Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Enhanced cellular uptake of iron oxide nanoparticles modified with 1,2-dimyristoyl-sn-glycero-3-phosphocholine
Lichao Su a,
Baolin Zhang
a,
Baolin Zhang *a,
Yinping Huanga,
Ziliang Fanb and
Yingzheng Zhao*b
*a,
Yinping Huanga,
Ziliang Fanb and
Yingzheng Zhao*b
aState Key Laboratory Breeding Base of Nonferrous Metals and Specific Materials Processing, School of Materials Science and Engineering, Guilin University of Technology, Jian Gan Road 12, Guilin, Guangxi 541004, China. E-mail: baolinzhang@ymail.com; zhangbaolin@glut.edu.cn
bCollege of Pharmaceutical Sciences, Wenzhou Medical University, Wenzhou, Zhejiang 325035, China. E-mail: pharmtds@163.com
First published on 2nd August 2017
Abstract
It is important for nanoparticles to enter cells to implement their biological applications. We synthesized and characterized superparamagnetic iron oxide nanoparticles (SPIONs) modified with poly(ethylene glycol) (PEG) and polyethyleneimine (PEI) (PEG/PEI-SPIONs), and further with 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) (DMPC-SPIONs) to improve their intracellular uptake. The nanoparticles internalized into PC-12 cells were observed by transmission electron microscopy (TEM) and quantified by inductively coupled plasma emission spectroscopy (ICP-OES). There was a much greater uptake of DMPC-SPIONs than PEG/PEI-SPIONs into PC-12 cells and remarkable amounts of accumulated nanoparticles were found in the lysosome, endoplasmic reticulum, mitochondria, vesicles, and around the nucleus, and some nanoparticles remained on the cell membrane. This can be attributed to the similarity between the chemical structures of DMPC and membrane phospholipids which result in the fusion of DMPC and cell membranes. Our results encourage further research on DMPC-SPIONs as drug carriers, transfection agents, MRI contrast agents for cells and hyperthermia agents.
1. Introduction
The cell membrane separates the interior of cells from the outside environment. The entrance of nanomaterials into cell membranes is inevitable for applications in drug delivery, magnetic hyperthermia, transfection and MRI imaging of cells.1–4 The interaction between nanoparticles and cell membranes is impacted by the surface modification, charge, size and shape of the nanoparticles.5–7 Phospholipids are important components of mammalian cell membranes.8 As a typical phospholipid, 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) has a long double carbon chain with the zwitterion head groups containing phosphate and choline. DMPCs are endogenous molecules and are completely biodegradable.9,10 The fusion of DMPC and cell membranes is able to enhance the interaction between the DMPC modified nanoparticles and cells, due to the phospholipid enhanced permeability and retention (EPR) effect.11–14 Phospholipids merged with the cell membrane, lipid exchange occurred and the gold nanoparticles modified with phospholipid could integrate within the membrane; phospholipids favor fusogenesis with the membrane of living cells.15–17 Strong interaction between poly(ethylene glycol) (PEG) stabilized positive liposomes and HeLa cells were found.16 Quantum dot-conjugated lipids were attached to cell membranes.17 However, the modification processes of phospholipids on nanoparticles in these works are relatively complex, and direct observation by transmission electron microscopy (TEM) of the distribution of nanoparticles in cellular organelles is absent. In the present work, PEG/PEI-SPIONs were prepared by the polyol method.18,19 PEG increases the colloidal stability of the nanoparticles in the high ionic strength cellular medium, and makes the nanoparticles biocompatible by reducing their cytotoxicity, and PEG is resistant against protein adsorption.15 Positive polyethyleneimine (PEI) is tending to be attracted to the negative membranes of cells and PEI provides conjugation with other functional molecules such DMPC. DMPC modified superparamagnetic iron oxide nanoparticles (DMPC-SPIONs) were very easily synthesized in this work. The uptake and distribution of DMPC-SPIONs in the PC-12 cells were investigated and compared with those of PEG/PEI-SPIONs. We have observed remarkable uptake of DMPC-SPIONs into the cells, this can make DMPC-SPIONs potential drug carriers, transfection agents, MRI contrast agents for cells and hyperthermia agents.2. Experimental
2.1. Synthesis of DMPC-SPIONs
The raw materials used and the synthesis of PEG/PEI-SPIONs were reported in our previous published work.18,19 DMPC-SPIONs were prepared by mixing 20 mg of DMPC with 20 mL of 1 mg mL−1 PEG/PEI-SPIONs dispersed in deionized water. After being shaken together at 4 °C for 5 h, and resting at 4 °C in refrigerator overnight, the mixture was dialyzed against deionized water for 120 h (MWCO 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 dialysis bag, Spectrum Labs, USA).
000 dialysis bag, Spectrum Labs, USA).
2.2. Characterization of the SPIONs
The morphology of the as-synthesized nanoparticles was observed under transmission electron microscopy (TEM, JEM-2100F). The zeta potential and hydrodynamic diameters of the nanoparticles were measured by a Zetasizer Nano ZS90 (Malvern Instruments). Fourier transform infrared spectroscopy (FTIR, Nicolet Nexus 470) and thermogravimetric analyses (TGA, TA Instruments Q500) were used to ascertain the DMPC on the SPIONs. The X-ray photoelectron spectroscopy (XPS) of the dried sample was performed in vacuum on Thermo ESCALAB 250 to further characterize the coating materials. For XPS analysis, particle dispersions were dropped on a silicon wafer and the solvent was air dried. The spectra were measured using a monochromatic Al Kα source (hν = 1486.6 eV) at 45° takeoff angle and pass energies of 20 eV and 100 eV for the high resolution and survey spectra, respectively. Calibration of the binding energy (BE) of the measured spectra was performed at the C 1s peak of the hydrocarbons (BE = 285 eV) in the coating layer as the internal reference. The saturation magnetizations of PEG/PEI-SPIONs and DMPC-SPIONs were determined by the superconducting quantum interference device (SQUID, MPMS XL-7, Quantum Design, USA).2.3. Cytotoxicity assay in vitro
The PC-12 cell line derived from the transplantable pheochromocytoma of the rat adrenal medulla was purchased from the Institute of Biochemistry and Cell Biology (Shanghai, China). In vitro cell viability of PC-12 cells exposed to the SPIONs was assessed by using an MTT cytotoxicity assay. MTT assay was used to evaluate cytotoxicities of nanoparticles against PC-12 cells. The cells were seeded in 96-well plates at 1 × 105 cells per well in 200 μL of complete DMEM and incubated at 37 °C in 5% CO2 atmosphere for 24 h. After removing the culture medium, different concentrations (0–200 μg mL−1) of PEG/PEI-SPIONs or DMPC-SPIONs in complete DMEM were added. Cell viabilities after incubation with DMPC-SPIONs or PEG/PEI-SPIONs for 24 h were evaluated by MTT assay.2.4. PC-12 cells cultured with the nanoparticles and the preparation of TEM samples
PC-12 cells were seeded onto 6-well culture plates at a density of 1 × 105 per well and cultured in complete DMEM for 24 h. The isolated cells were incubated for 12 h with fresh complete DMEM containing PEG/PEI-SPIONs (200 μg mL−1 Fe) or DMPC-SPIONs (200 μg mL−1 Fe). The PC-12 cells collected were fixed with 2.5% glutaraldehyde and 0.1 M sodium cacodylate for 1 h. The samples were then rinsed with sodium cacodylate, fixed again with 1% osmium tetroxide for 1 h. After dehydrated by rinsing with 50%, 70%, 95%, and 100% ethanol, the samples were impregnated in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of acetone and Epon 812 at room temperature for one night. Then the samples were embedded in Epon 812 resin at 60 °C. The embedded samples were cut into ultra-thin section with a PT-XL ultramicrotome (RMC, Boeckeler Instruments, USA) using the glass knife. Sections were then transferred to a copper grid coated with a carbon film. The samples were lightly stained with 0.5% uranyl acetate and lead citrate solution then subjected to the TEM examination (JEM-2100F and H-7650).
1 mixture of acetone and Epon 812 at room temperature for one night. Then the samples were embedded in Epon 812 resin at 60 °C. The embedded samples were cut into ultra-thin section with a PT-XL ultramicrotome (RMC, Boeckeler Instruments, USA) using the glass knife. Sections were then transferred to a copper grid coated with a carbon film. The samples were lightly stained with 0.5% uranyl acetate and lead citrate solution then subjected to the TEM examination (JEM-2100F and H-7650).
2.5. Preparation of the samples for ICP-OES
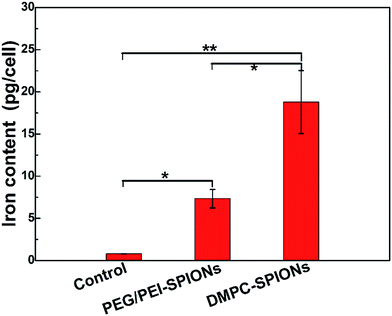
The iron content of PC-12 cells incubated with DMPC-SPIONs or PEG/PEI-SPIONs was assessed by inductively coupled plasma-optical emission spectroscopy (ICP-OES, Optima 8000). The cells growing on 6-well culture plates were washed twice with 5 mL of PBS (0.01 M, pH 7.4) to eliminate the iron oxide in the solution. After completely removing the PBS, the cells were frozen and the dishes were stored at −4 °C. Cells were pulverized by the ultrasonic cell breaker, 1 mL of concentrated nitric acid and 0.2 mL of concentrated hydrochloric acid were added to the sample in a tube and heated at 80 °C for 2 h, then 0.1 mL of Triton X-100 (Sinopharm Chemical Reagent, China) was added to the tube for 1 h. After digestion, the solution was diluted to 10 mL for ICP-OES tests.3. Results and discussion
3.1. Characterization of the nanoparticles
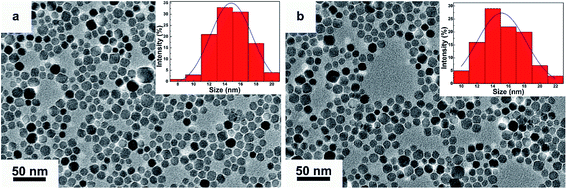
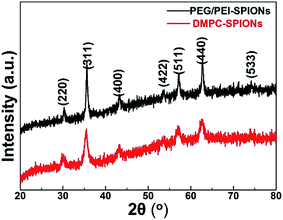
The TEM morphologies and size distributions of as-synthesized DMPC-SPIONs and PEG/PEI-SPIONs are shown in Fig. 1, these nanoparticles are well dispersed with average sizes of 15.3 ± 2.2 nm and 14.8 ± 1.9 nm, respectively. The XRD patterns of DMPC-SPIONs and PEG/PEI-SPIONs are shown in Fig. 2. The peaks at 30.09°, 35.44°, 43.07°, 53.43°, 56.96°, 62.55° and 74.00° match the crystal planes (220), (311), (400), (422), (511), (440) and (533) of magnetite (Fe3O4) crystal (JCPDS: 01-085-1436), respectively. The spinel crystal structure of magnetite is very similar to that of maghemite (γ-Fe2O3), and SPIONs are often a mixture of magnetite and maghemite.18,22 The hydrodynamic sizes of PEG/PEI-SPIONs and DMPC-SPIONs are 24 and 31 nm, and their zeta potential values are 15.4 and 12.5 mV, respectively (Table 1).| PEG/PEI-SPIONs | DMPC-SPIONs | |
|---|---|---|
| Size (nm) | 24.0 | 31.0 |
| Zeta potential (mV) | 15.4 | 12.5 |
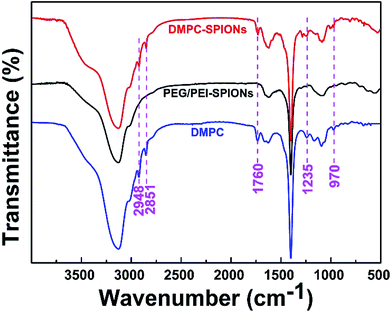
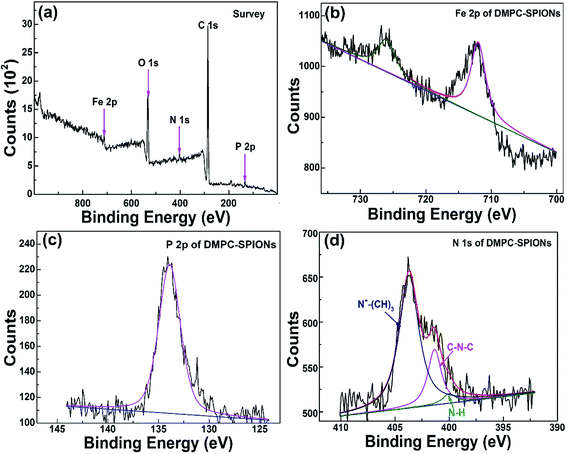
Fig. 3 shows the FTIR spectra of DMPC-SPIONs and PEG/PEI-SPIONs. The characteristic peaks around 970 cm−1 can be assigned to the CN+–(CH3)3 asymmetric vibrations of DMPC,19 which cannot be found in PEG/PEI-SPIONs. The absorption peaks at about 1235 cm−1 represent the vibrational modes of PO2− head groups, carbonyl (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) vibrations are around 1740 cm−1.20,21 To further characterize the surface modification of the DMPC-SPIONs, the X-ray photoelectron spectroscopy (XPS, ESCALAB 250Xi) measurement was carried out. The survey XPS spectrum of the DMPC-SPIONs is shown in Fig. 4a, and the peaks at 711 eV, 530 eV, 400 eV, 286 eV and 134.1 eV correspond to the Fe 2p, O 1s, N 1s, C 1s and P 2p, respectively. Fig. 4b shows the Fe 2p spectrum corresponding to Fe 2p3/2 (711.7 eV) and Fe 2p1/2 (725.6 eV) peaks.22 As shown in Fig. 4c, the binding energy at 134.1 eV belongs to P 2p of DMPC-SPIONs,23,24 the N 1s spectra of DMPC-SPIONs (Fig. 4d) can be curve fitted into three peaks by multiple Gaussians with an equal full width at half-maximum (FWHM) using a Shirley-type background, and the binding energies at about 400, 401.34, and 403 eV are attributable to the N–H, C–N–C, and N+–(CH3)3 groups of DMPC-SPIONs.25,26 Both of the results of FTIR and XPS reveal that DMPC is modified on the surface of the SPIONs. After deduction of the weight losses under 120 °C since these weight losses are due to the vaporization of water adsorbed into the DMPC-SPIONs and PEG/PEI-SPIONs, it can be calculated from the thermogravimetric analyses (Fig. 5a) that about 31.7 wt% of DMPC was coated on the SPIONs. These nanoparticles show excellent superparamagnetic properties (Fig. 5b).
O) vibrations are around 1740 cm−1.20,21 To further characterize the surface modification of the DMPC-SPIONs, the X-ray photoelectron spectroscopy (XPS, ESCALAB 250Xi) measurement was carried out. The survey XPS spectrum of the DMPC-SPIONs is shown in Fig. 4a, and the peaks at 711 eV, 530 eV, 400 eV, 286 eV and 134.1 eV correspond to the Fe 2p, O 1s, N 1s, C 1s and P 2p, respectively. Fig. 4b shows the Fe 2p spectrum corresponding to Fe 2p3/2 (711.7 eV) and Fe 2p1/2 (725.6 eV) peaks.22 As shown in Fig. 4c, the binding energy at 134.1 eV belongs to P 2p of DMPC-SPIONs,23,24 the N 1s spectra of DMPC-SPIONs (Fig. 4d) can be curve fitted into three peaks by multiple Gaussians with an equal full width at half-maximum (FWHM) using a Shirley-type background, and the binding energies at about 400, 401.34, and 403 eV are attributable to the N–H, C–N–C, and N+–(CH3)3 groups of DMPC-SPIONs.25,26 Both of the results of FTIR and XPS reveal that DMPC is modified on the surface of the SPIONs. After deduction of the weight losses under 120 °C since these weight losses are due to the vaporization of water adsorbed into the DMPC-SPIONs and PEG/PEI-SPIONs, it can be calculated from the thermogravimetric analyses (Fig. 5a) that about 31.7 wt% of DMPC was coated on the SPIONs. These nanoparticles show excellent superparamagnetic properties (Fig. 5b).
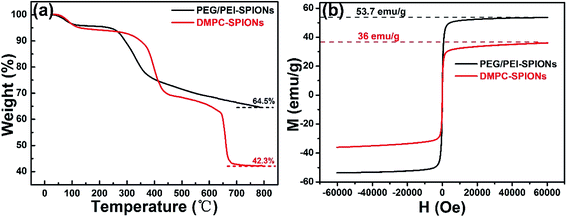 | ||
| Fig. 5 TGA curves from 25 to 800 °C of PEG/PEI-SPIONs and DMPC-SPIONs (a) and their hysteresis loops at 300 K (b). | ||
3.2. In vitro cytotoxicity of PEG/PEI-SPIONs and DMPC-SPIONs
Fig. 6 presents in vitro cytotoxicity of PEG/PEI-SPIONs and DMPC-SPIONs against PC-12 cells by MTT assay. It shows that the cell viabilities remain over 95% at different concentrations within 24 h for PEG/PEI-SPIONs and DMPC-SPIONs. These results indicate the low cytotoxicity of the PEG/PEI-SPIONs and DMPC-SPIONs.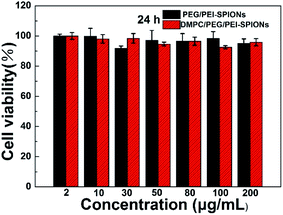 | ||
| Fig. 6 Cell viabilities of PC-12 cells after incubation with PEG/PEI-SPIONs or DMPC-SPIONs at various concentrations for 24 h. | ||
3.3. Subcellular distribution of the nanoparticles in PC-12 cells
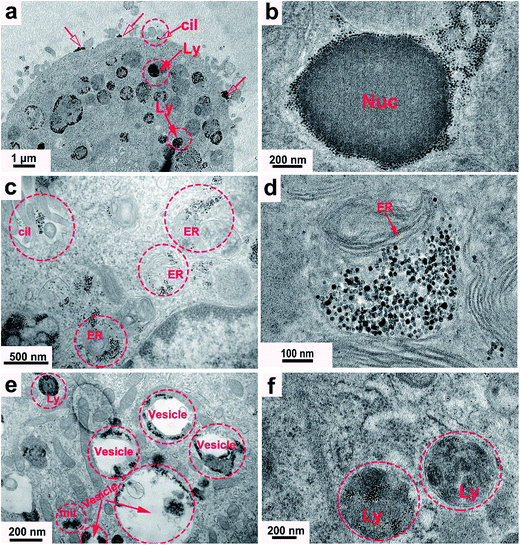
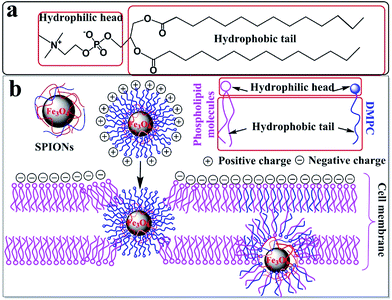
TEM results of the PC-12 cells incubated with the DMPC-SPIONs show that nanoparticles entered the cells, densely packed in organelles (Fig. 7a–e), small fraction of the nanoparticles stayed on the cell membranes around the cilia (red hollow arrows in Fig. 7a and c). In order to determine that the black clusters are iron oxide nanoparticles rather than other impurities or dyes, EDS (Energy Dispersive Spectrometer) via TEM 2100F was employed to identify the black clusters. The electron beam was directed to electronically dense black clusters to collect a representative X-ray spectrum showing the Fe Kα peak (Fig. 8a and b). The peaks of copper Kα, osmium Kα and uranium Kα were derived from the copper grid and chemicals for immobilization and staining of the cells. The result has confirmed the iron oxide composition. The lysosomes were filled with the nanoparticles (Fig. 7a). The internalized nanoparticles trafficking to lysosomes were often reported.27,28 It was also observed that the nanoparticles were accumulated around the nucleus (Nuc) (Fig. 7b) and in the endoplasmic reticulum (Fig. 7c and d). Fig. 7b and d reveal the well dispersed nanoparticles around the nucleus and in the endoplasmic reticulum with the same sizes as those of the synthesized DMPC-SPIONs (Fig. 1a). Iron oxide nanoparticles modified with dimercaptosuccinate near the nucleus in cultured astrocytes27 or iron oxide modified with glial fibrillary acidic protein located in the endoplasmic reticulum in cultured neuroglia of mouse29 were also reported. We also found that nanoparticles localized in the vesicles and in the mitochondrion (Fig. 7e), iron oxide nanoparticles modified with oligodeoxynucleotides could be accumulated by the vesicles in N9 cells as reported.30 By comparison, less iron oxide particles were found in the PC-12 cells incubated with PEG/PEI-SPIONs and few iron oxide nanoparticles were found on the cell membrane (Fig. 7f). It can be seen from the ICP-OES results (Fig. 9) that remarkably more iron content was present in PC-12 cells incubated with DMPC-SPIONs than in PC-12 cells incubated with PEG/PEI-SPIONs. Our work shows the crucial role of DMPC in enhancing the endocytotic uptake of SPIONs. This may be attributed to the similarity between the chemical structures of DMPC and membrane phospholipids which result in the fusion of DMPC and cell membranes (Scheme 1). And it seems that the function of DMPC was not compromised even in the cells as DMPC-SPIONs were tending to fuse with the membranes in cells and the nanoparticles were accumulated by cellular organelles. This work encourages further researches related to DMPC-SPIONs being used as high efficient agents for cancer treatment by hyperthermia in external magnetic field,31 drug delivery,32 transfection,33 and magnetic resonance imaging (MRI) of cells.34 Cell imaging and tracking is important for the applications such as stem cell therapy. MRI tracking of stem cells requires labeling of the cells with enough contrast agents which allow them to be distinguished from the background. DMPC-SPIONs with high magnetic properties and their enhanced cellular uptake can be used as T2 contrast agents35 for MRI cell tracking. The enhanced cellular uptake of DMPC-SPIONs can increase the intracellular delivery of drugs or transfection agents that DMPC-SPIONs have carried, and may also result in the heating of the concentrated SPIONs in cancer cells under alternating or radio frequency magnetic fields so as to implement efficient hyperthermia therapy.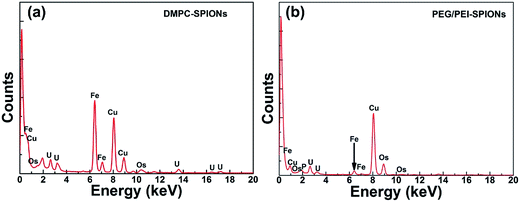 | ||
| Fig. 8 EDS analysis of electron-dense black clusters in cells incubated with DMPC-SPIONs (a) or PEG/PEI-SPIONs (b) showing the presence of Fe. | ||
4. Conclusions
DMPC had been easily modified on the PEG/PEI-SPIONs. DMPC greatly enhanced the endocytotic uptake of DMPC-SPIONs. Densely accumulated nanoparticles were found in the lysosome, endoplasmic reticulum, mitochondrion, vesicle, and around the nucleus, some nanoparticles remained on the cell membrane. DMPC-SPIONs have potential in MRI imaging of cells and developing multifunctional platforms for cancer treatment by hyperthermia, drug delivery and transfection.Conflict of interest
The authors declare no competing financial interests.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (No. 51562007).References
- M. Mahmoudi, J. Meng, X. Xue, X. J. Liang, M. Rahman, C. Pfeiffer, R. Hartmann, P. R. Gil, B. Pelaz and W. J. Parak, Biotechnol. Adv., 2014, 32, 679–692 CrossRef CAS PubMed.
- A. Verma and F. Stellacci, Small, 2010, 6, 12–21 CrossRef CAS PubMed.
- A. Ozdemir, M. S. Ekiz, A. Dilli, M. O. Guler and A. B. Tekinay, RSC Adv., 2016, 6, 45135–45146 RSC.
- M. Galli, A. Guerrini, S. Cauteruccio, P. Thakare, D. Dova, F. Orsini, P. Arosio, C. Carrara, C. Sangregorio, A. Lascialfari, D. Maggioni and E. Licandro, RSC Adv., 2017, 7, 15500–15512 RSC.
- M. Ibáñez, R. Zamani, W. Li, A. Shavel, J. Arbiol, J. R. Morante and A. Cabot, Cryst. Growth Des., 2015, 12, 1085–1090 Search PubMed.
- M. B. Cortie and A. M. Mcdonagh, Chem. Rev., 2011, 111, 3713–3735 CrossRef CAS PubMed.
- A. Lassenberger, A. Scheberl, A. Stadlbauer, A. Stiglbauer, T. Helbich and E. Reimhult, ACS Appl. Mater. Interfaces, 2017, 9, 3343–3353 CAS.
- S. Hong, A. U. Bielinska, A. Mecke, B. Keszler, J. L. Beals, X. Shi, L. Balogh, B. G. Orr and M. M. B. Holl, Interaction of Poly(amidoamine) Dendrimers with Supported Lipid Bilayers and Cells:
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Hole Formation and the Relation to Transport, Cambridge University Press, 2004 Search PubMed.
Hole Formation and the Relation to Transport, Cambridge University Press, 2004 Search PubMed. - M. Feng, Q. Cai, H. Huang and P. Zhou, Eur. J. Pharm. Biopharm., 2008, 68, 688–693 CrossRef CAS PubMed.
- S. K. Shukla, S. K. K. Shukla, P. P. Govender and N. G. Giri, RSC Adv., 2016, 6, 94235–94251 Search PubMed.
- P. Ai, H. Wang, K. Liu, T. Wang, W. Gu, L. Ye and C. Yan, RSC Adv., 2017, 7, 19954–19959 RSC.
- K. Yan, P. Li, H. Zhu, Y. Zhou, J. Ding, J. Shen, Z. Li, Z. Xu and P. Chu, RSC Adv., 2013, 3, 10598–10618 RSC.
- D. Ling, N. Lee and T. Hyeon, Acc. Chem. Res., 2015, 48, 1276–1285 CrossRef CAS PubMed.
- S. L. Jeon, M. K. Chae, E. J. Jang and C. Lee, Chem.–Eur. J., 2013, 19, 4217–4222 CrossRef CAS PubMed.
- H. Ba, J. Rodríguezfernández, F. D. Stefani and J. Feldmann, Nano Lett., 2010, 10, 3006–3012 CrossRef CAS PubMed.
- C. R. Miller, B. Bondurant, S. D. Mclean, K. A. Mcgovern and D. F. O'Brien, Biochemistry, 1998, 37, 12875–12883 CrossRef CAS PubMed.
- M. J. Murcia, D. E. Minner, G. M. Mustata, K. Ritchie and C. A. Naumann, J. Am. Chem. Soc., 2008, 130, 15054–15062 CrossRef CAS PubMed.
- B. L. Zhang, Z. J. Tu, F. Y. Zhao and J. Wang, Appl. Surf. Sci., 2013, 266, 375–379 CrossRef CAS.
- F. Y. Zhao, B. L. Zhang, J. Wang and Z. J. Tu, J. Nanosci. Nanotechnol., 2013, 13, 6793–6797 CrossRef PubMed.
- C. Jiang, A. Alla Gamarnik and C. P. Tripp, J. Phys. Chem. B, 2005, 109, 4539–4544 CrossRef CAS PubMed.
- J. Giri, S. G. Thakurta, J. Bellare, A. K. Nigam and D. Bahadur, J. Magn. Magn. Mater., 2005, 293, 62–68 CrossRef CAS.
- L. Minati, V. Micheli, B. Rossi, C. Migliaresi, L. Dalbosco, G. Bao, S. Hou and G. Speranza, Appl. Surf. Sci., 2011, 257, 10863–10868 CrossRef CAS.
- C. Yam, L. Zheng, M. Salmain, C. Pradier, P. Marcus and G. Jaouen, Colloids Surf., B, 2001, 21, 317–327 CrossRef CAS PubMed.
- P. H. Lo, W. T. Tsai, J. T. Lee and M. P. Hung, Surf. Coat. Technol., 1994, 67, 27–34 CrossRef CAS.
- J. R. Oliveira, M. C. L. Martins, L. Mafra and P. Gomes, Carbohydr. Polym., 2012, 87, 240–249 CrossRef CAS.
- K. R. Gopidas and P. V. Kamat, Langmuir, 1989, 5, 22–26 CrossRef CAS.
- C. Petters, K. Thiel and R. Dringen, Nanotoxicology, 2016, 10, 332–342 CrossRef CAS PubMed.
- E. M. Luther, C. Petters, F. Bulcke, A. Kaltz, K. Thiel, U. Bickmeyer and R. Dringen, Acta Biomater., 2013, 9, 8454–8465 CrossRef CAS PubMed.
- C. H. Liu, J. Yang, J. Q. Ren, C. M. Liu, Z. You and P. K. Liu, FASEB J., 2013, 27, 712–724 CrossRef CAS PubMed.
- E. E. White, A. Pai, Y. Weng, A. K. Suresh, H. D. Van, T. Pailevanian, D. Alizadeh, A. Hajimiri, B. Badie and J. M. Berlin, Nanoscale, 2015, 7, 7780–7789 RSC.
- Z. Nemati, J. Alonso, H. Khurshid, M. H. Phan and H. Srikanth, RSC Adv., 2016, 6, 38697–38702 RSC.
- J. Li, J. Mao, J. Tang, G. Li, F. Fang, Y. Tang and J. Ding, RSC Adv., 2017, 7, 22954–22963 RSC.
- B. Neuhaus, B. Tosun, O. Rotan, A. Frede, A. M. Westendorf and M. Epple, RSC Adv., 2016, 6, 18102–18112 RSC.
- G. Yang, W. Q. Ma, B. L. Zhang and Q. Xie, Mater. Sci. Eng., C, 2016, 62, 384–390 CrossRef CAS PubMed.
- J. Wang, B. L. Zhang, L. Wang, M. Wang and F. B. Gao, Mater. Sci. Eng., C, 2015, 48, 416–423 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2017 |