Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Kinetic resolution of (RS)-1-chloro-3-(4-(2-methoxyethyl)phenoxy)propan-2-ol: a metoprolol intermediate and its validation through homology model of Pseudomonas fluorescens lipase†
Surbhi Sonia,
Bharat P. Dwivedeeb,
Vishnu K. Sharmac and
Uttam C. Banerjee *b
*b
aDepartment of Biotechnology, National Institute of Pharmaceutical Education and Research, S.A.S. Nagar, 160062, Punjab, India
bDepartment of Pharmaceutical Technology (Biotechnology), National Institute of Pharmaceutical Education and Research, S.A.S. Nagar 160062, Punjab, India. E-mail: ucbanerjee@niper.ac.in
cDepartment of Pharmacoinformatics, National Institute of Pharmaceutical Education and Research, S.A.S. Nagar 160062, Punjab, India
First published on 24th July 2017
Abstract
In the present study Pseudomonas fluorescens lipase (PFL) was screened as a time efficient biocatalyst for the kinetic resolution of a racemic intermediate [(RS)-1-chloro-3-(4-(2-methoxyethyl)phenoxy)propan-2-ol] of metoprolol, an important selective β1-blocker drug. PFL selectively acylated the R-form of this racemic intermediate in a short duration of 3 h. Different reaction parameters were optimized to achieve maximum enantioselectivity. It was found that at 30 °C, enzyme activity of 400 units and substrate concentration of 10 mM gave a high enantioselectivity and conversion in an optimum time of 3 hours (C = 50.5%, eep = 97.2%, ees = 95.4%, E = 182). To validate these experimental results, the 3D structure of PFL was built using homology modelling. Validation of the model through Ramachandran plot (92.7% in favored region), Errat plot (overall quality factor, 79.27%), Verify-3D score (86.19) and ProSA_Z score (−6.24) depicted the overall good quality of the model. Molecular docking of the R- and S-enantiomers of the intermediate, which was performed on this model, demonstrated a strong H-bond interaction (1.6 Å) between the hydroxyl group of the R-enantiomer and Arg54, a key binding residue of the catalytic site of PFL, while no significant interaction with the S-enantiomer was observed. Thus, the outcome of this docking study was in agreement with the experimental data, clarifying that PFL preferentially catalysed the transesterification of the R-enantiomer into the corresponding ester, leaving the S-enantiomer intact.
Introduction
The growing demand of optically pure compounds for applications in pharmaceutical, chemical, agricultural and cosmetic industries is well understood. The increased interest in these compounds can be clearly attributed to the better understanding of chirality and its importance in synthetic reactions.1,2 As the predominant outlook of the industry is always on economically expedient, dependable and scalable processes with minimal waste generation, biocatalysis has become a cornerstone for the synthesis of chiral intermediates.3 Biocatalysis involves the application of enzymes in a suitable form (whole-cell, immobilized or commercial preparation) to catalyze chemical reactions.4,5 Lipases as biocatalysts have earned a distinguished position in the field of chiral synthesis due to their excellent chemo-, regio- and enantioselectivity.6 Lipases obtained from different sources belong to the α/β-hydrolase folding group. Specific sequences of the α-helices and β-strands are responsible for the hydrolyzing activity of lipases.7 Lipases are widely applied in kinetic resolution, where the lipase discriminates between two enantiomers of the racemic mixture and selectively catalyzes a faster conversion of one enantiomer to the product. This is a convenient method for separation of both enantiomers from a racemic mixture.8 The rationality of lipase catalyzed biocatalysis as a compatible method for enantiopure synthesis is further augmented by the incorporation of computational strategies.9 Computational chemistry provides an effective set of tools to simulate the biocatalytic reactions in silico and help in the assessment of the effectiveness of the process, thereby validating the experimentation.10 One key computational methodology is the docking of small molecules into the active site of lipase.11 The utilization of docking studies enables the fast evaluation of enantioselectivity and is of prime interest to suggest a plausible biotransformation reaction. This enables expensive experimentation to be directed and focused, thereby speeding up bioprocess development.12β-Aryloxyalcohols are useful building blocks in the synthesis of several biologically active compounds. Metoprolol, 1-[4-(2-methoxyethyl)phenoxy]-3-(propan-2-ylamino)propan-2-ol, a selective β1-adrenergic blocking agent, is used in the treatment of angina and hypertension. It is well established that the desirable therapeutic activity of metoprolol resides mainly in the (S)-enantiomer.13 A number of chemical methods have been described in the literature for the synthesis of enantiopure metoprolol.14–22 However, most of the chemical synthesis methods suffer from some disadvantages, such as poor enantioselectivity, long reaction time, hazardous reaction conditions, formation of by products, and higher cost. Biocatalytic approach has also been used for the synthesis of chiral intermediates of metoprolol.23
To the best of our knowledge, herein, we report for the first time the homology model of PFL and its application to validate the R-selectivity of PFL in the kinetic resolution of a racemic intermediate of metoprolol. In the present study, the racemic intermediate [(RS)-1-chloro-3-(4-(2-methoxyethyl)phenoxy)propan-2-ol] of metoprolol was chemically synthesized. Various commercial lipase preparations were screened for the kinetic resolution of this racemic intermediate. Among them, PFL was selected as the most suitable lipase. The reaction parameters were optimized using ‘one-factor at a time’ approach. Furthermore, to validate the experimental results, a homology model of PFL was built since it was not available. The developed homology model was utilized for the molecular docking analysis wherein experimental observations were validated by docking the R and S form of intermediate into the active site of PFL.
Results and discussion
Chemical synthesis of racemic intermediate, (RS)-4
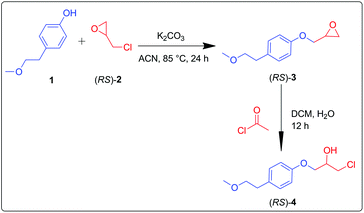
β-Aryloxyalcohols are key intermediates in the preparation of metoprolol. The fundamental structure of metoprolol comprises aryloxypropanolamine. The aromatic ring substituents govern the hydrophobicity of the molecule. Branched alkyl amine substituents present in metoprolol are essential for the β-antagonist activity.24 A convenient protocol for the preparation of β-aryloxyalcohols requires the synthesis of epoxide intermediates with phenols25–27 and subsequent ring-opening of the epoxides.28,29 The racemic intermediate (RS)-4 of metoprolol was prepared via a chemical route. Detailed discussion regarding the synthetic schemes has been furnished in the Experimental section.Screening of suitable biocatalysts using experimental approach
Lipases are interfacial enzymes and display prodigious stereo-, chemo-, and regioselectivity.30 In the present study, (RS)-4 was subjected to lipase catalyzed kinetic resolution (Scheme 1). Five commercially available lipases [Thermomyces lanuginosus (TLL), Candida rugosa (CRL), Pseudomonas fluorescens (PFL), Candida antarctica lipase A (CALA) and Candida antarctica lipase B (CALB)] were screened for the kinetic resolution of (RS)-4 using vinyl acetate as an acyl donor and toluene as a solvent. Interestingly, Pseudomonas fluorescens lipase (PFL) was found to be the most suitable biocatalyst (Table 1). PFL resolved (RS)-4 to the corresponding (R)-ester and (S)-alcohol products, displaying good enantioselectivity and conversion. Moreover, PFL catalyzed the kinetic resolution of the racemate in a short time. This behaviour of PFL towards the rapid enantioselective conversion may be due to the favourable interactions between the amino acid residues in the active site and the ligands.| S. no. | Lipase | % Cb | eesc | eepd | Ee |
|---|---|---|---|---|---|
a Conditions: (RS)-4 (20 mM) in toluene (1 mL) was treated with vinyl acetate (5.4 mmol) at 30 °C in the presence of different lipases (300 IU mL−1).b % conversion were calculated from the enantiomeric excess (ee) of (S)-4 and (R)-5 as follows: conversion (C) = ees/(ees + eep).c Enantiomeric excess of (S)-4 was determined by HPLC analysis (Daicel Chiralcel OD-H column) 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10; hexane 10; hexane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) IPA, flow rate of 1.0 mL min−1, detected at 254 nm.d Enantiomeric excess of (R)-5 was determined by HPLC analysis (Daicel Chiralcel OD-H column) 90 IPA, flow rate of 1.0 mL min−1, detected at 254 nm.d Enantiomeric excess of (R)-5 was determined by HPLC analysis (Daicel Chiralcel OD-H column) 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10; hexane 10; hexane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) IPA, flow rate of 1.0 mL min−1, detected at 254 nm.e E value were calculated using the formula: E = ln[eep(1 − ees)/(eep + ees)]/ln[eep(1 + ees)/(eep + ees)].35 IPA, flow rate of 1.0 mL min−1, detected at 254 nm.e E value were calculated using the formula: E = ln[eep(1 − ees)/(eep + ees)]/ln[eep(1 + ees)/(eep + ees)].35 |
|||||
| 1 | Thermomyces lanuginosus (TLL) | 50.0 ± 1.21 | 74.4 ± 2.95 | 74.3 ± 2.31 | 14.9 ± 3.51 |
| 2 | Candida antarctica immobilized (CAL B) | 50.3 ± 2.98 | 70.8 ± 3.23 | 69.9 ± 1.99 | 11.7 ± 3.78 |
| 3 | Candida antarctica immobilized (CAL-A) | 51.2 ± 1.27 | 93.7 ± 3.01 | 89.5 ± 2.08 | 62.9 ± 1.76 |
| 4 | Pseudomonas fluorescens (PFL) | 50.0 ± 1.58 | 96.2 ± 2.69 | 96.1 ± 3.26 | 200 ± 2.65 |
| 5 | Candida rugosa (CRL) | 53.5 ± 2.25 | 95.2 ± 2.84 | 82.8 ± 2.76 | 39.3 ± 1.82 |
Optimization of reaction parameters for kinetic resolution
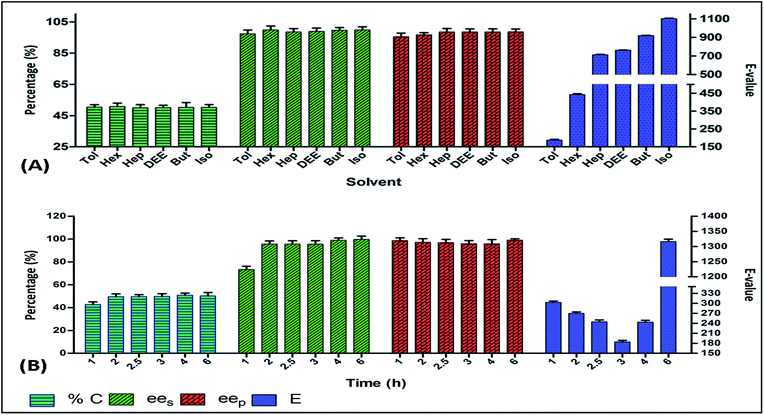
To achieve a successful and efficient lipase catalyzed reaction, conditions favouring rapid progress and irreversibility must be taken into account. Various reaction parameters such as type of solvents, reaction time and temperature, type of acyl donors, substrate and enzyme concentrations etc. were optimized. The substrate concentration (10 mM), enzyme concentration (400 IU), solvent (toluene), acyl donor (vinyl acetate), temperature (30 °C), and reaction time (3 h) were optimized on the basis of high enantiomeric excess (ees & eep), high conversion rate (>49%) and enantiomeric ratio (E ≤ 200) [detailed discussion on each parameter is given in the next section]. Under these optimized conditions, PFL performed the acylation of (R)-enantiomer of (RS)-4 with a final result of C = 50.5%, eep = 97.2%, ees = 95.4%, E = 182.Effect of solvents
Several examples of improved regioselectivity and enantioselectivity of lipases in organic reactions have been observed. Lipases show good stability in organic solvents.31 One of the important reasons for such a behaviour is that hydrophilic solvents have a higher tendency to strip the tightly bound water, which is necessary for the catalytic activity of lipase. Organic solvents have a lesser tendency for this, making them a better reaction medium for lipase catalyzed reactions.32 Considering these facts, different organic solvents with a diverse range of log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values were examined for the kinetic resolution of (RS)-4 with vinyl acetate using PFL. The best result was obtained using toluene with the highest percentage conversion (50.5%), enantiomeric excess (97.4% ees & 95.5% eep) and E-value 188 (Fig. 1A).
P values were examined for the kinetic resolution of (RS)-4 with vinyl acetate using PFL. The best result was obtained using toluene with the highest percentage conversion (50.5%), enantiomeric excess (97.4% ees & 95.5% eep) and E-value 188 (Fig. 1A).
Effect of acyl donors
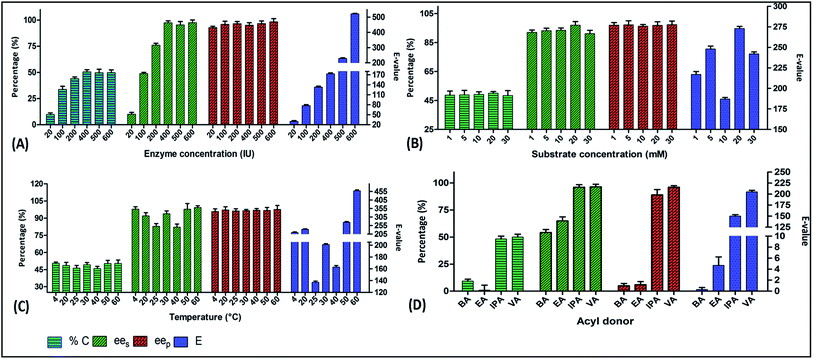
Enantioselectivity in the lipase mediated reactions originates during the deacylation step with the nucleophilic attack of the alcohol leading to the formation of the first acyl–enzyme complex.33 Hence, the type of acyl donor also has a pivotal role in the progress of the reaction. Among the different acyl donors [BA = benzyl acetate, EA = ethyl acetate, IPA = isopropenyl acetate, VA = vinyl acetate], vinyl acetate showed the best results in the kinetic resolution of (RS)-4. Vinyl acetate is an enol ester that forms unstable enol as a by-product and rapidly tautomerizes to form the corresponding aldehydes or ketones making the reaction completely irreversible.34 The highest percentage conversion (50.1%), enantiomeric excess (96.4% ees & 96.1% eep) and E-value (205) of PFL catalysed kinetic resolution of (RS)-4 in toluene were obtained with vinyl acetate as the acyl donor (Fig. 2D).Effect of reaction time
The most interesting part of the study is the short duration in which PFL catalyzes the enantioselective reaction of (RS)-4. In the reaction of (RS)-4, PFL took 3 h for 50.0% conversion with 95.5% ees, 96.0% eep, and E-value of 183.5 (Fig. 1B). PFL started to display good enantioselectivity and conversion rate very early in the reaction, in only over 30 min to deliver the best result. Although the values of enantioselective ratio and percentage conversion along with enantiomeric excess were low at the early onset of the reaction, PFL exhibited the best results promptly. Such high reactivity of PFL towards the intermediate is justified with the interaction of the (R)-enantiomer at the active site of the enzyme, as later validated by the docking study. During the course of the reaction, the enantioselectivity gradually decreased and PFL exhibited non-selective behaviour. In a recent study on kinetic resolution of racemic atenolol, PFL has been reported to give (S)-enantiomer in 24 h.36 In another study, PFL catalyzed the kinetic resolution of some novel antifungal N-substituted benzimidazole derivatives in more than 72 h.37 Thus, the rapid action of PFL on (RS)-4 with high degree of enantioselectivity and rate of conversion reported here is suggestive of strong interaction between (RS)-4 and PFL, thereby revealing a better chemoenzymatic process that could be used for the synthesis of enantiopure metoprolol in a shorter time.Effect of substrate concentration
The effect of substrate concentration on the enantioselectivity and conversion of an enzymatic reaction has been extensively studied in the literature.38 Thus, we performed the reaction using a range of substrate concentration from 1 to 30 mM. The optimal substrate concentration was 10 mM, at which PFL displayed good conversion rate of 49.2% along with the ees, eep and E-value of 93.4%, 96.3% and 187, respectively (Fig. 2B).Effect of enzyme concentration
A biocatalytic transformation occurs at the active sites of the enzyme. Owing to the kinetic consideration of the enzymes, an optimal enzyme concentration, where all the active sites are acquired by substrate molecules, should be identified. This has a direct effect on the cost effectiveness of the process.39 Thus, we optimized the enzyme concentration to obtain the highest enantioselectivity. The reaction of (RS)-4 was carried out using different concentrations (100, 200, 300, 400, 600 IU) of PFL in toluene. At the concentration of 400 IU, PFL showed the highest conversion 50.6% and ees, eep and E-value were found to be 97.4%, 95.1% and 173, respectively (Fig. 2A). Further increase in enzyme concentration reduced the enantioselectivity of the reaction, probably due to the non-selective interaction of the enzymes with the substrates at higher enzyme concentration.Effect of temperature
The effect of temperature on the degree of enantioselectivity and percentage conversion was investigated by performing the reactions at a range of temperatures (4, 20, 25, 30, 40, 50 and 60 °C). In the reactions of (RS)-4, PFL catalyzed the conversion with the highest enantioselectivity at 30 °C. The increase in temperature showed a gradual demise in the enantioselective resolution of the (R) and (S) forms of the racemic alcohol. For (RS)-4, the conversion at 30 °C was 49.3%, with 93.8% ees, 96.5% eep and 201 E-value after 3 h of reaction (Fig. 2C).Validation of suitable biocatalyst using computational approach
The structural information of several lipases is well exploited to give an insight into the catalytic mechanism and enantiopreference of the biocatalytic reaction.40 During the experimental study, PFL exhibited biocatalytic reaction in only three hours. This outcome encouraged us to explore the interaction between PFL and the substrate. Homology modeling and docking studies of two enantiomers into the active site of PFL were carried out to substantiate the experimental outcomes of this study (Table 2).| Physicochemical parameter | Optimum parameter | C (%)a | ees (%)b | eep (%)c | Ed |
|---|---|---|---|---|---|
| a a, b, c, d are the reaction conditions and equations described in Table 1. | |||||
| Solvent | Toluene | 50.5 ± 2.80 | 97.4 ± 2.50 | 95.5 ± 2.33 | 188 ± 3.79 |
| Acyl donor | Vinyl acetate | 50.1 ± 2.21 | 96.4 ± 2.17 | 96.1 ± 2.35 | 205 ± 2.65 |
| Reaction time | 3 h | 49.9 ± 2.00 | 95.5 ± 2.17 | 96.0 ± 3.07 | 184 ± 3.51 |
| Temperature | 30 °C | 49.3 ± 2.34 | 93.8 ± 2.16 | 96.5 ± 2.03 | 201 ± 4.04 |
| Substrate conc. | 10 mM | 49.2 ± 3.00 | 93.4 ± 2.06 | 96.3 ± 1.91 | 187 ± 3.61 |
| Enzyme conc. | 400 IU (20 mg mL−1) | 50.6 ± 2.54 | 97.4 ± 2.40 | 95.1 ± 2.27 | 173 ± 3.06 |
To validate the quality of the developed PFL homology model, various online validation tools like ERRAT plot, Ramachandran plot, ProSA, ProQ and Verify-3D etc. were utilized. In the Ramachandran contour plot42 of PFL (Fig. 3A) 92.4% residues were in favored regions, 5.6% in additionally allowed regions and 2.0% residues in outlier regions. The modeled structures were also validated by ERRAT plot,43 which showed an overall quality factor of 79.27 as illustrated in Fig. 3C. This was close to the overall quality factor of the template protein (81.85). Verify-3D program was used to validate the stereochemical quality of the model.44 The residues with a scoring function over 0.2 are considered to be reliable. Furthermore, 86.2% of the residues had a score over 0.2. This validated the good quality of the predicted model45 as shown in Fig. 3D. Furthermore the structure was analyzed using the ProSA Z-score to assess the quality of the model. The ProSA Z-score value for PFL was −6.24, which falls within the range for native set of proteins having the same size (Fig. 3B). If the Z-score of a model structure is located outside the range characteristic for native proteins from different sources (X-ray and NMR), it indicates an erroneous structure.46 Thus ProSA Z-score indicated a good correlation between the modeled PFL and the native structure (PDB-2Z8X) in almost of the parts of the sequence. The ProQ LG score, which also predicts the quality of a model protein, was 3.73. It indicated that the developed model is of good quality (LG score > 3). Comparative analysis of template protein (PDB code 2Z8X) and target protein (PFL) is given in Table 3. Finally, energy minimization of the homology model of PFL was performed using Prime module of Schrodinger. The overall results from these model validation tools revealed good quality of the model for PFL.
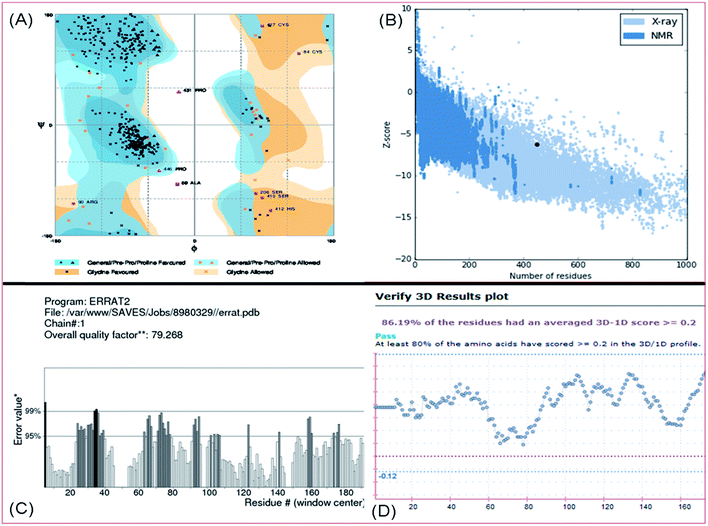 | ||
| Fig. 3 Validation of homology model of PFL: (A) Ramachandran plot (B) ProSA Z-score (C) Errat plot (D) Verify-3D plot. | ||
| Validation parameters | 2Z8X | PFL |
|---|---|---|
| Ramachandran plot | ||
| Favoured | 98.4% | 92.4% |
| Allowed | 1.5% | 5.6% |
| Outliers | 0.2% | 2.0% |
| Errat plot (overall quality factor) | 81.85 | 79.27 |
| Verify-3D | 98.54 | 86.19 |
| ProSA_Z score | −10.66 | −6.24 |
| ProQ server_LG score | 5.85 | 3.73 |
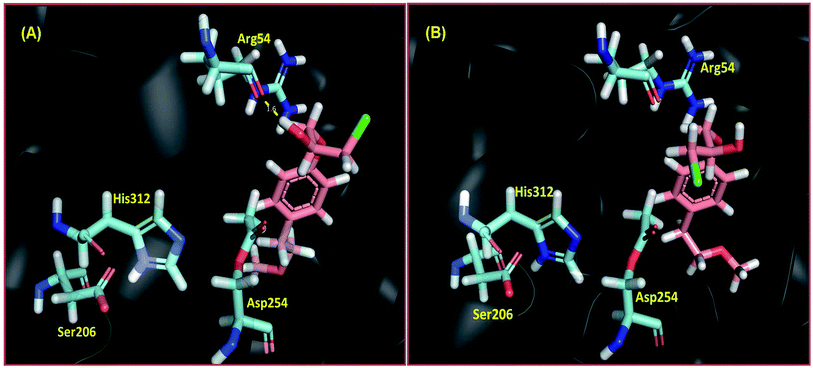
It is followed by the nucleophilic attack by an alcohol, leading to the formation of an ester.50 The acyl–enzyme intermediate was prepared by the acylation of the hydroxyl group of Ser206. Molecular docking of R and S enantiomers of (RS)-4 was performed into the acylated enzyme active site, using Schrodinger's Glide module.51 It has been well established that the R and S enantiomers of the substrate interact differently with the residues at the active site of the lipase.26 Stereochemical rules have been proposed for the determination of enantioselectivity. A large number of studies have established an empirical rule that states the R-preference of the stereochemical trend observed for lipase catalyzed kinetic resolution of secondary alcohols.52 The R-enantiomer (R)-4 showed strong interaction with Arg54 residue, which forms a part of the oxyanion hole of PFL, with a H-bond distance of 1.6 Å (Fig. 4A), while the S enantiomer (S)-4 showed no significant interactions needed for the lipase catalyzed reaction (Fig. 4B). The G score for the R-enantiomer was −5.33, while the S-enantiomers had a lower score of −4.50. This outcome of the docking study is in agreement with experimental data, clarifying that PFL preferentially catalyzed the transesterification of the R-alcohol. However, this selectivity was time dependent. Initially, PFL catalyzed the reaction very fast in favor of transesterification of the R-enantiomer. The maximum conversion of (RS)-4 into the corresponding ester (R)-5 was achieved in 3 h. The result obtained in 3 h was C = 50.0%, ees = 95.5%, eep = 96.0%, and E = 183.5.
Conclusions
The methodology of complementing a biocatalytic reaction with in silico studies, which is corroborated by this work, is a time-efficient and reliable process. The present study involves screening of various lipases and selecting Pseudomonas fluorescens lipase as an expeditious biocatalyst, performing the kinetic resolution of the racemic intermediate of metoprolol in a short duration of three hours. The biocatalytic reaction yields the (S)-alcohol, which can be directly utilized in the synthesis of (S)-metoprolol. A suitable homology model for PFL was successfully developed, validated and used for docking to analyse the difference in interaction of two enantiomers with PFL. The R-enantiomer presented a higher docking score and stronger H-bond interaction than the S-enantiomer. Both approaches proved to be advantageous to select a suitable lipase for the kinetic resolution. Under the optimized conditions, PFL exhibited selective acylation of the (R)-enantiomer of (RS)-4 [C = 50.5%, eep = 97.2%, ees = 95.4%, E = 182]. The study presents a versatile biocatalyst giving speedy and good result for the enantiopure synthesis of the metoprolol intermediate. This simple, cost effective and green approach may likewise be explored for the synthesis of various enantiopure drugs using Pseudomonas fluorescens lipase as a biocatalyst.Experimental
Materials
4-(2-Methoxyethyl)phenol, (RS)-epichlorohydrin, and (R)-epichlorohydrin were purchased from Sigma-Aldrich Corporation (St. Louis, MO, USA). Anhydrous Na2SO4 and K2CO3 were obtained from Merck (Germany). HPLC grade solvents were obtained from Merck (Germany). Candida antarctica immobilized (CALA and CALB), Candida rugosa lipase, Thermomyces lanuginosus lipase and Pseudomonas fluorescens lipase were purchased from SIGMA (St. Louis, Missouri, USA). TLC plates were purchased from Merck (Germany). Silica gel (60–120 mesh) for column chromatography was obtained from SRL (India).Synthesis of 1-chloro-3-(4-(2-methoxyethyl)phenoxy)propan-2-ol (RS)-4
4-(2-Methoxyethyl)phenol (1) (10 mM, 1 eq.) and K2CO3 (10 mM, 2 eq.) were mixed and refluxed at 85 °C in acetonitrile. After 30 min, (RS)-epichlorohydrin [(RS)-2] (10 mM, 1.5 eq.) was added dropwise into the reaction mixture. Reaction progression was constantly monitored through TLC. The reaction was completed in 24 h. Recovered product (RS)-3 was acetylated using acetyl chloride (6.48 mM, 1.5 eq.) in dichloromethane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (1
water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) mixture. The reaction was completed in 12 h. Recovered product (RS)-4 was analyzed by NMR spectroscopy (Scheme 2). Using the same method, the compound 1 was treated with (R)-2 to afford the epoxide intermediate (S)-3. Upon ring opening, using acetyl chloride, (S)-3 afforded (S)-4.
1) mixture. The reaction was completed in 12 h. Recovered product (RS)-4 was analyzed by NMR spectroscopy (Scheme 2). Using the same method, the compound 1 was treated with (R)-2 to afford the epoxide intermediate (S)-3. Upon ring opening, using acetyl chloride, (S)-3 afforded (S)-4.
(RS)-2-((4-(2-Methoxyethyl)phenoxy)methyl)oxirane (RS-3), 1H NMR (400 MHz; MeOD; Me4Si): δ (ppm): 2.87 (2H, dd), 2.95 (1H, dd), 3.37 (3H, s), 3.6 (2H, t) 4.38 (2H, d), 6.91 (2H, ddd), 7.28 (2H, ddd). 13C NMR (100 MHz, CDCl3): δ (ppm): 35.30, 43.40, 50.10, 58.68, 68.78, 68.86, 114.18, 114.80, 129.58, 130.10, 131.60, 156.99.
(RS)-1-Chloro-3-(4-(2-methoxyethyl)phenoxy)propan-2-ol (RS-4), 1H NMR (400 MHz; MeOD; Me4Si) δ (ppm): 2.87–2.83 (2H, dd), 3.37 (3H, s), 3.68–3.57 (2H, d), 3.79–3.70 (2H, d), 4.08 (2H, d), 4.2 (1H, tt), 6.80 (2H, ddd), 7.11 (2H, dd). 13C NMR (100 MHz, CDCl3): δ (ppm): 35.21, 46.02, 58.65, 69.67, 69.85, 73.81, 114.67, 114.86, 129.62, 130.15, 156.80.
Analysis
All biocatalytic reactions were incubated in an incubator shaker (Kuhner, Switzerland) at 200 rpm. The products from these chemical reactions were analyzed using 1H and 13C NMR with Bruker DPX 400 (1H 400 MHz and 13C 100 MHz) in MeOD and CDCl3, respectively. Chemical shift values were expressed in δ (ppm) units relative to the tetra methyl silane (TMS) as an internal standard. The chiral intermediates were analyzed by HPLC (Shimadzu, Japan) using a Chiralcel OD-H column (4.6 mm × 250 mm, Daicel Chemical, Japan). Mobile phase consisted of hexane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2-propanol at 90
2-propanol at 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 (v/v) with a flow rate of 1.0 mL min−1. At 254 nm, the substrate and product were detected. At the abovementioned condition, the retention times of (R)-4 and (S)-4 were 12.2 and 13.3 min, respectively. All samples were run under the same conditions as stated previously.
10 (v/v) with a flow rate of 1.0 mL min−1. At 254 nm, the substrate and product were detected. At the abovementioned condition, the retention times of (R)-4 and (S)-4 were 12.2 and 13.3 min, respectively. All samples were run under the same conditions as stated previously.
Enantioselectivity was expressed as E value and calculated by eqn (1), substrate enantiomeric excess (ees) was calculated by eqn (2), and substrate conversion (C) by eqn (3).
| E = [ln(1 − C(1 + eep))]/[ln(1 − C(1 − eep))] | (1) |
| ees = (S − R)/(S + R) | (2) |
| C = ees/(ees + eep) | (3) |
General procedure for the enantioselective transesterification of (RS)-4
(RS)-4 (20 mmol, 1 eq.) was subjected to transesterification with vinyl acetate (acyl donor) (100 μL) in toluene (900 μL) using commercial lipase preparations. All lipase preparations were individually added; flasks were tightly capped and placed in the shaker at 30 °C and 200 rpm. After completion, the reaction mixture was extracted with toluene and centrifuged, and the supernatant was evaporated under vacuum. Samples for HPLC analyses were filtered and added to 2-propanol, and the enantiomeric excess and ratio were determined.Optimization of process parameters for kinetic resolution
The influence of various physicochemical parameters on the percentage conversion and enantioselectivity of the lipase catalyzed reaction was investigated. The effect of various organic solvents on the conversion and enantiomeric excess of lipase catalyzed resolution of (RS)-4 was investigated. Kinetic resolution investigation was carried out in 1 mL of the organic solvent at the optimum values of all other reaction parameters. The effect of reaction time on enantioselectivity and percentage conversion was studied by collecting samples at different time intervals till maximum conversion was achieved. The optimization of the acyl donor was performed using various acyl donors (benzyl acetate, ethyl acetate, isopropenyl acetate, and vinyl acetate) for the kinetic resolution of (RS)-4. While studying the effect of acyl donors, all other reaction parameters were kept at their optimum values. The percentage conversion and enantiomeric excess of the product and residual substrate were determined by chiral HPLC. The optimum substrate concentrations for the resolution of (RS)-4 through lipase catalyzed transesterification, using vinyl acetate/hexane and vinyl acetate/toluene, respectively, were determined at different concentrations of substrate (1 to 30 mM). The effect of enzyme concentration on the resolution of (RS)-4 was investigated using different concentrations of lipases (100 to 600 IU mL−1) for the reaction. Conversion and enantiomeric excess of the product and the residual substrate were determined by chiral HPLC. The determination of the optimum temperature for the transesterification of (RS)-4 was carried out at different temperatures from 4 °C to 60 °C. Samples were collected from the reaction mixture at regular time intervals and conversion and enantiomeric excess was determined using chiral HPLC. All other reaction parameters were kept at their optimum values as obtained in earlier experiments.Methodology for molecular docking
Glide module of Schrödinger software incorporated with the maestro graphical user interface (GUI) was utilized for the molecular docking study. PFL structure was prepared using protein preparation wizard incorporating OPLS_2005 force field. The grid for PFL was generated using the key binding residues mentioned in the literature. Moreover, (R)-4 and (S)-4 ligand structures were drawn using ChemDraw tools and saved in MOL format. These MOL files were opened and prepared using LigPrep module of Schrödinger software and the possible ionization states were generated at neutral pH (7.0 ± 0.5). Finally, prepared ligands were docked in the active site of the lipase using the standard precision (SP) protocol from the glide module of Schrodinger software and 20 poses per ligand were generated. Finally, the best docked conformations (poses) were selected on the basis of G score.Conflict of interest
There are no conflicts of interest to declare.Acknowledgements
SS would like to thank the Department of Science and Technology, Govt. of India for providing DST-INSPIRE fellowship. BPD and VKS acknowledge NIPER, Mohali for providing fellowship. Authors acknowledge the Department of Pharmacoinformatics, National Institute of Pharmaceutical Education and Research, S.A.S. Nagar 160062, Punjab, India for providing facilities for in silico study.References
- B. M. Nestl, B. A. Nebel and B. Hauer, Curr. Opin. Chem. Biol., 2011, 15, 187–193 CrossRef CAS PubMed.
- D. Muñoz Solano, P. Hoyos, M. J. Hernáiz, A. R. Alcántara and J. M. Sánchez-Montero, Bioresour. Technol., 2012, 115, 196–207 CrossRef PubMed.
- A. Schmid, J. S. Dordick, B. Hauer, A. Kiener, M. Wubbolts and B. Witholt, Nature, 2001, 409, 258–268 CrossRef CAS PubMed.
- S. Sayin, E. Akoz and M. Yilmaz, Org. Biomol. Chem., 2014, 12, 6634–6642 CAS.
- C. M. Kisukuri and L. H. Andrade, Org. Biomol. Chem., 2015, 13, 10086–10107 CAS.
- B. P. Dwivedee, J. Bhaumik, S. K. Rai, J. K. Laha and C. Banerjee, Bioresour. Technol., 2017, 239, 464–471 CrossRef CAS PubMed.
- B. Joseph, P. W. Ramteke and G. Thomas, Biotechnol. Adv., 2008, 26, 457–470 CrossRef CAS PubMed.
- A. Kamal, M. A. Azhar, T. Krishnaji, M. S. Malik and S. Azeeza, Coord. Chem. Rev., 2008, 252, 569–592 CrossRef CAS.
- T. Ema, Y. Nakano, D. Yoshida, S. Kamata and T. Sakai, Org. Biomol. Chem., 2012, 10, 6299 CAS.
- A. Tafi, A. Van Almsick, F. Corelli, M. Crusco, K. E. Laumen, M. P. Schneider and M. Botta, J. Org. Chem., 2000, 65, 3659–3665 CrossRef CAS PubMed.
- D. B. Kitchen, H. Decornez, J. R. Furr and J. Bajorath, Nat. Rev. Drug Discovery, 2004, 3, 935–949 CrossRef CAS PubMed.
- G. Sin, J. M. Woodley and K. V. Gernaey, Biotechnol. Prog., 2009, 25, 1529–1538 CrossRef CAS PubMed.
- J. Agustian, A. H. Kamaruddin and S. Bhatia, Process Biochem., 2010, 45, 1587–1604 CrossRef CAS.
- A. V. R. Rao, M. K. Gurjar and S. V. Joshi, Tetrahedron: Asymmetry, 1990, 1, 697–698 CrossRef CAS.
- M. K. Gurjar, K. Sadalapure, S. Adhikari, B. V. N. B. S. Sarma, A. Talukdar and M. S. Chorghade, Heterocycles, 1998, 48, 1471–1476 CrossRef CAS.
- S. H. Jung, T. L. Pham, H. K. Lim, H. J. Kim, K. H. Kim and J. S. Kang, Arch. Pharmacal Res., 2000, 23, 226–229 CrossRef CAS.
- M. Muthukrishnan, D. R. Garud, R. R. Joshi and R. A. Joshi, Tetrahedron, 2007, 63, 1872–1876 CrossRef CAS.
- S. Cheng, X. Liu, P. Wang, X. Li, W. He and S. Zhang, Lett. Org. Chem., 2012, 9, 516–519 CrossRef CAS.
- H. U. Shetty and W. L. Nelson, J. Med. Chem., 1988, 31, 55–59 CrossRef CAS PubMed.
- H. Sasai, T. Suzuki, N. Itoh and M. Shibasaki, Appl. Organomet. Chem., 1995, 9, 421–426 CrossRef CAS.
- S. Cheng, X. Liu, P. Wang, X. Li, W. He and S. Zhang, Lett. Org. Chem., 2012, 9, 516–519 CrossRef CAS.
- T. Roy, S. Barik, M. Kumar, R. I. Kureshy, B. Ganguly, N. H. Khan, S. H. R. Abdi and H. C. Bajaj, Catal. Sci. Technol., 2014, 4, 3899–3908 CAS.
- A. Kaler, V. S. Meena, M. Singh, B. Pujala, A. K. Chakraborti and U. C. Banerjee, Tetrahedron Lett., 2011, 52, 5355–5358 CrossRef CAS.
- D. Zelaszczyk and K. Kiec, Curr. Med. Chem., 2007, 14, 53–65 CrossRef CAS PubMed.
- S. P. Panchgalle, R. G. Gore, S. P. Chavan and U. R. Kalkote, Tetrahedron: Asymmetry, 2009, 20, 1767–1770 CrossRef CAS.
- J. Y. Zhang, H. M. Liu, H. W. Xu and L. H. Shan, Tetrahedron: Asymmetry, 2008, 19, 512–517 CrossRef CAS.
- N. S. Thakur, J. Bhaumik, B. Sooram, L. Banoth and U. C. Banerjee, ChemistrySelect, 2016, 1, 871–876 CrossRef CAS.
- B. P. Dwivedee, S. Ghosh, J. Bhaumik, L. Banoth and U. C. Banerjee, RSC Adv., 2015, 5, 15850–15860 RSC.
- L. Banoth, N. S. Thakur, J. Bhaumik and U. C. Banerjee, Chirality, 2015, 27, 382–391 CrossRef CAS PubMed.
- S. Hari Krishna and N. G. Karanth, Catal. Rev., 2002, 44, 161–4940 Search PubMed.
- A. Zaks and A. M. Klibanov, J. Biol. Chem., 1988, 263, 3194–3201 CAS.
- A. M. Klibanov, Nature, 2001, 409, 9–11 CrossRef PubMed.
- M. Paravidino and U. Hanefeld, Green Chem., 2011, 13, 2651–2657 RSC.
- J. E. Puskas, K. S. Seo and M. Y. Sen, Eur. Polym. J., 2011, 47, 524–534 CrossRef CAS.
- A. J. J. Straathof and J. A. Jongejan, Enzyme Microb. Technol., 1997, 21, 559–571 CrossRef CAS.
- J. Agustian, A. H. Kamaruddin and H. Y. Aboul-Enein, Chirality, 2017, 1–10 Search PubMed.
- E. Ł. Chojnacka, M. Staniszewska, M. Bondaryk, J. K. Maurin and M. Bretner, Chirality, 2016, 28, 347–354 CrossRef PubMed.
- M. D. Romero, L. Calvo, C. Alba and A. Daneshfar, J. Biotechnol., 2007, 127, 269–277 CrossRef CAS PubMed.
- L. E. Janes, R. J. Kazlauskas and S. S. West, J. Org. Chem., 1997, 3, 4560–4561 CrossRef.
- V. Ferrario, C. Ebert, L. Knapic, D. Fattor, A. Basso, P. Spizzo and L. Gardossi, Adv. Synth. Catal., 2011, 353, 2466–2480 CrossRef CAS.
- http://www.uniprot.org/uniprot/P26504.
- J. M. T. R. A. Laskowski, M. W. MacArthur and D. S. Moss, J. Appl. Crystallogr., 1993, 283–291 CrossRef.
- C. Colovos and T. O. Yeates, Protein Sci., 1993, 2, 1511–1519 CrossRef CAS PubMed.
- J. U. B. D. Eisenberg and R. Lothy, Methods Enzymol., 1997, 277, 396–404 Search PubMed.
- J. Bowie, R. Luthy and D. Eisenberg, Science, 1991, 253, 164–170 CAS.
- M. Wiederstein and M. J. Sippl, Nucleic Acids Res., 2007, 35, 407–410 CrossRef PubMed.
- K. K. Kim, H. K. Song, D. H. Shin, K. Y. Hwang and S. W. Suh, Structure, 1997, 5, 173–185 CrossRef CAS PubMed.
- C. Angkawidjaja, D. Ju You, H. Matsumura, K. Kuwahara, Y. Koga, K. Takano and S. Kanaya, Fed. Eur. Biochem. Soc., Lett., 2007, 581, 5060–5064 CrossRef CAS PubMed.
- A. Ghanem, Tetrahedron, 2007, 63, 1721–1754 CrossRef CAS.
- D. Guieysse, J. Cortés, S. Puech-Guenot, S. Barbe, V. Lafaquière, P. Monsan, T. Siméon, I. André and M. Remaud-Siméon, ChemBioChem, 2008, 9, 1308–1317 CrossRef CAS PubMed.
- C. Llc, S. S. Francisco and A. Guttman, Chem. Genomics, 2004, 59–60 Search PubMed.
- M. Höhne, S. Schätzle, H. Jochens, K. Robins and U. T. Bornscheuer, Nat. Chem. Biol., 2010, 6, 807–813 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra06499c |
| This journal is © The Royal Society of Chemistry 2017 |