Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Boron- and nitrogen-doped photoluminescent polymer carbon nanoparticles as nanosensors for imaging detection of Cu2+ and biothiols in living cells†
Buhong Gao a,
Fengyi Zhaob,
Yingchun Miaoa,
Huihua Mina,
Li Xu
a,
Fengyi Zhaob,
Yingchun Miaoa,
Huihua Mina,
Li Xu *ac and
Chaobo Huang*d
*ac and
Chaobo Huang*d
aAdvanced Analysis & Testing Center, Nanjing Forestry University, Nanjing 210037, China. E-mail: gaobuhong@126.com; xuliby@njfu.edu.cn
bCollege of Forestry, Nanjing Forestry University, Nanjing, 210037, China
cCollege of Science, Nanjing Forestry University, Nanjing, 210037, China
dCollege of Chemical Engineering, Nanjing Forestry University, Nanjing 210037, China
First published on 9th October 2017
Abstract
Boron and nitrogen co-doped polymer carbon nanoparticles (BNPCNPs) were synthesized by a facile hydrothermal treatment using uric acid as a nitrogen source, and phenylboronic acid as a boron source for the first time. The nanoparticles with sizes in the range of 90 to 180 nm show excellent and stable fluorescence properties. Moreover, these BNPCNPs showed highly efficient fluorescence quenching ability in the presence of copper (Cu2+) ions due to the formed nonfluorescent metal complexes via robust Cu2+–O or Cu2+–N interactions with the O and N of fluorescent BNPCNPs, which allowed the analysis of Cu2+ ions in the range of 0.0033 to 80 μM. Besides Cu2+ sensing, when biothiols were added, the quenched BNPCNPs-Cu2+ system could be regained via the effective coordination/chelation interactions between Cu2+ ions and the plentiful mercapto and amino groups of biothiols. In the light of this theory, simple biothiol sensors were fabricated without complicated, costly and time-consuming operations. The linear range and the limit of detection of the BNPCNPs-Cu2+ system were 0.0078–80 μM and 2.1 nM for Lcy, 0.0085–85 μM and 2.7 nM for Hcy, and 0.013–89 μM and 4.2 nM for GSH, respectively. Especially, the nanoprobe exhibits good cell membrane permeability and excellent biocompatibility by HeLa cells assay, which is favorable for bioimaging applications. So this BNPCNPs probe can be further used for imaging of biothiols in living cells.
Introduction
Fluorescent carbon nanomaterials (FCNMs) have attracted substantial attention in many areas, due to their unique combination of many distinct merits, such as high photostability, excellent biocompatibility, low cytotoxicity, especial physicochemical properties, good dispersibility and chemical inertness.1–4 However, the pristine FCNMs have a high density of defect states, which lead to their low fluorescence efficiency and selectivity.5,6 Fortunately, recent research has demonstrated that doping with different heteroatoms, such as nitrogen (N), boron (B), sulfur (S), phosphorous (P) and fluorine (F), provides an excellent way to tailor the surface defects, tune their intrinsic properties and explore new and interesting phenomena for sensing and bioimaging applications.7–11 Among in these heteroatoms, B and N, nearby elements of carbon in the periodic table, possess similar atomic radii to carbon, which make it possible to effectively modify the properties of FCNMs after doping. Up to now, many state-of-the-art techniques have been applied to incorporate N and B groups into the carbon framework. For example, the Shi's group has synthesized fluorescent carbon dots by using 3-aminophenylboronic acid as the single starting material through one-pot hydrothermal method.12 Fei et al. reported the synthesis of B and N co-doped graphene quantum dots/graphene hybrid nanoplatelets as efficient electrocatalysts for oxygen reduction by annealing at high temperature.13 Jahan et al. demonstrated the facile hydrothermal oxidative synthetic route to develop highly fluorescent boron/nitrogen co-doped carbon nanodots.14 Liu et al. has been synthesized shell/core structural boron and nitrogen co-doped graphitic carbon/nanodiamond non-noble metal catalyst by a simple one-step heat-treatment.15 Despite several successes in this area, it is still highly desired to develop simple, low-cost, and green methods for the synthesis of B and N co-doped FCNMs with uniform morphology and high sensitivity in next work.In addition to Fe3+ and Zn2+, it is well-known that copper ion (Cu2+) is the third most abundant heavy metal in human body and plays an important role in various physiological processes.16 However, it can be harmful to human health in high concentrations due to damaging to liver, renal tubules, the brain, and other organs.17 Hence, the development of highly efficient methods for the sensitive and selective detection of trace amounts of Cu2+ is in ever-increasing demand. Currently reported methods for the detection of Cu2+ include fluorescence, atomic absorption, inductively coupled plasma mass spectrometry, colorimetry and stripping voltammetry.18–21 Among them, fluorescence chemosensors are widely regarded as an alternative method for Cu2+ detection due to high sensitivity, fast analysis, good selectivity, simplicity and less cell damage. Several fluorescence probes, such as organic dyes, semiconductor nanocrystals, carbon nanomaterials, are widely used to detect a variety ions and biomolecules. While organics dyes involve in disadvantages like poor photo-stability, narrow excitation spectrum, broad absorption bands with red tailing.22 Semiconductor nanocrystals suffer from intrinsic limitations such as complicated synthesis routes, biotoxicity from heavy metals and environmental hazards.23 In light of these drawbacks, the FCNMs stand out as a good candidate due to chemical inertness, no optical blinking, low cytotoxicity, low photobleaching and excellent biocompatibility. Although the FCNMs hold great promise in detection Cu2+, much work is still of great urgency to explore this system and extend its applications.
Biothiols, including Lcy, Hcy and GSH, play an essential role in biological processes such as reversible redox reactions and cellular functions.24 Many diseases are related to the abnormal levels of them, such as slowed growth, edema, lethargy, liver damage and so on.25 Therefore, it is highly desired to detect biothiols level for the diagnosis of related diseases as well as various biochemical investigations. Fluorescence probes are attractive for sensing of biothiols since they are simple, rapid and sensitive. At present, many fluorescence sensor based on FCNMs are fabricated.26–28
Herein, a facile and green synthesis of boron and nitrogen co-doped polymer carbon nanoparticles (BNPCNPs) was reported through one-pot hydrothermal method by using uric acid and phenylboronic acid as precursors, without any additives. Our results show that the resultant BNPCNPs possess a uniform ellipsoidal particles with an average diameter of ca. 120 nm, biocompatibility and cellular labeling capability. Furthermore, the fluorescence BNPCNPs are applied to exhibit an amusing “ON–OFF–ON” three-state emission with the stepwise addition of Cu2+ ions and biothiols (Scheme 1), demonstrating potential applications as a biofunctional sensing platform. Additionally, this probe was successfully explored for the imaging detection of Cu2+ and biothiols in cells.
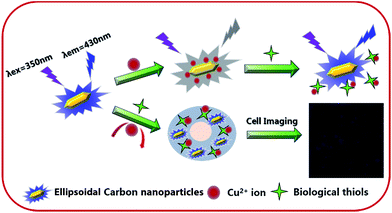 | ||
| Scheme 1 Schematic diagram of the mechanism of the detection of Cu2+ and biothiols by the fluorescent BNPCNPs. | ||
Experimental
Reagents and chemicals
Uric acid (UA) and phenylboronic acid were purchased from Sigma-Aldrich. Copper sulfate pentahydrate, Lcy, Hcy and GSH were purchased from Shanghai Sinopharm Chemical Reagent Co. Ltd. (Shanghai, China). Dulbecco's Modified Eagle Medium (DMEM) and 3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide (MTT) were purchased from Nanjing Keygen Biotech. Co. LTD. All other chemicals and solvents were of analytical grade and were used without further purification. Double distilled water was used throughout.Apparatus
The morphologies of BNPCNPs samples were characterized by transmission electron microscopy (TEM) using a JSM-7600f TEM facility (JEOL, Japan). The TEM samples were prepared by drying a droplet of the BNPCNPS solution on a Cu grid. Atomic force microscopy (AFM) images were collected on a Bruker Dimension Edge system with tapping mode. Dynamic light scattering (DLS) measurements were carried out at 25 °C on Malven Zetasizer NanoZS instrument (Malvern Instruments Ltd., UK). X-ray photoelectron spectroscopy (XPS) characterizations were conducted by using AXIS Ultra DLD instrument (Kratos, UK). UV-vis absorption spectra were recorded on a Lambda 950 (PE, USA). Fourier transform infrared (FT-IR) spectra were obtained using a VERTEX 80V spectrometer (Bruker, Germany) at resolution of 4 cm−1 in the range of 600–4000 cm−1. The fluorescence spectra were recorded on a LS55 (PE, USA). The cellular fluorescence images were recorded by LSM 710 (Zeiss, Germany).Preparation of BNPCNPs
The BNPCNPs were prepared by hydrothermal treatment of UA and phenylboronic acid. In a typical experiment, UA (0.11 g) and phenylboronic acid (0.11 g) was dissolved in deionized water (2.5 mL) and ethanol (2.5 mL) to form a homogeneous suspension solution under sonication. Then, the as-prepared solution (5 mL) was transferred into an autoclave and heated at 180 °C for 8 h, next, cooled naturally to room temperature. The resulting solution was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 30 min to remove the large tracts, and a pale yellow aqueous solution of BNPCNPs was obtained.
000 rpm for 30 min to remove the large tracts, and a pale yellow aqueous solution of BNPCNPs was obtained.
Fluorescence assay of Cu2+ and biothiols
The detection of Cu2+ was conducted as follows. In a typical run, 7 μL of BNPCNPs (2 mg mL−1), 350 μL PBS solution (pH 7.4, 100 mM) and various concentrations of Cu2+ were added with shaking into a 1.5 mL centrifuge tube and the final volume of the mixtures was diluted to 700 μL with double distilled water. The FL emission spectra were recorded after reaction for 2 min at room temperature with the wavelength range of 360 nm to 600 nm at 350 nm excitation.Then, we investigated the effect of Lcy, Hcy and GSH on the FL recovery of the BNPCNPs-Cu2+ system. Herein, different amounts of analytes were added to the BNPCNPs-Cu2+ solution, then the solution was shaken thoroughly for 30 min before the fluorescent spectra were measured with the excitation at 350 nm. The excitation and emission slit widths respectively were 15 nm and 3 nm.
Cell viability studies and cellular imaging
The standard MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide) assays were conducted using HeLa cells. Initially, HeLa cells were seeded onto a 96-well plate at a density of 4000 cells per well and incubated at 37 °C for 24 h in 100 μL of DMEM medium with 10% fetal bovine serum under a 5% CO2 atmosphere. The different concentrations of BNPCNPs (10, 20, 50, 100, 200, 300, 400, 500 μg mL−1) were incubated with HeLa cells for 24 h prior to incubated with the freshly prepared MTT solution (50 mL, 5 mg mL−1) for 4 h. After discarding the culture medium, HeLa cells conjugated BNPCNPs were treated with 150 μL of DMSO and then vortexed for 15 min at the room temperature. The cell viability was estimated as: cell viability (%) = ODtreated/ODcontrol × 100%, where ODcontrol and ODtreated were obtained in the absence and presence of BNPCNPs, respectively. All the records were explained as average data with error bars.Human epithelial carcinoma (HeLa) cells were cultured in DMEM with 10% FBS and 1% penicillin streptomycin with 5% CO2 as media at 37 °C. The cells were pre-cultured with 2 mL of media, and mixed with 20 μL of BNPCNPs at the final concentration of 60 μg mL−1 and incubated for 6 h prior to cell imaging. The incubated cells were washed thrice with PBS, and cell imaging were captured using confocal laser scanning microscope (Carl Zeiss 710 LSM).
Results and discussions
Characterization of BNPCNPs
Water-soluble fluorescent BNPCNPs were facilely prepared by hydrothermal treatment through using UA as nitrogen source and phenylboronic acid as boron source, and ethanol was used as stabilizer, without any other toxic reducing agents, which makes the whole experimental process green and environmental friendly. The morphology and structure of the as-prepared BNPCNPs were characterized by TEM and AFM. The TEM images show that they were non-spherical particles, and unique one kind of particles with a well-defined ellipsoidal shape (Fig. 1A and B). Meanwhile, the Fig. 1C shows the selected-area electron diffraction (SAED) patter of the BNPCNPs, which reveals the crystal structure of the prepared BNPCNPs, namely amorphous. The corresponding particle size distribution histogram obtained by counting about 30 nanoparticles (inset of Fig. 1A), reveals that these BNPCNPs have diameters ranging from 90 to 180 nm with an average diameter of around 120 nm, which is further evidenced by the DLS measurements. The DLS pot (Fig. S1, ESI†) further demonstrates the monodispersity of the BNPCNPs, which possess an average dynamic size of ca. 200 nm, and the results have some differences with the TEM results. The slight overestimation in the DLS measured sizes is due to the fact that DLS measures the hydrodynamic diameter/radius and the BNPCNPs are suspended in water with loose nanostructure; consequently, the range of average size of BNPCNPs is large. Relatively uniform and unique ellipsoidal shape with almost no aggregation can be seen in AFM images (Fig. 1E), which was consistent with TEM. The height of these particles are between 40 and 68 nm (Fig. 1F), as derived from topographical height profile (section analysis), demonstrating that most of the BNPCNPs aggregates maybe have a multilayered structure.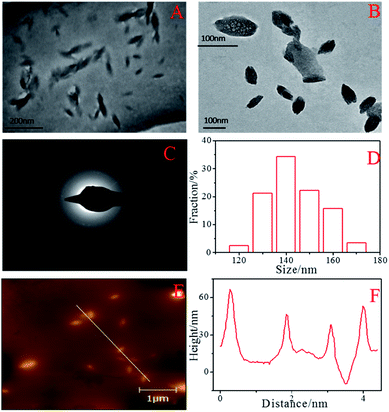 | ||
| Fig. 1 TEM images (A, B) at different magnifications, SAED image (C), the diameter distribution of the BNPCNPs (D), AFM image (E) and height distributions of the BNPCNPs (F). | ||
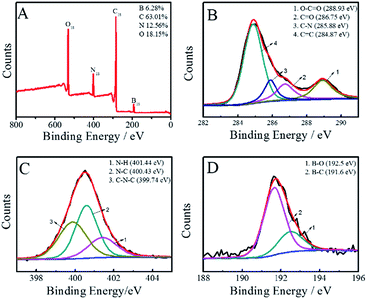
FT-IR spectra (Fig. S2, ESI†) and XPS (Fig. 2) were performed to examine the surface states and functional groups of the BNPCNPs. The absorption bands for O–H, N–H and carbonyl groups vibrations appeared at 3212 and 1645 cm−1, respectively. These groups make BNPCNPs hydrophilic and improve their stability and dispersibility in water.29 The strong absorption for B–O stretching vibration centers at 1451 cm−1 and the B–C absorption band appears at 1189 cm−1. The absorption peak at 1038 cm−1 is assigned to the bonding of B![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O–C. The spectrum shows additional bands due to B–N bonding (urea serves as the nitrogen source).30 For sp2-bonded BN, there are two typical peaks at 1367 and 798 cm−1, which are attributed to the in-plane B–N bond stretching vibration and the out-of-plane B–N–B bending vibration, respectively. BN with sp3 bonding, on the other hand, has an FT-IR active transverse optical mode at 1098 cm−1. For a better insight, XPS characterization were carried out. As shown in Fig. 2A, the full range XPS analysis of the BNPCNP clearly shows four distinct peaks at 191.6, 285.1, 400.5 and 532.0 eV, which are attributed to B1s, C1s, N1s and O1s, respectively. Furthermore, the high-resolution spectrum of C1s shows four peaks at 288.93, 286.75, 285.88 and 284.87 eV, which were attributed to the O–C
O–C. The spectrum shows additional bands due to B–N bonding (urea serves as the nitrogen source).30 For sp2-bonded BN, there are two typical peaks at 1367 and 798 cm−1, which are attributed to the in-plane B–N bond stretching vibration and the out-of-plane B–N–B bending vibration, respectively. BN with sp3 bonding, on the other hand, has an FT-IR active transverse optical mode at 1098 cm−1. For a better insight, XPS characterization were carried out. As shown in Fig. 2A, the full range XPS analysis of the BNPCNP clearly shows four distinct peaks at 191.6, 285.1, 400.5 and 532.0 eV, which are attributed to B1s, C1s, N1s and O1s, respectively. Furthermore, the high-resolution spectrum of C1s shows four peaks at 288.93, 286.75, 285.88 and 284.87 eV, which were attributed to the O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, C
O, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, C–N and C
O, C–N and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C groups, respectively, indicating the as-prepared are rich in hydroxyl, carbonyl, and carboxylic acid groups on the surfaces.31,32 It is noteworthy that the signal at 281.8 eV associated with C–B group is not observed due to the relatively low boron content.33 In the high-resolution N1s spectrum, three fitted peaks at 401.44, 400.43 and 399.74 eV could be assigned to N–H, N–C and C–N–C, respectively, and indicated that the BNPCNPs are rich in C
C groups, respectively, indicating the as-prepared are rich in hydroxyl, carbonyl, and carboxylic acid groups on the surfaces.31,32 It is noteworthy that the signal at 281.8 eV associated with C–B group is not observed due to the relatively low boron content.33 In the high-resolution N1s spectrum, three fitted peaks at 401.44, 400.43 and 399.74 eV could be assigned to N–H, N–C and C–N–C, respectively, and indicated that the BNPCNPs are rich in C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, C–N and –NH2 groups on the surfaces, as observed in the case of the C1s and FT-IR spectra. The signal at the lower binding energy around 398.2 eV related to the N–B bonding is not observed.34 From the B1s spectrum, the two peaks at 192.5 and 191.6 eV were ascribed to B–C and B–O, respectively. Meanwhile, according to XPS spectrum results, the amounts of C, N, O and N elements are 63.01, 12.56, 18.15 and 6.28%, respectively.
N, C–N and –NH2 groups on the surfaces, as observed in the case of the C1s and FT-IR spectra. The signal at the lower binding energy around 398.2 eV related to the N–B bonding is not observed.34 From the B1s spectrum, the two peaks at 192.5 and 191.6 eV were ascribed to B–C and B–O, respectively. Meanwhile, according to XPS spectrum results, the amounts of C, N, O and N elements are 63.01, 12.56, 18.15 and 6.28%, respectively.
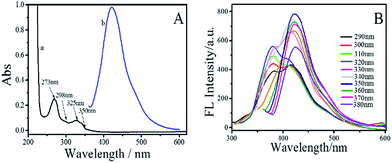
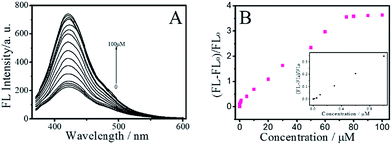
To explore the optical properties of the BNPCNPs, UV-vis absorption and FL spectra on its aqueous solution were investigated. The BNPCNPs in aqueous solution exhibits two characteristic UV-vis absorption peaks at 273 nm and 350 nm. The absorption centered at around 273 nm was attributed to the π–π* transition of aromatic sp2 domains and indicated the existence of aromatic heterocycle, which results in almost no observed FL signal.35 The other absorption peak at about 350 nm with strong emission was resulted from the trapping of excited-state energy by the surface states.36–38 Meanwhile, it is amusing that the UV-vis shows other two absorption peaks at 298 nm and 325 nm (Fig. 3A). The latter peak looks like two prominent bands with unsymmetrical shape and adjoining shoulders, and these absorption bands may be ascribed to that deriving from the N atoms of UA to aromatic polyimides and/or to amide intramolecular charge transfer states.39,40 The emission spectrum of the BNPCNPs demonstrated that the maximum emission peak centered at 430 nm for the excitation at 350 nm, with a Stokes shift of 80 nm (Fig. 3B). The strong FL emission peak at 430 nm may be due to the synergistic effect of the carbogenic nucleus and the surface/molecular state of the BNPCNPs.28,41 Similar to other carbon dots reported previously, the excitation-dependent emission was surveyed with the red-shifted emission peaks from the long excitation (Fig. 3B), which indicate the multicolor properties of the BNPCNPs and characteristics of carbon materials.42
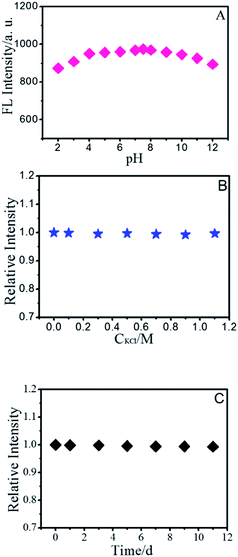
As the optical properties of BNPCNPs are related to particle surface states, thus different pH values may have apparent impact on the FL intensity of BNPCNPs. As shown in Fig. 4A, the FL intensity of BNPCNPs were displayed to slightly decay in both strong acidic and basic environments, which may be attributed to changes in the surface charge induced by protonation–deprotonation, similar to the observed performance of other carboxyl-grafted carbon dots in the literature.43,44 The BNPCNPs were very stable under high ironic strength condition (Fig. 4B), indicating the great potential application in salt solution. Meanwhile, it is noteworthy that these BNPCNPs in suspension were very stable for eight weeks without precipitation (Fig. 4C). To prove good stability of BNPCNPs, ζ-zeta experiment was performed. The results indicate that BNPCNPs show a small negative surface charge (ζ = −3.32 mV), that is, the prepared BNPCNPs may be amphipathic polymer, probably due to the both presence of N–H and carbonyl groups (Fig. S3, ESI†). These results indicated the excellent stability of the BNPCNPs, which is attributed to the electrostatic repulsions between the negatively charged BNPCNPs resulting in electrosteric stabilization. All the above features make the BNPCNPs beneficial for practical applications in bioimaging or other biomedical applications.
The sensing of Cu2+ by BNPCNPs as the fluorescence sensor
To evaluate the specificity for Cu2+ ions, the changes of FL intensity of the BNPCNPs in the presence of other metal ions (Such as Co2+, Cr3+, Ba2+, Fe2+, Pb2+, Hg2+, Mn2+, Ag+, Cd2+, Ca2+, Mg2+, K+, Na+) were measured. The common metal ions at a concentration of 400 μM observed no obvious or small influence on the FL intensity of the BNPCNPs with the exception of Cu2+ (Fig. S4, ESI†), which noticeably quenched the FL of the BNPCNPs. These results indicated that the BNPCNPs probe presented a highly selective sensor of Cu2+, which can be attributed to that Cu2+ is a paramagnetic ion with an unfilled d shell, and as a result, quenched the FL of BNPCNPs via electron or energy transfer.45 To elucidate the FL quenching mechanism further between the BNPCNPs and Cu2+, the FL decays were applied. Owing to differences in the distribution of complex luminescent pathways resulting from multiple BNPCNPs species and/or sites,41 the FL intensity of BNPCNPs obeys dual-exponential decay kinetics, and two lifetimes were obtained (Fig. 5). The analysis of the resulting data without Cu2+ as a dual-exponential decay gained an average lifetime of 4.77 ns (Fig. 5B), and the average lifetime time reduced to 2.81 ns with the addition of Cu2+ to BNPCNPs solution (Fig. 5B). The reduced lifetime and the nonlinear Stern–Volmer plot (Fig. 6B) demonstrate that the FL quenching of BNPCNPs by Cu2+ does not obey a simple static or dynamic quenching mechanism, further inferred that the FL change of BNPCNPs is caused by both dynamic and static quenching processes in this sensor system, rather than through other possible routes.46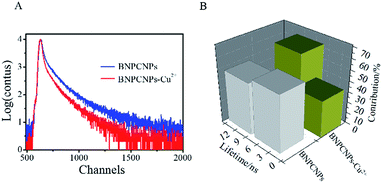 | ||
| Fig. 5 (A) Decay of the FL of BNPCNPs in the absence and presence of Cu2+ (80 μM). (B) FL lifetimes of GLNPCNs in the absence and presence of Cu2+ (80 μM). | ||
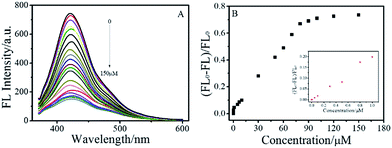
In order to maximize the detection sensitivity of Cu2+, the experimental conditions were optimized. The fluorescence response to Cu2+ of the BNPCNPs was highest under the pH 7 (Fig. S5, ESI†). As increasing the pH value, partial copper ions was chelated with hydroxide and it generated the copper hydroxide precipitate, thus further increasing the pH value caused a low quenching efficiency. Considering the potential application in biological systems, the pH 7.4 was selected as the optimum value for the detection of Cu2+ in this work. As described in Fig. S5 (ESI†), the reaction time also impacts the quenching effects. Overall, the interactions can reach the balance as Cu2+ was added into the BNPCNPs solution 2 min later. As a consequence, 2 min of reaction time was appropriate for the sensing. Under the optimized conditions discussed above, the linear response range of the sensing system was investigated. As shown in Fig. 6A, the fluorescence intensities of the BNPCNPs at 430 nm decrease as the concentration of the Cu2+ is increased. There was a good linear relationship (R2 = 0.998) between fluorescence intensities and concentrations of Cu2+ in the range from 0.003 to 80 μM with a limit detection of 1.5 nM (signal-to-noise ratio of 3), which is lower than that of the previously reported methods based on these FL nanomaterials (Table S1, ESI†), and is also lower than the maximum allowable level (MAL) of Cu2+ in drinking water set by the U.S. Environmental Protection Agency (EPA) and the World Health Organization (WHO) (∼20 and 30 μmol L−1, respectively).
The BNPCNPs-based FL probe were also successfully applied to detect Cu2+ in water samples obtained from Xuanwu lake of Nanjing. The collected samples were filtered through a 0.22 μm membrane and then centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 20 min. The standard addition method was used to analyze real samples, and recovery experiments were carried out using lake water samples as a matrix to validate the method. The results are listed in Table S2 (ESI†). It was found that the recoveries of the Cu2+ from spiked samples were in range of 95.35–100.75% with the relative standard deviation (RSD) of 4.95–5.78%. These results indicated that the developed BNPCNPs was applicable for the detection of Cu2+ in real water samples.
000 rpm for 20 min. The standard addition method was used to analyze real samples, and recovery experiments were carried out using lake water samples as a matrix to validate the method. The results are listed in Table S2 (ESI†). It was found that the recoveries of the Cu2+ from spiked samples were in range of 95.35–100.75% with the relative standard deviation (RSD) of 4.95–5.78%. These results indicated that the developed BNPCNPs was applicable for the detection of Cu2+ in real water samples.
The sensing of biothiols by BNPCNPs-Cu2+ as the fluorescence sensor
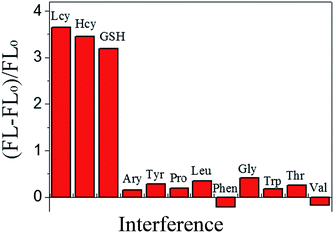
The fluorescence emission intensity of the BNPCNPs was weakened upon conjugation to the Cu2+ via a fluorescence “turn off” process usually occasioned by the chelation of Cu2+ with N and O of BNPCNPs brought them into close proximity with each other, leading to substantial FL quenching. Conversely, the FL intensity of BNPCNPs-Cu2+ was gradually restored upon the addition of different concentrations of Lcy, Hcy or GSH, respectively (Fig. S6, ESI†). It is conceivable that the biothiols as strong metal chelators were able to remove Cu2+ from Cu2+-functionalized BNPCNPs through the formation of Cu–S bonds, and FL recovery occurs from the induced. To evaluate the sensing system's specificity for biothiols detection, the fluorescence analyses of the BNPCNPs-Cu2+ were carried out in the present of other amino acids including arginine (Arg), tyrosine (Tyr), proline (Pro), leucine (Leu), phenylalanine (Phen), glycine (Gly), tryptophane (Trp), threonine (Thr) and valine (Val). As shown in Fig. 7, the quenched fluorescence intensity was restored upon the addition of Lcy, Hcy, and GSH into the system of BNPCNPs-Cu2+, whereas no obvious signal change was observed for other substances. It is noteworthy to consider that the excellent selectivity results obtained herein stems from the rapid and specific reactions propensity of the biothiols as strong metal chelators and Cu2+, which expediently removed Cu2+ from Cu2+-functionalized BNPCNPs through the formation of Cu–S bonds.In order to evaluate the fluorescence “turn on” sensitivity of BNPCNPs-Cu2+ based probe, the recovered fluorescence intensities in the absence (FL0) and presence (FL) of Lcy, Hcy or GSH were evaluated using eqn (1).
| ΔFL/FL0 = 1 + Kc | (1) |
Human serum (5%) respectively spiked with Lcy, Hcy and GSH at the concentration of 5.0 μM and 10 μM were detected the BNPCNPs-Cu2+ system (Table S3, ESI†). The recoveries of the BNPCNPs-Cu2+ system for the detection of Lcy, Hcy and GSH in spiked serum samples ranged between 97.9% and 107.4% with the RSD in range of 4.98–7.13%. These results indicated the potential applicability of the BNPCNPs-Cu2+ system for the test of Lcy, Hcy and GSH in serum samples.
Cell imaging of HeLa cells using the BNPCNPs as probe
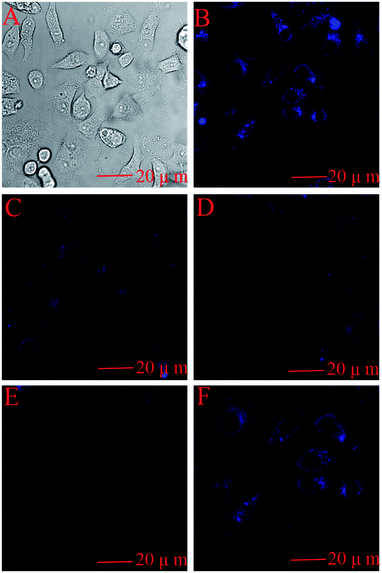
In order to demonstrate that the pure BNPCNPs can be taken up efficiently by HeLa cells, since the cellular uptake of BNPCNPs by HeLa cells is influenced by BNPCNPs size, surface properties, and dispersed medium. Cells were incubated with the BNPCNPs (60 μg mL−1) for 6 h. After incubation, the cells were washed with PBS solution and their optical properties were investigated by confocal fluorescence microscopy. As shown in Fig. 9B, the obvious blue emission was observed from HeLa cells when excited at 350 nm owing to the strong fluorescence from the BNPCNPs. This result indicated the BNPCNPs could penetrate the cells membrane and translocate into cells by endocytosis. Fig. 9C–E shows the intracellular FL intensity in these HeLa cells was getting weaker with the time increase from 5 min to 30 min, treated by adding 50 μM Cu2+. It can be elucidated that Cu2+ diffused into the cell and connected with the BNPCNPs. Moreover, when the cells loaded with BNPCNPs-Cu2+ were further treated with Lcy, Hcy or GSH, strong blue fluorescence was restored (Fig. 9F). All these results clearly demonstrated that the synthesized BNPCNPs can be potentially applied to high selective visual detection of Cu2+ and biothiols in living cells.Conclusions
In summary, we have reported a simple route to synthesize B and N co-doped photoluminescent polymer carbon nanoparticles with a uniform size of ca. 120 nm on a large scale through hydrothermal treatment of uric acid and phenylboronic acid. Optical property characterization and cytotoxicity experiments demonstrate that these BNPCNPs display excellent fluorescent property and low biotoxicity, which have been successfully applied in imaging living cell. Furthermore, these BNPCNPs can be utilized as fluorescent nano-sensor for highly sensitive and selective detection of Cu2+ and biothiols (Lcy, Hcy and GSH) in serum samples. Thus, we expect that this new type of heteroatom co-doped polymer carbon nanoparticles can be used widely both in bioimaging and bioanalytical applications.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by Jiangsu key lab of biomass-based energy and materials (JSBEM2016011), National Natural Science Foundation of China (No. 21644004) and Natural Science Key Project of the Jiangsu Higher Education Institutions (16KJA220006).Notes and references
- K. A. Shiral Fernando, S. Sahu, Y. M. Liu, W. K. Lewis, E. A. Guliants, A. Jafariyan, P. Wang, C. E. Bunker and Y. P. Sun, ACS Appl. Mater. Interfaces, 2015, 7, 8363–8376 Search PubMed.
- L. Cao, X. Wang, M. J. Meziani, F. S. Lu, H. F. Wang, P. G. Luo, Y. Lin, B. A. Harruff, L. M. Veca, D. Murray, S. Y. Xie and Y. P. Sun, J. Am. Chem. Soc., 2007, 129, 11318–11319 CrossRef CAS PubMed.
- Y. L. Tai and G. Lubineau, Sci. Rep., 2016, 6, 19632 CrossRef CAS PubMed.
- M. Ganiga and J. Cyriac, Sens. Actuators, B, 2016, 225, 522–528 CrossRef CAS.
- H. Liu, T. Ye and C. Mao, Angew. Chem., Int. Ed., 2007, 119, 6593–6595 CrossRef.
- S. L. Hu, K. Y. Niu, J. Sun, J. Yang, N. Q. Zhao and X. W. Du, J. Mater. Chem., 2009, 19, 484–488 RSC.
- P. F. Shen and Y. R. Xia, Anal. Chem., 2014, 86, 5323–5329 CrossRef CAS PubMed.
- Z. X. Wang and S. N. Ding, Anal. Chem., 2014, 86, 7436–7445 CrossRef CAS PubMed.
- H. F. Wu, J. H. Jiang, X. T. Gu and C. L. Tong, Microchim. Acta, 2017, 184, 2291–2298 CrossRef CAS.
- R. J. Liu, J. J. Zhao, Z. R. Huang, L. L. Zhang, M. B. Zou, B. F. Shi and S. L. Zhao, Sens. Actuators, B, 2017, 240, 604–612 CrossRef CAS.
- S. Kundu, R. M. Yadav, T. N. Narayanan, M. V. Shelke, R. Vajtai, P. M. Ajayan and V. K. Pillai, Nanoscale, 2015, 7, 11515–11519 RSC.
- Z. B. Qu, X. G. Zhou, L. Gu, R. M. Lan, D. D. Sun, D. J. Yu and G. Y. Shi, Chem. Commun., 2013, 49, 9830–9832 RSC.
- H. L. Fei, R. Q. Ye, G. L. Ye, Y. J. Gong, A. W. Peng, X. J. Fan, E. L. G. Samuel, P. M. Ajayan and J. M. Tour, ACS Nano, 2014, 8, 10837–10843 CrossRef CAS PubMed.
- S. Jahan, F. Mansoor, S. Naz, J. P. Lei and S. Kanwal, Anal. Chem., 2013, 85, 10232–10239 CrossRef CAS PubMed.
- X. X. Liu, Y. H. Wang, L. Dong, X. Chen, G. X. Xin, Y. Zhang and J. B. Zang, Electrochim. Acta, 2016, 194, 161–167 CrossRef CAS.
- W. Y. Liu, H. Y. Li, H. S. Lv, B. X. Zhao and J. Y. Miao, Spectrochim. Acta, Part A, 2012, 95, 658–663 CrossRef CAS PubMed.
- R. R. Ward, D. T. Dexter and R. J. Ward, Coord. Chem. Rev., 2008, 252, 1189–1199 CrossRef.
- C. C. Zhang, Y. J. Gong, Y. Yuan, A. L. Luo, W. J. Zhang, J. Zhang, X. Y. Zhang and W. H. Tan, Anal. Methods, 2014, 6, 609–614 RSC.
- Z. Ajtony, N. Laczai, G. Dravecz, N. Szoboszlai, A. Marosi, B. Marlok, C. Streli and L. Bencs, Food Chem., 2016, 213, 799–805 CrossRef CAS PubMed.
- M. Faraji, Y. Yamini and S. Shariati, J. Hazard. Mater., 2009, 166, 1383–1388 CrossRef CAS PubMed.
- I. Ustundag, A. Erkal, T. Koralay, Y. K. Kadioglu and S. Jeon, J. Anal. Chem., 2016, 71, 685–695 CrossRef CAS.
- I. Costas-Mora, V. Romero, I. Lavilla and C. Bendicho, Anal. Chem., 2014, 86, 4536–4543 CrossRef CAS PubMed.
- X. Wang, L. Cao, S.-T. Yang, F. Lu, M. J. Meziani, L. Tian, K. W. Sun, M. A. Bloodgood and Y. P. Sun, Angew. Chem., Int. Ed., 2010, 49, 5310–5314 CrossRef CAS PubMed.
- Y. L. Tai, Y. X. Wang, Z. G. Yang and Z. Q. Chai, Surf. Interface Anal., 2011, 43, 1480–1485 CrossRef CAS.
- Z. Huang, F. Pu, Y. Lin, J. Ren and X. Qu, Chem. Commun., 2011, 47, 3487–3489 RSC.
- L. Zhou, Y. H. Lin, Z. Z. Huang, J. S. Ren and X. G. Qu, Chem. Commun., 2012, 48, 1147–1149 RSC.
- S. M. Xu, Y. Liu, H. Yang, K. Zhao, J. G. Li and A. P. Deng, Anal. Chim. Acta, 2017, 964, 150–160 CrossRef CAS PubMed.
- M. H. Lan, J. F. Zhang, Y. S. Chui, H. Wang, Q. D. Yang, X. Y. Zhu, H. X. Wei, W. M. Liu, J. C. Ge, P. F. Wang, X. F. Chen, C. S. Lee and W. J. Zhang, J. Mater. Chem. B., 2015, 3, 127–134 RSC.
- S. Qu, X. Wang, Q. Lu, X. Liu and L. Wang, Angew. Chem., Int. Ed., 2012, 51, 12215–12218 CrossRef CAS PubMed.
- C. Rohr, J. Boo and W. Ho, Thin Solid Films, 1998, 322, 9–13 CrossRef CAS.
- Y. Li, Y. Hu, Y. Zhao, G. Shi, L. Deng, Y. Hou and L. Qu, Adv. Mater., 2011, 23, 776–780 CrossRef CAS PubMed.
- Y. Li, Y. Zhao, H. Cheng, Y. Hu, G. Shi, L. Dai and L. Qu, J. Am. Chem. Soc., 2012, 134, 15–18 CrossRef CAS PubMed.
- Z. H. Sheng, H. L. Gao, W. J. Bao, F. B. Wang and X. H. Xia, Mater. Chem., 2012, 22, 390–395 RSC.
- Y. Zhao, L. J. Yang, S. Chen, X. Z. Wang, Y. W. Ma, Q. Wu, Y. F. Jiang, W. J. Qian and Z. Hu, J. Am. Chem. Soc., 2013, 135, 1201–1204 CrossRef CAS PubMed.
- G. Eda, Y. Lin, C. Mattevi, H. Yamaguchi, H. Chen, I. Chen, C. Chen and M. Chhowalla, Adv. Mater., 2010, 22, 505–508 CrossRef CAS PubMed.
- X. Wang, L. Cao, S. T. Yang, F. Lu, M. J. Meziani, L. Tian, K. W. Sun, M. A. Bloodgood and Y. P. Sun, Angew. Chem., Int. Ed., 2010, 49, 5310–5314 CrossRef CAS PubMed.
- P. Anilkumar, X. Wang, L. Cao, S. Sahu, J. H. Liu, P. Wang, K. Korch, K. Tackett, A. Parenzan and Y. P. Sun, Nanoscale, 2011, 3, 2023–2027 RSC.
- Y. P. Sun, X. Wang, F. Lu, L. Cao, M. J. Meziani, P. G. Luo, L. Gu and L. M. Veca, J. Phys. Chem. C, 2008, 112, 18295–18298 CAS.
- J. Wakita, S. Inoue, N. Kawanishi and S. Ando, Macromolecules, 2010, 43, 3594–3605 CrossRef CAS.
- S. A. Sydlik, Z. H. Chen and T. M. Swager, Macromolecules, 2011, 44, 976–980 CrossRef CAS.
- R. Liu, D. Wu, X. Feng and K. Mellen, J. Am. Chem. Soc., 2011, 133, 15221–15223 CrossRef CAS PubMed.
- Y. Xu, M. Wu, X. Z. Feng, X. B. Yin, X. W. He and Y. K. Zhang, Chem.–Eur. J., 2013, 19, 6282–6288 CrossRef CAS PubMed.
- B. F. Shi, L. L. Zhang, C. Q. Lan, J. J. Zhao, Y. B. Su and S. L. Zhao, Talanta, 2015, 142, 131–139 CrossRef CAS PubMed.
- Z. X. Wang, F. Y. Kong and W. Wang, Chem.–Eur. J., 2017, 23, 665–675 CrossRef CAS PubMed.
- Y. Rahimi, A. Goulding, S. Shrestha, S. Mirpuri and S. K. Deo, Biochem. Biophys. Res. Commun., 2008, 370, 57–61 CrossRef CAS PubMed.
- S. Liu, J. Tian, L. Wang, Y. Zhang, X. Qin, Y. Luo, A. M. Asiri, A. O. AlYoubi and X. Sun, Adv. Mater., 2012, 24, 2037–2041 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra07683e |
| This journal is © The Royal Society of Chemistry 2017 |