Tian, J. X. Zhao, Y. Q. Chen, H. Zhang and W. Chen, Food Control, 2014, 35, 65–72 CrossRef CAS.
- A. Parte, W. Whitman, M. Goodfellow, P. Kämpfer, H.-J. Busse, M. Trujillo, W. Ludwig and K.-i. Suzuki, Bergey's Manual of Systematic Bacteriology: Volume 5: The Actinobacteria, Springer Science & Business Media, 2012 Search PubMed.
- G. H. van Geel-Schutten, F. Flesch, B. ten Brink, M. R. Smith and L. Dijkhuizen, Appl. Microbiol. Biotechnol., 1998, 50, 697–703 CrossRef CAS.
- G. Reuter, Curr. Issues Intest. Microbiol., 2001, 2, 43–53 CAS.
- G. Solís, C. G. de los Reyes-Gavilan, N. Fernández, A. Margolles and M. Gueimonde, Anaerobe, 2010, 16, 307–310 CrossRef PubMed.
- I. Adlerberth, E. Lindberg, N. Aberg, B. Hesselmar, R. Saalman, I. L. Strannegard and A. E. Wold, Pediatr. Res., 2006, 59, 96–101 CrossRef PubMed.
- F. Turroni, C. Peano, D. A. Pass, E. Foroni, M. Severgnini, M. J. Claesson, C. Kerr, J. Hourihane, D. Murray, F. Fuligni, M. Gueimonde, A. Margolles, G. De Bellis, P. W. O'Toole, D. van Sinderen, J. R. Marchesi and M. Ventura, PLoS One, 2012, 7, e36957 CAS.
- L. Bry, P. G. Falk, T. Midtvedt and J. I. Gordon, Science, 1996, 273, 1380–1383 CAS.
- A. D. Welman and I. S. Maddox, Trends Biotechnol., 2003, 21, 269–274 CrossRef CAS PubMed.
- M. Tieking, S. Kaditzky, R. Valcheva, M. Korakli, R. F. Vogel and M. G. Ganzle, J. Appl. Microbiol., 2005, 99, 692–702 CrossRef CAS PubMed.
- M. Tieking, M. Korakli, M. A. Ehrmann, M. G. Ganzle and R. F. Vogel, Appl. Environ. Microbiol., 2003, 69, 945–952 CrossRef CAS PubMed.
- M. L. Werning, I. Ibarburu, M. T. Duenas, A. Irastorza, J. Navas and P. Lopez, J. Food Prot., 2006, 69, 161–169 CrossRef CAS PubMed.
- C. Hidalgo-Cantabrana, B. Sanchez, C. Milani, M. Ventura, A. Margolles and P. Ruas-Madiedo, Appl. Environ. Microbiol., 2014, 80, 9–18 CrossRef CAS PubMed.
- F. Stingele, J. R. Neeser and B. Mollet, J. Bacteriol., 1996, 178, 1680–1690 CrossRef CAS PubMed.
- J. K. Morona, R. Morona and J. C. Paton, J. Bacteriol., 1999, 181, 5355–5364 CAS.
- C. Hidalgo-Cantabrana, I. Ordoñez, P. Ruas-Madiedo and A. Margolles, Benefic. Microbes, 2015, 6, 553–562 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2017 |