Open Access Article
Open Access ArticleInfluence of culture conditions on extracellular polysaccharide production and the activities of enzymes involved in the polysaccharide synthesis of Nostoc flagelliforme
Pei-pei Han†
 *,
Shun-yu Yao†,
Rong-jun Guo,
Rong-rong Yan,
Yi-kai Wu,
Shi-gang Shen and
Shi-ru Jia*
*,
Shun-yu Yao†,
Rong-jun Guo,
Rong-rong Yan,
Yi-kai Wu,
Shi-gang Shen and
Shi-ru Jia*
Key Laboratory of Industrial Fermentation Microbiology, Ministry of Education, College of Biotechnology, Tianjin University of Science and Technology, Tianjin 300457, P. R. China. E-mail: pphan@tust.edu.cn; jiashiru@tust.edu.cn; Fax: +86 22 60602298; Tel: +86 22 60601598
First published on 21st September 2017
Abstract
Culture conditions significantly influence extracellular polysaccharide (EPS) production of Nostoc flagelliforme, however, the key enzyme controlling EPS synthesis has not been fully explored yet. The influence of different culture conditions including light quality, carbon source and nitrogen source on EPS production of N. flagelliforme and activities of EPS synthesis enzymes was investigated. Three experimental groups produced higher amounts of EPS than the control group, including the carbon source group with 1.26 g L−1 NaHCO3, the nitrogen source group with 0 g L−1 NaNO3 and the light quality group with blue light. Activities of seven related enzymes phosphoglucose isomerase (PGI), fructose-1,6-bisphosphatase (FBPase), UDP-glucose pyrophosphorylase (UGPase), UDP-galactose-4-epimerase (UGE), UDP-glucose dehydrogenase (UGDH), phosphomannose isomerase (PMI), and phosphofructokinase (PFK) were significantly influenced by culture conditions. Partial least-squares analysis and correlation analysis methods were used to analyze the relationship between the activities of these enzymes and EPS production, and a correlation between the production of EPS and the activities of PGI, PMI, FBPase, UGDH, and UGPase was found under different culture conditions. Subsequent analysis of the transcription level of genes encoding the five enzymes showed genes pgi and fbp1 in three experimental groups were significantly up regulated. The results revealed PGI and FBPase might be important enzymes positively influencing the biosynthesis of N. flagelliforme EPS. The findings would be helpful to further understand the pathway of EPS biosynthesis aimed to improve the EPS production of N. flagelliforme.
Introduction
The cyanobacteria extracellular polysaccharide can be divided into two main forms: the capsule polysaccharide (CPS) fraction bound to the cell surface and the extracellular polysaccharide (EPS) fraction released into the surrounding environment.1,2 The EPS shows advantages over the polysaccharides from plants or microalgae, given its characteristic easy recovery from the culture medium and its physicochemical properties.3,4 The future looks bright for its potential applications in the food, pharmaceutical and environmental industries, consequently, the interest toward EPS has significantly increased in recent years.5–7Nostoc flagelliforme is a species of edible terrestrial filamentous cyanobacteria, distributed throughout arid and semi-arid areas.8,9 The properties of N. flagelliforme EPS, including antivirus, antioxidant and anti-tumor, have been intensively investigated.10–12 Considerable research efforts have been devoted to find the optimum culture medium and culture condition to obtain high EPS production yields.13–17 Light quality, carbon and nitrogen sources have been reported as important factors controlling N. flagelliforme EPS synthesis.15,17–21 As a group of photoautotrophic microorganisms, several studies reported that light quality especially red-light and blue-light could promote the accumulation of N. flagelliforme EPS.17,20–25 Most of cyanobacteria have efficient carbon concentration mechanisms (CCM) which could transport and accumulate inorganic carbon actively (Ci; HCO3−, and CO2).26–28 and could utilize very different forms of nitrogen, including combined nitrogen source or atmospheric nitrogen,2,29–31 which was helpful for increasing EPS production.
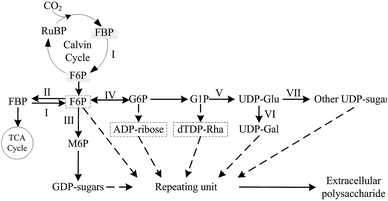
In previous studies, various complex EPS biosynthetic mechanisms were investigated and described in many bacterial, fungal and microalgae species, but the necessary pathways and enzymes involved in typical EPS biosynthesis, such as UDP-glucose pyrophosphorylase and UDP-glucose dehydrogenase, were relatively conserved, which played important roles in EPS production.32–41 Based on previous reports,42–44 and KEGG pathway, a simplified biosynthesis pathway for biosynthesis of cyanobacterial EPS was summarized (Fig. 1). In the complex pathway, the key enzyme that control EPS production in N. flagelliforme remains still unclear, and little is known about the correlation between related enzymes activities in N. flagelliforme EPS biosynthesis and the production of EPS under these different culture conditions.
In the study, the effects of culture conditions including different light quality, NaHCO3 concentration, nitrogen source type and nitrogen concentration on EPS production and activities of EPS biosynthesis enzymes were investigated, and the correlation between them was subsequently analyzed for key enzyme affecting EPS production. The selected conditions (including different concentrations of NaHCO3 and NaNO3, nitrogen sources and light qualities) have been found to significantly influence cell growth and EPS production in our previous study. Furthermore, expression level of genes encoding key EPS synthesis enzymes were determined for improved understanding EPS synthesis process in N. flagelliforme.
Materials and methods
Strain and culture conditions
The N. flagelliforme cells (TCCC11757) utilized in liquid suspension cultures were obtained from the Tianjin Key Lab of Industrial Microbiology (Tianjin, China). The cells were inoculated into a 500 mL shake-flask containing 200 mL BG-11 medium, and cultured at 25 °C under continuous white fluorescent light at a photon flux density of 60 mol photon per (m2 per s) for 16 days, which was set as the basic group. Differing values of three culture conditions (carbon source, nitrogen source, and light quality) were evaluated in submerged fermentation of N. flagelliforme. For carbon source condition, initial concentrations of NaHCO3 were set at 0 g L−1, 1.26 g L−1 (equivalent to 15 mmol L−1) and 2.94 g L−1 (equivalent to 35 mmol L−1), respectively, with fixed NaNO3 concentration at 1.50 g L−1. For nitrogen source condition, one group set initial concentrations of NaNO3 at 0 g L−1, 1.50 g L−1 and 4.50 g L−1 respectively, with no exogenous addition of NaHCO3. The other group set the initial concentrations of NH4Cl, CO(NH2)2 and NaNO3 at 0.95 g L−1, 0.53 g L−1 and 1.50 g L−1 respectively, with the same concentration of N element. For light quality condition, the same photon flux density of monochromatic red (660 nm) and blue (460 nm) were used instead of the white fluorescent light as the treatment group. The half-band widths are 5 nm for each monochromatic light, according to manufacturer's instruction (Shenzhen federal heavy Secco electronic Co. LTD., China).The measurement of cell growth and EPS production
The cell growth was measured by weighing the cell dry weight. The total EPS (T-EPS) production was determined via phenol-sulfuric acid method.45 The unit quality EPS (U-EPS) production was calculated as follows: U-EPS (mg g−1) = T-EPS per cell dry weight.Enzyme extract
The enzyme extract method is modified by the method of Looijesteijn et al.33 and Tamoi et al.46 The N. flagelliforme cells were harvested by centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 10 min at 4 °C, washed twice with cell extract buffer pre-cooled at 4 °C, which was 20 mM phosphate buffer containing 50 mM NaCl, 10 mM MgCl2, and 1 mM dithiothreitol at pH 6.5. The cells pellet resuspended in the extract buffer were disrupted ultrasonically at 20 kHz for 12 min on ice using an ultrasonic processor (SCIENTZ-IID sonicator, Ningbo Scientz Biotechnology Inc., China). Cell debris was removed by centrifuged at 12
000 × g for 10 min at 4 °C, washed twice with cell extract buffer pre-cooled at 4 °C, which was 20 mM phosphate buffer containing 50 mM NaCl, 10 mM MgCl2, and 1 mM dithiothreitol at pH 6.5. The cells pellet resuspended in the extract buffer were disrupted ultrasonically at 20 kHz for 12 min on ice using an ultrasonic processor (SCIENTZ-IID sonicator, Ningbo Scientz Biotechnology Inc., China). Cell debris was removed by centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 15 min at 4 °C, and the resulting supernatant (cell extract) was used for the enzyme activity assay. The protein content of the cell extract was determined by the method of Bradford.47
000 × g for 15 min at 4 °C, and the resulting supernatant (cell extract) was used for the enzyme activity assay. The protein content of the cell extract was determined by the method of Bradford.47
Enzyme assays
Activity of EPS synthesis enzymes were measured spectrophotometrically by kinetic assay in a final volume of 250 μL at 24 °C. Reactions were initiated by adding 30 μL cell extract to various reaction mixtures depending on the enzyme. The enzyme activity was determined by monitoring the formation or disappearance of NAD(P)H by measuring the absorbance at 340 nm (ε340 = 6220 M−1 cm−1), and specific activities were expressed in μmol NAD(P)H (mg protein)−1 min−1. The blank for each enzyme was the same reaction condition with the same volume of cell extract that had been heated at 100 °C for 5 min. In all of the assays, the reaction velocity was linearly proportional to the amount of cell extract. The reaction rate (Δabsorbance/min) calculated from the slope of linear plots (R2 = 0.95–1.0) from 0–3 min or 0–5 min that was normalized to the control. The enzyme activities were calculated using the following equations:48| Enzyme activity (U, μmol min−1) = (ΔA340/Δt) × V1 × D/(V2 × ε340 × d) |
| Specific activities = enzyme activity/protein content |
Phosphoglucose isomerase (PGI)37 reaction mixture contained 50 mM potassium phosphate buffer (pH 6.8), 5 mM MgCl2, 4 U glucose-6-phosphate dehydrogenase, 0.4 mM NADP+, 10 mM fructose-6-phosphate.
Fructose-1,6-bisphosphatase (FBPase)37 reaction mixture contained 100 mM Tris–HCl buffer (pH 8.0), 0.4 mM NADP+, 20 mM MgCl2, 20 mM DTT, 0.5 U phosphoglucose isomerase, 0.5 U glucose-6-phosphate dehydrogenase, 2 mM fructose-1,6-bisphosphate.
UDP-glucose pyrophosphorylase (UGPase)49 reaction mixture contained 50 mM Tris–HCl buffer (pH 7.8), 0.4 mM UDP-glucose, 14 mM MgCl2, 4 U glucose-6-phosphate.
UDP-galactose-4-epimerase (UGE)37 reaction mixture contained 50 mM Tris–HCl buffer (pH 8.5), 5 mM MgCl2, 0.015 U UDP-glucose dehydrogenase, 0.5 mM NAD, 0.2 mM UDP-galactose.
UDP-glucose dehydrogenase (UGDH)50 reaction mixture contained 100 mM Tris–HCl buffer (pH 7.5), 5 mM UDP-glucose, 1 mM dithiothreitol, 1 mM MgCl2, 1 mM NADP+.
Phosphomannose isomerase (PMI)51 reaction mixture contained 50 mM MOPS buffer (pH 7.0), 1 mM CoCl2, 1 mM NADP+, 10 mM mannose-6-phosphate, 4 U phosphoglucose isomerase, 4 U glucose-6-phosphate dehydrogenase.
Phosphofructokinase (PFK)33,37 reaction mixture contained 50 mM Tris–HCl buffer (pH 7.5), 5 mM MgCl2, 50 mM KCl, 1.25 mM ATP, 0.15 mM NADH, 4.5 U aldolase, 18 U triose-phosphate isomerase, 6.2 U glycerol-3-phosphate dehydrogenase, 5 mM fructose-6-phosphate.
Total RNA extraction
Total RNA of N. flagelliforme cells was prepared using MiniBEST Universal RNA Extraction Kit (TaKaRa, Japan) according to the manufacturer's instruction. For cDNA synthesis of total RNA was transcribed with the PrimeScript™ RT reagent Kit (TaKaRa, Japan) according to the manufacturer's instruction. Total RNA was quantified through the absorbance spectrum at wavelengths 260 nm and 280 nm using UV-mini1246 spectrophotometer (SHIMADZU, Japan). Total RNA solution was loaded on 2% agarose gel to assess the integrity of total RNA bands. The DNA maker was DL2000 maker (TaKaRa, Japan).Primer designing and real-time quantitative PCR analysis
Expression levels of pgi, pmi, fbp1, fbp2, ugp and ugdh (encoding enzymes PGI, PMI, FBPase, UGPase and UGDH, respectively) were analyzed by qRT-PCR using SYBR® Premix Ex Taq™(Tli RNaseH Plus) Kit (TaKaRa, Japan) according to the manufacturer's instructions with Bio-Rad IQ5 (Bio-Rad, American). Primers used for qRT-PCR were listed in Table 1. GAPDH was used as the reference gene to normalize the expression levels of different genes. For each gene, the reference sample (the control group) was defined as 1.0, and the 2−ΔΔCt method was applied to calculate other gene expression level which was expressed as the fold of the reference sample.52,53 Negative controls (reactions without cDNA template) were included to detect the presence of any DNA contamination. All the reactions of these samples were done in triplicate to determine the accuracy and repeatability of the qRT-PCR reaction.| Enzyme | Target gene | Primer sequences (5′ → 3′) |
|---|---|---|
| GAPDH | GAPDH | Forward: GGTTTGCGGCTCCTTCGTAG |
| Reverse: TCATTCCTTGCGTGTCGGTG | ||
| PMI | pmi | Forward: GTAGTCTCAGGGACAGCTAGGGTAAC |
| Reverse: AACACCAAGGGAATCACGCC | ||
| PGI | pgi | Forward: CCACGAGGGATTGGGACTGT |
| Reverse: GCTTGCTCTGGGAGGATGAATAG | ||
| FBPase | Fbp1 | Forward:GCTTAGTGTGTCGCCTCGCTTC |
| Reverse: TCGGTTGCCTTACCATCGCT | ||
| FBPase | Fbp2 | Forward: CGGGAGCGGATGAATAAAAT |
| Reverse: GCGGGAGGTGCTGCTAACTT | ||
| UGPase | ugp | Forward: TCTATGAAAAGCCTACGGTTGAG |
| Reverse: TCCCTCTTCCTGACACAATCTATC | ||
| UGDH | ugdh | Forward: ATTGGTTGGGGTGGTTCTTG |
| Reverse: GTCCGACTGTTTTGCCTTTGAG |
Data analysis and statistical analysis
Experimental data in this study was obtained from three replicates for each treatment, and values were shown as mean ± standard deviation. Supervised partial least-squares (PLS) analysis was used to verify the relative enzymes which were significant response to the EPS production for different culture conditions. Supervised PLS was performed by SIMCA 11.5 software (Umetrics, Umea, Sweden). Statistical significance of correlations between enzyme activities and EPS production was based on correlation test, which were analyzed based on data obtained under various culture conditions by SPSS statistics vision 2.0. Correlation coefficient (R2 > 0.5) and significant effects (P < 0.05) were considered statistically significant.Results
Effects of culture conditions on EPS production
In order to optimize the culture condition with maximum production of N. flagelliforme EPS, the effects of carbon source, nitrogen source, and light quality on EPS production were studied. As shown in Fig. 2A, the productions of U-EPS were similar and high under both blue-light and red-light condition, which were 48.01 ± 3.70 and 45.84 ± 0.68 mg g−1 DCW, respectively. The U-EPS productions under blue-light and red-light condition were 1.52 and 1.45 times compared to that of white-light. Because of the highest biomass, the T-EPS obtained the highest production (37.32 ± 1.75 mg L−1) under blue-light, which was 1.78 times to that of white-light.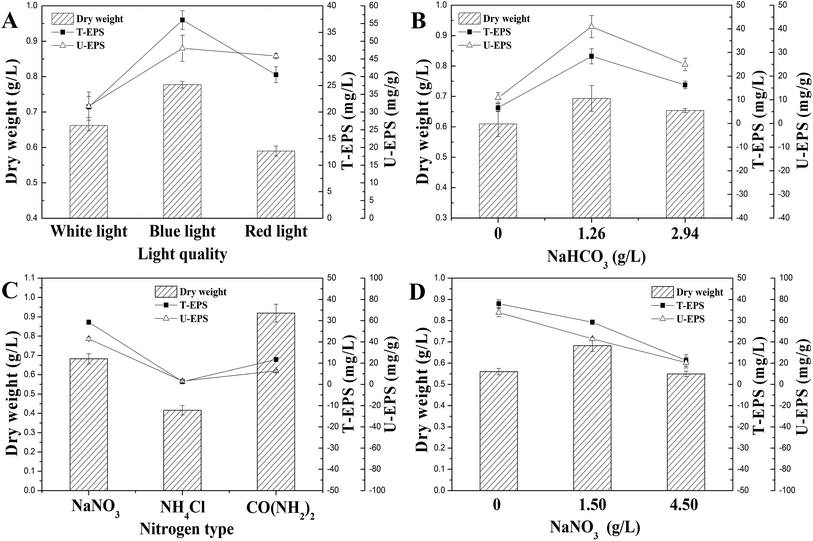 | ||
| Fig. 2 Effects of culture conditions on EPS production. (A) Light quality; (B) NaHCO3 concentration; (C) nitrogen source type; (D) NaNO3 concentration. | ||
The carbon sources used for submerged fermentation of N. flagelliforme were NaHCO3 with different initial concentrations, and the profiles of EPS production were showed in Fig. 2B. The highest T-EPS (28.38 ± 3.19 mg L−1) and U-EPS (40.93 ± 4.70 mg g−1 DCW) production were both observed in the medium containing 1.26 g L−1 NaHCO3, which were 4.27 and 3.75 times compared to that of the medium containing 0 g L−1 NaHCO3 respectively.
The effects of nitrogen source type and initial concentration of NaNO3 on EPS production by N. flagelliforme were shown in Fig. 2C and D, respectively. Using EPS production as maker, we tested three nitrogen source types (Fig. 2C), including NaNO3, NH4Cl and CO(NH2)2. It showed that NaNO3 was the best nitrogen source for accumulating EPS (29.27 ± 0.37 mg L−1 T-EPS and 42.87 ± 1.52 mg g−1 DCW U-EPS) among these three nitrogen source types. The low level of EPS production was caused by adding NH4Cl and CO(NH2)2 as nitrogen source, especially in the medium containing NH4Cl as solo nitrogen source with few EPS production (1.32 ± 0.27 mg L−1 T-EPS and 3.17 ± 0.81 mg g−1 DCW U-EPS) detected, although NH4+ was considered to be the preferred reduced N form for cyanobacteria uptake in general.30,31 As shown in Fig. 2D, the changes of initial NaNO3 concentration had different effects on EPS production. The highest EPS production (37.95 ± 1.87 mg L−1 T-EPS and 67.73 ± 3.98 mg g−1 DCW U-EPS) was observed in the media without exogenous addition of NaNO3, and EPS production was reduced when initial NaNO3 concentration was increased.
Effects of culture conditions on enzyme activities involved in N. flagelliforme EPS synthesis
Activities of seven enzymes were evaluated under different culture conditions (Fig. 3), which were involved in N. flagelliforme EPS synthesis. Fig. 3A showed the profiles of the activity levels of seven enzymes measured under light quality condition. The activities of PGI, PMI, UGDH, UGE and UGPase were significantly improved under blue-light and red-light condition. The order of activity of FBPase was red-light > white-light > blue-light condition. As can be seen from Fig. 3B, it was interesting that for the five enzymes, the lowest activity was obtained at 1.26 g L−1 NaHCO3 and then their activities were increased at 2.94 g L−1 NaHCO3, which was an opposite trend with EPS production. However, FBPase activity was increased with the increase of NaHCO3 concentration.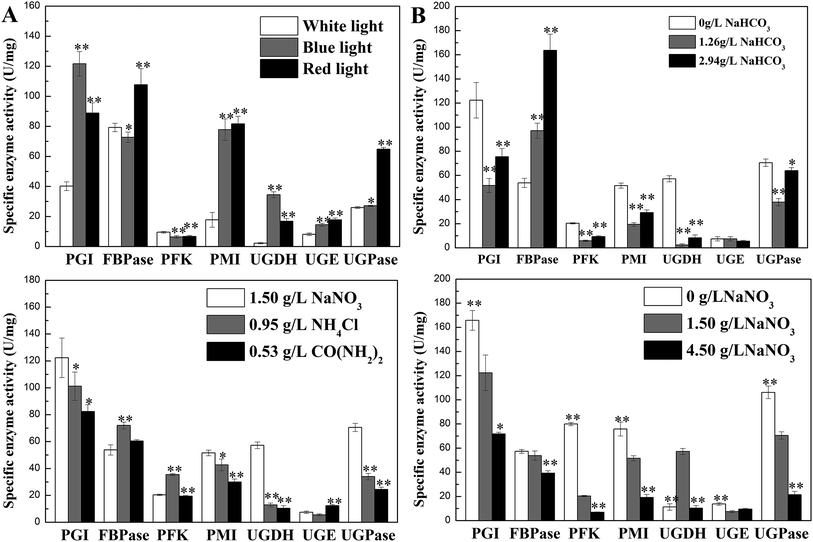 | ||
| Fig. 3 Effects of culture conditions on enzyme activities involved in N. flagelliforme EPS synthesis. (A) Light quality; 59 (B) NaHCO3 concentration; 59 (C) nitrogen source type; (D) NaNO3 concentration.59 * indicates p < 0.05 and ** indicates p < 0.01. | ||
For different nitrogen source types, the activities of PGI, PMI, UGDH, UGPase in Fig. 3C had similar trend with EPS production, which was declined after adding NH4Cl and CO(NH2)2 as nitrogen source, except FBPase and PFK. For different initial NaNO3 concentrations (Fig. 3D), increased concentration of NaNO3 leaded to the reduction of enzyme activity, which showed similar trend with EPS production, except UGDH. Therefore, the activity of PGI, PMI, and UGPase had the same trend with the changes of EPS production regardless of nitrogen source type and the initial NaNO3 concentration used.
Relationship between activities of enzymes and EPS production under differing culture conditions
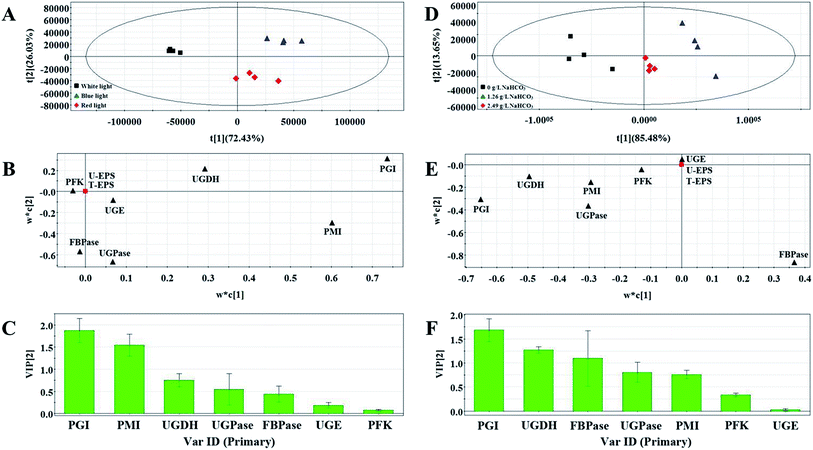
To study relationship between activities of enzymes and EPS production under differing culture conditions and avoid the impact of collinearity, multivariate statistical analysis was performed by using supervised partial least-squares (PLS) analysis and correlation analysis. The PLS was used to validate the differences between the different groups. The t1/t2 scatter plot used to explain the applicability of the PLS Method. The score plot displayed the space formed by the two predictive components as first predictive component (t[1]) and second predictive component (t[2]), respectively. These models were all well-constructed and possessed excellent fit and satisfactory predictive capability (Table 2). The first two components accounted for >95% of the total variance for each culture conditions. In the PLS score plots, the confidence interval is defined by Hotelling's T2 ellipse (95% confidence interval), and observations outside the confidence ellipse are considered outliers.| Culture condition | PLS | |||||
|---|---|---|---|---|---|---|
| R2X | R2Y | Q2 | t [1] | t [2] | t[1] + t[2] | |
| Light quality | 0.985 | 0.971 | 0.958 | 72.43% | 26.03% | 98.46% |
| NaHCO3 concentration | 0.991 | 0.947 | 0.913 | 85.48% | 13.65% | 99.13% |
| Nitrogen source type | 0.972 | 0.993 | 0.990 | 86.48% | 10.74% | 97.22% |
| NaNO3 concentration | 0.993 | 0.998 | 0.997 | 85.48% | 13.78% | 99.26% |
Variable Importance Plot (VIP) is one of the important plots used to explain or determine the fitting effect of PLS model and VIP coefficients reflect the contribution of each variables to the PLS models. In this study, VIP values ≥ 0.5 were considered as an index indicating that these were possible key enzymes which had contribution to EPS production.
Three groups clearly separated in each profiles of PLS score plots (Fig. 4A and D and Fig. 5A and D). These results showed that the activities of enzymes involved in EPS biosynthesis of N. flagelliforme differed significantly. PLS loading plots and VIP were used to analyze the contribution of each enzyme to the EPS production. The results from Fig. 4B and C showed that PGI, PMI, UGDH and UGPase were possible key enzymes affecting EPS production under light quality condition. As shown in Fig. 4E and F, PGI, UGDH, FBPase, UGPase and PMI might be the potential major drivers of EPS accumulation under carbon conditions. The order of enzyme contribution was UGDH, UGPase, PGI, FBPase, PFK and PMI under various nitrogen source conditions (Fig. 5B and C), while PGI, UGPase, PFK and PMI might be the possible key enzymes in the medium containing different concentration of NaNO3 (Fig. 5E and F).
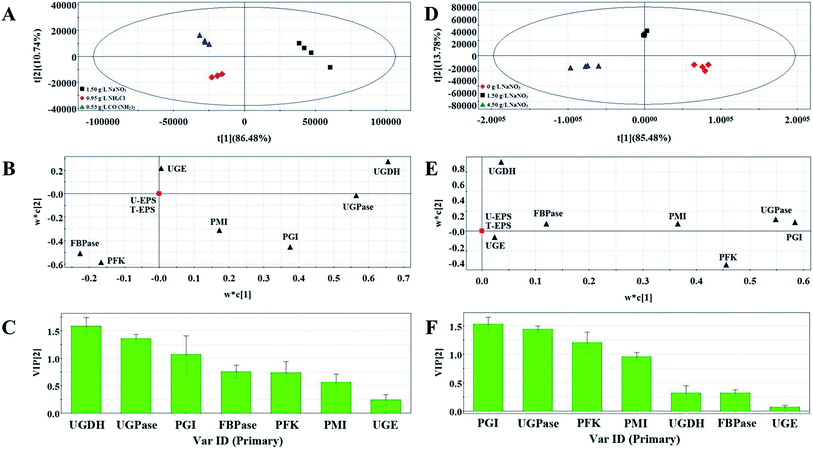 | ||
| Fig. 5 PLS score plot (A), loading plot (B) and VIP (C) results of samples with the addition of different nitrogen source types. PLS score plot (D), loading plot (E) and VIP (F) results of samples. | ||
The correlation test was performed by correlation coefficient (R2) > 0.5 and the results of linear correlation analysis showed in Table 3. The result of correlation analysis between T-EPS and enzyme activities was in accord with that between U-EPS and enzyme activities under different culture conditions, except light quality condition. PGI and UGDH showed positive correlation with both T-EPS and U-EPS under light quality condition. PGI, UGDH, UGPase, PMI and PFK were negatively associated with EPS (T-EPS and U-EPS) under carbon source condition. The effect of UGDH and UGPase was positive on EPS, while that of FBPase was negative under various nitrogen source conditions. The positive correlations were observed between five enzymes (PGI, FBPase, UGPase, PFK and PMI) and EPS production when different concentrations of NaNO3 were added.
| Culture condition | EPS type | Related enzyme | ||||||
|---|---|---|---|---|---|---|---|---|
| PGI | FBPase | PFK | PMI | UGDH | UGE | UGPase | ||
| a ++: significant positive, p < 0.01; +: positive, p < 0.05; −: negative, p < 0.05; −−: significant negative, p < 0.01; +/−: uncorrelated with the addition of different NaNO3 concentrations. | ||||||||
| Light quality | T-EPS | ++ | +/− | +/− | +/− | + + | +/− | +/− |
| U-EPS | ++ | +/− | −− | ++ | ++ | ++ | +/− | |
| NaHCO3 concentration | T-EPS | −− | +/− | −− | −− | −− | +/− | −− |
| U-EPS | −− | +/− | −− | −− | −− | +/− | −− | |
| Nitrogen source type | T-EPS | +/− | −− | +/− | +/− | ++ | +/− | ++ |
| U-EPS | +/− | −− | +/− | +/− | ++ | +/− | ++ | |
| NaNO3 concentration | T-EPS | ++ | ++ | ++ | ++ | +/− | +/− | ++ |
| U-EPS | ++ | ++ | ++ | ++ | +/− | +/− | ++ | |
Combing the results of PLS analysis and correlation analysis, by choosing the enzymes showing significant correlation with EPS production under all tested conditions, PGI, PMI, UGDH, UGPase and FBPase were preliminarily selected as the potential important enzymes influencing EPS production.
Transcription levels of genes encoding key EPS synthesis enzymes
To evaluate the effects of culture conditions on expression of key enzymes, qRT-PCR was used to analyze the transcription levels of genes including pgi (encoding PGI), pmi (encoding PMI), fbp1 and fbp2 (encoding FBPase), ugdh (encoding UGDH) and ugpg (encoding UGPase) in control group and three experimental groups with high-yield production of EPS consisting of carbon source group with 1.26 g L−1 NaHCO3, nitrogen source group with 0 g L−1 NaNO3, and light quality group with blue-light.The relative quantitative results of the above genes under four culture conditions were shown in Fig. 6. The mRNA levels were normalized against GAPDH level of control group. In three experimental groups, pgi and fbp1 genes were highly expressed compared with that of control group, while the transcription levels of other four genes were decreased in three experimental groups.
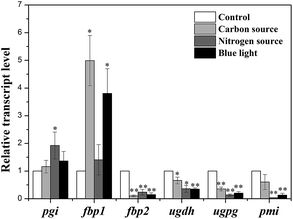 | ||
| Fig. 6 Transcription levels of key enzymes in N. flagelliforme under four culture conditions. * indicates p < 0.05 and ** indicates p < 0.01. | ||
Discussion
In recent two decades, various efforts have been devoted to increase N. flagelliforme EPS production, which mostly focus on optimization of culture condition.14–18,20,21 So far, little information is available about the effects of culture conditions on activities of enzymes involved in EPS production. Therefore, in this study, the possible relationship between N. flagelliforme EPS production and enzymes activities related to EPS biosynthesis was evaluated.In this study, the results showed that the EPS production of N. flagelliforme was significantly influenced by culture conditions, which were consistent with previous reports.16–18,20,21 In the PLS analysis, PGI was the only enzyme with VIP value > 1 under all four culture conditions. The results of correlation analysis showed that PGI was significantly correlated with EPS production, except under the nitrogen source condition. PGI catalyzes the reversible isomerization of glucopyranose-6-phosphate and fructofuranose-6-phosphate playing a key role in glycolysis and gluconeogenesis pathway.37,54,55 The highest EPS production was also associated with the highest activity of PGI under four culture conditions, except under the condition of adding different concentration of NaHCO3. The gene pgi encoding PGI was highly expressed in three experimental groups verified the results of correlation analysis, referring that pgi might be one important gene influencing EPS production under different culture conditions.
FBPase can catalyze the conversion of fructose-1,6-diphosphate into fructose-6-phosphate, which occurs in two essential steps: one involved in gluconeogenesis and the other in Calvin cycle.33,46 The transcription analysis of genes encoding FBPase revealed that two genes (fbp1 and fbp2) encoding two forms of FBPase (FBPase-I and FBPase-II) were detected in N. flagelliforme, which were mainly involved in Calvin cycle and involved in gluconeogenesis respectively.46,62–64 The observed significant up-regulation of fbp1 and down-regulation of fbp2 under experimental conditions indicated the adjustment of intracellular carbon metabolism to promote EPS synthesis and corresponded well with the enhanced EPS production, which suggested that gene fbp1 rather than fbp2 was the potential gene influencing EPS synthesis in N. flagelliforme, coinciding with the previous reports.46,62–64 Interestingly, the activity of FBPase was significantly enhanced under red-light, and was obviously increased with increasing initial NaHCO3 concentration and decreasing NaNO3 concentration, but the FBPase activity was more closely associated with EPS production under nitrogen condition, which might indicate different regulation mechanisms were involved in different culture conditions.
UGPase was involved in the production of UDP-glucose, which was a key precursor to polysaccharide synthesis.34,37,48 UGDH was required for the synthesis of the precursor UDP-glucuronic acid,56 which was an important intermediate in polysaccharide synthesis pathway.57,58 In this study, both partial least-squares analysis and correlation analysis showed the activities of UGPase and UGDH had strong correlation with EPS production under most of culture conditions. However, the activities were not all increased in experimental groups with higher EPS production. In addition to that, the transcription levels of corresponding genes ugdh and ugpg were decreased. Therefore it might be deducted that UGPase and UGDH were not directly associated with EPS yield. In our previous study, it was found that culture conditions showed significant influences on monosaccharide composition of EPS, and UGPase and UGDH were correlated with monosaccharide ratio under different culture conditions.59 Thus it might be speculated that changes of UGPase and UGDH activities induced by culture conditions had greater influence on EPS monosaccharide composition than yield.
In many cyanobacteria, PMI possess mannose-6-phosphate isomerase activity and nucleotidyltransferase activity which can catalyze the reversible isomerization of fructose-6-phosphate from central metabolism into mannose-6-phosphate and forming GDP-mannose, respectively.60,61 The correlation between the activity of PMI and the amount of EPS produced was observed, indicating that EPS production might be affected by activity of PMI. However, in contrast to the increased activity, down-regulation of gene pmi expression level was detected. The reason for discrepancy between encoding gene transcription and activity levels remains unclear and needs to be further investigation.
In addition, UGE, the Leloir pathway key enzyme catalyzing the interconversion of UDP-galactose and UDP-glucose, was not significantly changed, except light quality condition, which seemed to be not closely associated with EPS synthesis. PFK was the first committed step of glycolysis catalyzing the phosphorylation of fructose-6-phosphate to fructose 1,6-bisphosphate by ATP, which seemed not to play an important role in controlling N. flagelliforme EPS production.
In this study, PGI and FBPase were found to be more important enzymes positively influencing the EPS biosynthesis than the other five enzymes, however, through comparing the relationship between enzyme activities and EPS production under four kinds of culture conditions, the results showed some discrepancies that the activities of enzymes had positive effects on EPS production under some culture conditions but had negative effects under other culture conditions. It might be due to the complex EPS biosynthesis process and regulation mechanism. Different influence patterns were found under different culture conditions indicating the utilization of distinct regulation mechanism for different culture conditions, which might explain the discrepancies, and further indicate the existence of coordinated regulation of several enzymes in the EPS production, which still requires further investigation.
Conclusions
The effects of culture conditions on N. flagelliforme EPS production and related enzymes were investigated in the study. Culture conditions showed significant influences on EPS production, and related enzyme activities. Both PLS analysis and correlation analysis were used to analyze the relationship between activities of seven enzymes and EPS production. The results showed that the activities of PGI, PMI, FBPase, UGDH and UGPase were correlated with EPS production under different culture conditions. Subsequent analysis of the transcription level of genes encoding the five enzymes showed pgi and fbp1 in three experimental groups was significantly up regulated, which implied PGI and FBPase might be the important enzymes positively influencing EPS production of N. flagelliforme. The findings would improve the understanding of EPS synthesis process in N. flagelliforme.Conflicts of interest
There are no conflicts of interest to declare.Abbreviations
| dTDP-Rha | dTDP-rhamnose |
| F6P | Fructose-6-phosphate |
| FBP | Fructose-1,6-bisphosphatase |
| FBPase | Fructose-1,6-biphosphate |
| G1P | Glucose-1-phosphate |
| G6P | Glucose-6-phosphate |
| M6P | Mannose-6-phosphate |
| PFK | Phosphofructokinase |
| PGI | Phosphoglucose isomerase |
| PMI | Phosphomannose isomerase |
| T-EPS | Total EPS |
| UDP-Gal | UDP-galactose |
| UDP-Glu | UDP-glucose |
| U-EPS | Unit quality EPS |
| UGDH | UDP-glucose dehydrogenase |
| UGE | UDP-galactose-4-epimerase |
| UGPase | UDP-glucose pyrophosphorylase |
Acknowledgements
The authors are very grateful for the financial support from the National Natural Science Foundation of China (Grant No. 31671842).References
- B. H. A. Rehm, Nat. Rev. Microbiol., 2010, 8(8), 578–592 CrossRef CAS PubMed.
- S. Pereira, A. Zille, E. Micheletti, P. Moradas-Ferreira, R. De Philippis and P. Tamagnini, FEMS Microbiol. Rev., 2009, 33(5), 917–941 CrossRef CAS PubMed.
- I. W. Sutherland, Trends Biotechnol., 1998, 16(1), 41–46 CrossRef CAS PubMed.
- R. De Philippis, C. Sili, R. Paperi and M. Vincenzini, J. Appl. Phycol., 2001, 13(4), 293–299 CrossRef CAS.
- R. De Philippis, M. C. Margheri, R. Materassi and M. Vincenzini, Appl. Environ. Microbiol., 1998, 64(3), 1130–1132 CAS.
- R. De Philippis and M. Vincenzini, FEMS Microbiol. Rev., 2006, 22(3), 151–175 CrossRef.
- R. De Philippis, G. Colica and E. Micheletti, Appl. Microbiol. Biotechnol., 2011, 92(4), 697–708 CrossRef PubMed.
- K. X. Qian, H. R. Zhu and S. G. Chen, Acta Phytoecol. Geobot. Sin., 1989, 13, 97–105 Search PubMed.
- H. Takenaka, Y. Yamaguchi, S. Sakaki, K. Watarai, N. Tanaka and M. Hori, Food Chem. Toxicol., 1998, 36, 1073–1077 CrossRef CAS PubMed.
- K. Kanekiyo, J. B. Lee, K. Hayashi, H. Takenaka, Y. Hayakawa, S. Endo and T. Hayashi, J. Nat. Prod., 2005, 68(7), 1037–1041 CrossRef CAS PubMed.
- K. Kanekiyo, K. Hayashi, H. Takenaka, J. B. Lee and T. Hayashi, Biol. Pharm. Bull., 2007, 30(8), 1573–1575 CAS.
- K. Hayashi, K. Kanekiyo, Y. Ohta, J. B. Lee, H. Takenaka and T. Hayashi, Planta Med., 2008, 74(9), 946 Search PubMed.
- J. Moreno, M. A. Vargas, H. Olivares, J. Rivas and M. G. Guerrero, J. Biotechnol., 1998, 60(3), 175–182 CrossRef CAS.
- J. Su, S. R. Jia, X. Chen and H. Yu, J. Appl. Phycol., 2008, 20(3), 213–217 CrossRef CAS.
- H. F. Yu, Y. S. Dong, and S. R. Jia, International Conference on Biomedical Engineering and Informatics, IEEE, 2010, 5, pp. 2052–2056 Search PubMed.
- Z. Ding, S. R. Jia, P. P. Han, N. Yuan and N. Tan, J. Appl. Phycol., 2013, 25(4), 1017–1021 CrossRef CAS.
- P. P. Han, S. G. Shen, H. Y. Wang, S. Y. Yao, Z. L. Tan, C. Zhong and S. R. Jia, J. Appl. Phycol., 2016, 1–11 Search PubMed.
- H. Yu, S. Jia and Y. Dai, Appl. Biochem. Biotechnol., 2010b, 160(2), 552–560 Search PubMed.
- P. P. Han, S. G. Shen, H. Y. Wang, Y. Sun, Y. J. Dai and S. R. Jia, Algal Res., 2015, 9, 143–150 CrossRef.
- P. P. Han, Y. Sun, X. Y. Wu, Y. J. Yuan, Y. J. Dai and S. R. Jia, Appl. Biochem. Biotechnol., 2014a, 172(1), 36–49 Search PubMed.
- P. P. Han, Y. Sun, S. R. Jia, C. Zhong and Z. L. Tan, Carbohydr. Polym., 2014b, 105(1), 145–151 Search PubMed.
- W. Kowallik, Plant Physiol., 1982, 33, 51–72 CrossRef CAS.
- M. Atta, A. Idris, A. Bukhari and S. Wahidin, Bioresour. Technol., 2013, 148(11), 373 CrossRef CAS PubMed.
- L. Zhang, G. Ma, K. Yamawaki, Y. Ikoma, H. Matsumoto, T. Yoshioka, O. Satoshi and K. Masaya, J. Plant Physiol., 2015, 188, 58–63 CrossRef CAS PubMed.
- E. Bland and L. T. Angenent, Bioresour. Technol., 2016, 216, 579–586 CrossRef CAS PubMed.
- T. Ogawa, American Society of Plant Physiologists, Rockville, MD, 1993 Search PubMed.
- M. R. Badger and G. D. Price, J. Exp. Bot., 2003, 54(383), 609–622 CrossRef CAS PubMed.
- G. D. Price, M. R. Badger, F. J. Woodger and B. M. Long, J. Exp. Bot., 2008, 59(7), 1441–1461 CrossRef CAS PubMed.
- A. Otero and M. Vincenzini, J. Biotechnol., 2003, 102(2), 143–152 CrossRef CAS PubMed.
- J. M. García-Fernández and J. Diez, Res. Microbiol., 2004, 155(10), 795–802 CrossRef PubMed.
- P. M. Glibert, F. P. Wilkerson, R. C. Dugdale, J. A. Raven, C. L. Dupont, P. R. Leavitt, A. E. Parker, J. M. Burkholder and T. M. Kana1, Limnology & Oceanography, 2016, 61(1), 284–300 Search PubMed.
- B. L. Horecker, Annu. Rev. Microbiol., 1966, 20(20), 253–290 CrossRef CAS PubMed.
- P. J. Looijesteijn, I. C. Boels, M. Kleerebezem and J. Hugenholtz, Appl. Environ. Microbiol., 1999, 65(11), 5003–5008 CAS.
- B. Degeest and L. de Vuyst, Appl. Environ. Microbiol., 2000, 66(8), 3519–3527 CrossRef CAS PubMed.
- C. Petit, J. P. Grill, N. Maazouzi and R. Marczak, Appl. Microbiol. Biotechnol., 1991, 36(2), 216–221 CrossRef CAS.
- A. Escalante, C. Wacher-Rodarte, M. García-Garibay and A. Farrés, J. Appl. Microbiol., 1998, 84(1), 108–114 CrossRef CAS PubMed.
- G. J. Grobben, M. R. Smith, J. Sikkema and J. A. M. de Bont, Appl. Microbiol. Biotechnol., 1996, 46(3), 279–284 CrossRef CAS.
- H. Li, H. Xu, H. Xu, S. Li and P. K. Ouyang, Appl. Microbiol. Biotechnol., 2010, 86(1), 295–303 CrossRef CAS PubMed.
- S. Pan, D. Yao, J. Chen and S. Wu, Carbohydr. Polym., 2013, 92(1), 629 CrossRef CAS PubMed.
- Z. Y. Zhu, X. C. Liu, F. Y. Dong, M. Z. Guo, X. T. Wang, Z. Wang and M. Z. Yong, Appl. Microbiol. Biotechnol., 2016, 100(9), 3909–3921 CrossRef CAS PubMed.
- F. Mozzi, D. G. G. Savoy and D. V. G. Font, J. Appl. Microbiol., 2003, 94(2), 175–183 CrossRef CAS PubMed.
- A. J. Smith, Ann. Inst. Pasteur/Microbiol., 1983, 134(1), 93–113 CrossRef.
- L. J. Stal and R. Moezelaar, FEMS Microbiol. Rev., 1997, 21(2), 179–211 CrossRef CAS.
- V. L. Knowles and W. C. Plaxton, Plant Cell Physiol., 2003, 44(7), 758–763 CrossRef CAS PubMed.
- M. Dubois, K. A. Gilles and J. K. Hamilton, Anal. Chem., 1956, 28, 350–353 CrossRef CAS.
- M. Tamoi, A. Murakami, T. Takeda and S. Shigeoka, Biochim. Biophys. Acta, 1998, 1383(2), 232–244 CrossRef CAS.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS PubMed.
- H. Bisswanger, Practical enzymology, Wiley VCH Verlag GmbH & Co, KGaA, New York, 2004 Search PubMed.
- R. L. Bernstein and P. W. Robbins, J. Biol. Chem., 1965, 240(1), 391–397 CAS.
- R. J. Hung, H. S. Chien, R. Z. Lin, C. T. Lin, J. Vatsyayan, H. L. Peng and H. Y. Chang, J. Biol. Chem., 2007, 282(24), 17738–17748 CrossRef CAS PubMed.
- D. Shinabarger, A. Berry, T. B. May, R. Rothmel, A. Fialho and A. M. Chakrabarty, J. Biol. Chem., 1991, 266(4), 2080–2088 CAS.
- K. J. Livak and T. D. Schmittgen, Methods, 2001, 25, 402–408 CrossRef CAS PubMed.
- E. Szekeres, C. Sicora, N. Dragoş and B. Drugă, FEMS Microbiol. Lett., 2014, 359(1), 102–109 CrossRef CAS PubMed.
- L. Peng, J. Li, Y. Liu, Z. Xu, J. Y Wu, Z. Ding, Z. Gu, L. Zhang and G. Shi, RSC Adv., 2016, 6(45), 39284–39291 RSC.
- L. Peng, S. Qiao, Z. Xu, F. Guan, Z. Ding, Z. Gu, L. Zhang and G. Shi, Carbohydr. Polym., 2015, 133, 104–109 CrossRef CAS PubMed.
- A. T. Granja, A. Popescu and A. R. Marques, Appl. Microbiol. Biotechnol., 2007, 76(6), 1319–1327 CrossRef CAS PubMed.
- A. Kereszt, I. ErnöK, B. L. Reuhs, R. W. Carlson, I. Ádám Kondoros and P. Péter, J. Bacteriol., 1998, 180, 5426–5431 CAS.
- B. Seitz, C. Klos, M. Wurm and R. Tenhaken, Plant J., 2000, 21, 537–546 CrossRef CAS PubMed.
- P. P. Han, S. Y. Yao, R. J. Guo, S. G. Shen, R. R. Yan, Z. L. Tan and S. R. Jia, Carbohydr. Polym., 2017, 174, 111–119 CrossRef CAS PubMed.
- F. Sousa, R. Joana, H. Jorge and S. A. Leitão, Biotechnology of Biopolymers, InTech, 2011, pp. 257–274 Search PubMed.
- S. F. Ana, N. Inês Silva, H. O. Vítor, C. Raquel and M. M. Leonilde, Front. Cell. Infect. Microbiol., 2011, 1(12), 16 Search PubMed.
- M. Tamoi, T. Takeda and S. Shigeoka, Plant Cell Physiol., 1999, 40(2), 257–261 CrossRef CAS.
- W. Ma, D. Shi, Q. Wang, L. Wei and H. Chen, J. Appl. Phycol., 2005, 17(3), 273–280 CrossRef CAS.
- J. Becker, C. Klopprogge, O. Zelder, E. Heinzle and C. Wittmann, Appl. Environ. Microbiol., 2005, 71, 8587–8596 CrossRef CAS PubMed.
Footnote |
| † These two authors made equal contributions to the study. |
| This journal is © The Royal Society of Chemistry 2017 |