Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
“Blinking” silica nanoparticles for optical super resolution imaging of cancer cells
Jia Lia,
Shenfei Zongb,
Zhuyuan Wangb and
Yiping Cui *b
*b
aDepartment of Ultrasonography, Zhongda Hospital, Medical School Southeast University, Nanjing 210009, China
bAdvanced Photonics Center, Southeast University, Nanjing 210096, Jiangsu, China. E-mail: cyp@seu.edu.cn
First published on 17th October 2017
Abstract
Fluorophores for single molecule localization microscopy (SMLM) must exhibit continuous signal fluctuations (i.e. blinking). Here, we present a strategy to make silica nanoparticles (NPs) blink using Alexa Fluor 647 (A647), an organic dye widely used in SMLM. We find that encapsulating A647 inside the silica NPs produces non-blinking silica NPs, while decorating A647 onto the outer surface of silica NPs generates blinking silica NPs. The morphology and fluorescence properties of the blinking silica NPs are studied. Besides, by attaching cancer specific antibodies, the blinking silica NPs can specifically target the cancer cells. Cells incorporated with the blinking silica NPs are imaged using confocal laser scanning microscopy and SMLM. With the optical super resolution imaging ability, this kind of cancer specific blinking silica NPs can have a great potential in detailed investigation of the interaction between cancer cells and NPs.
Introduction
Due to the diffraction limit, the spatial resolution of conventional optical microscopies is approximately half of the illumination wavelength (typically 200–300 nm), which makes them unsuitable for the investigation of nanoscale objects such as cell organelles or nanoparticles (NPs). During the last decade, scientists have invented several optical super resolution microscopies with greatly improved spatial resolution.1,2 For example, stimulated emission depletion microscopy (STED), structured illumination microscopy (SIM) and single molecule localization microscopy (SMLM).3–10 The best spatial resolution is provided by SMLM, which is about 20–30 nm. However, SMLM requires that the fluorophores must exhibit signal fluctuations or blinking to realize precise localization. Zhuang et al. have evaluated a serious of organic dyes and found the ones suitable for SMLM.11 Among different dyes, Alexa Fluor 647 (A647) is one of the fine choice and thus it is widely used in SMLM imaging experiments.Since SMLM can provide a quite high spatial resolution, it is a powerful tool for the investigation of nanoscale particles and their interaction with cells. As one of the representative NPs, silica NPs are commonly employed in in vitro experiments owing to their excellent biocompatibility and feasible surface functionalization.12–15 The silica surface can be easily modified with functional groups or targeting molecules, such as antibodies, peptides and aptamers, forming biologically functionalized nanoprobes.16–18
When studying the interaction between cells and silica NPs using optical microscopies (for example, tracking the intracellular locations of silica NPs), a pre-requisite is to encode optical signals into these silica NPs. Surface enhanced Raman scattering and fluorescence are two typical kinds of optical signals used in encoding silica NPs.19–21 Up to now, most SMLM experiments utilize fluorescence signals. Hence, in the present work, we proposed a way to make silica NPs fluorescent and blink using the organic dye molecule (A647). By decorating cancer cell specific antibodies, these fluorescent silica NPs can realize cancer cell targeting. Moreover, after taken up by the cells, the blinking silica NPs can also be employed in intracellular SMLM imaging, which indicates that the blinking silica NPs are fine candidates for the study of intracellular nanomaterials when using SMLM.
Materials and methods
Materials
Alexa Fluor 647 (A647) NHS ester (http://www.thermofisher.com/order/catalog/product/A20006), PKH26 and SYTO™ 9 Green were purchased from ThermoFisher Scientific. Polyethylenimine (PEI, branched, MW ∼25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) was purchased from Sigma-Aldrich. Tetraethoxysilane (TEOS), glutaraldehyde (GA, 50%), dimethyl sulfoxide (DMSO) and (3-aminopropyl)triethoxysilane (APTES) were purchased from Alfa Aesar. Ammonia water (25%) and absolute ethanol (EtOH) were purchased from Nanjing Chemical Reagent Co., Ltd. Rabbit anti-human HER2 antibody, bovine serum albumin (BSA) and phosphate buffered saline (PBS, 10 mM, pH 7.4) were purchased from Beijing Biosynthesis Biotechnology Co., Ltd. All the reagents were used as received. Deionized water (Millipore Milli-Q grade) with a resistivity of 18.2 MΩ cm−1 was used in all the experiments.
000) was purchased from Sigma-Aldrich. Tetraethoxysilane (TEOS), glutaraldehyde (GA, 50%), dimethyl sulfoxide (DMSO) and (3-aminopropyl)triethoxysilane (APTES) were purchased from Alfa Aesar. Ammonia water (25%) and absolute ethanol (EtOH) were purchased from Nanjing Chemical Reagent Co., Ltd. Rabbit anti-human HER2 antibody, bovine serum albumin (BSA) and phosphate buffered saline (PBS, 10 mM, pH 7.4) were purchased from Beijing Biosynthesis Biotechnology Co., Ltd. All the reagents were used as received. Deionized water (Millipore Milli-Q grade) with a resistivity of 18.2 MΩ cm−1 was used in all the experiments.
Fabrication of the fluorescent silica NPs
First, A647 was conjugated to APTES as follows. 30 μL of 1 mM A647 (in DMSO) and 200 μL of APTES were mixed in 1 mL of EtOH. The mixture was shaken under room temperature for 2 h to allow the reaction. Then the mixture was kept at 4 °C for further use. Silica NPs were fabricated according to previously published literature with slight modification.22 For blinking silica NPs, 1 mL of TEOS was mixed with 50 mL of EtOH and vigorously stirred, followed by the addition of 10 mL of ammonia water. After stirring for 20 min, the silica NPs are formed and collected by centrifugation, the final sediments were suspended in 20 mL of EtOH. Next, 200 μL of deionized water and 200 μL of the APTES-A647 conjugates were added into 4 mL of the as-formed silica NPs. The mixture was stirred at room temperature for 12 h. Finally, the A647 modified silica NPs were collected by centrifugation and washed repetitively with EtOH to remove excess APTES-A647. The final products (i.e. the blinking silica NPs) were suspended in 4 mL of EtOH.For non-blinking silica NPs, 1 mL of TEOS, 200 μL of the APTES-A647 conjugates, 10 mL of ammonia water and 50 mL of EtOH were mixed together and vigorously stirred for 20 min. Then the non-blinking silica NPs were collected by centrifugation and washed repetitively with EtOH to remove excess reagents. The final sediments were suspended in 20 mL of EtOH.
To attach HER2 antibodies onto the surface of blinking silica NPs, first, PEI was added into 500 μL of the blinking silica NPs with a final concentration of 0.1%. The mixture was shaken for 20 min, then excess PEI was removed by centrifugation. The precipitates were suspended in 500 μL of deionized water. After that, GA was added into the PEI modified blinking silica NPs with a final concentration of 0.5% and shaken for 1 h. Excess GA was removed by centrifugation and the precipitates were suspended in 500 μL of PBS. Next, 5 μL of 1 mg mL−1 rabbit anti-human HER2 antibody was added into the GA modified blinking silica NPs. The mixture was shaken for 2 h under room temperature and then incubated at 4 °C for 12 h. After that excess antibodies were removed by centrifugation. The precipitates were suspended in 500 μL of PBS containing 0.1% BSA and stored at 4 °C for further usage. The estimated amount of antibodies on each silica NPs is about 200 per nanoparticle.
Evaluation of the fluorescence “blinking” behavior
Blinking or non-blinking silica NPs were adsorbed onto the bottom of an eight chambered Nunc™ Lab-Tek™ II Chamber Slide™ as follows. First, the as-prepared blinking or non-blinking silica NPs were diluted for 20 times using EtOH. Then, the diluted silica NPs were added into two chambers of the slide, respectively. After incubating for 20 min, some of the silica NPs deposited onto the bottom of the chamber slide and excess silica NPs were removed and washed away with EtOH. Next, imaging buffer prepared according to previously published literature was added into the chambers and the chamber slide was mounted onto a Zeiss Elyra P.1 microscope for blinking test.11 The lasers used are 405 nm (activation laser) and 642 nm (illumination laser).Cell culture and cell imaging
Human breast cancer cells (SKBR3, MDA-MB-231) were purchased from China Type Culture Collection. Cells were kept under standard cell culture condition (5% CO2, 37 °C). The culture media (RPMI1640) were supplemented with 10% fetal bovine serum (GIBCO) and 1% penicillin–streptomycin (Nanjing KeyGen Biotech. Co., Ltd.). For fluorescence imaging, SKBR3 cells were seeded into two chambers of the Nunc™ Lab-Tek™ II Chamber Slide™ and incubated for 24 h before use. Next, 50 μL of antibody modified blinking silica NPs and 50 μL of bare blinking silica NPs (without antibodies) were added into the two chambers containing SKBR3 cells, respectively. After incubation for 2 h at 37 °C, the culture media were discarded and the cells were washed gently with PBS. After that, cells were fixed, the cell nucleus was stained by SYTO™ 9 Green and imaging buffer was added into the cell chamber. Then, the cells were imaged using confocal laser scanning microscopy (CLSM) and SMLM microscopy.Instruments
Transmission electron microscope (TEM) images were obtained with an FEI Tecnai G2T20 electron microscope operating at 200 kV. CLSM images and fluorescence spectrum were collected using a confocal laser scanning microscopy (Olympus, FV1000) with a 60× oil immersion objective. SMLM images were acquired using a Zeiss Elyra P.1 microscope equipped with 405 nm (50 mW) and 640 nm (150 mW) lasers. SMLM images was recorded using a 100×/1.46 oil immersion objective and an Andor EM-CCD camera (iXon DU897). SMLM imaging data were analyzed using the Zeiss ZEN 2012 software.Results and discussion
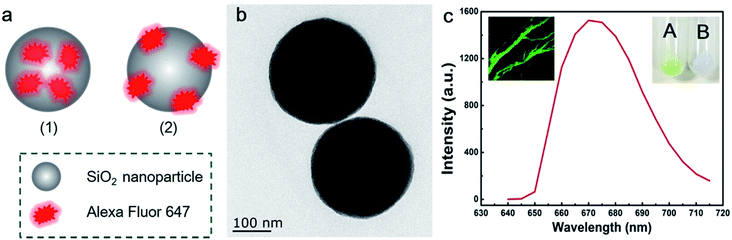
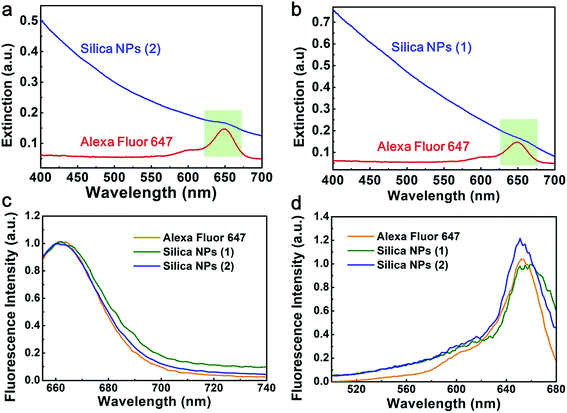
In the experiment, we fabricated two kinds of silica NPs as shown in Fig. 1a. The first kind, which is denoted as silica NPs (1), is non-blinking. The second kind, which is denoted as silica NPs (2), is blinking. Detailed fabrication procedures of these two kinds of silica NPs are presented in the Materials and methods section. The difference in the two kinds of silica NPs is that A647 molecules are encapsulated inside silica NPs (1) while they are attached outside the surface of silica NPs (2). Fig. 1b shows the TEM image of silica NPs (2). As can be seen, spherical like silica NPs are successfully fabricated. The diameter of silica NPs (2) is approximately 260 nm. Since the A647 molecules cannot be observed in the TEM images, the TEM image of silica NPs (1) is not provided because it is similar to that of the silica NPs (2). To examine whether A647 has been successfully modified to the silica NPs, we measured the fluorescence spectra of the silica NPs using the lambda scan function of CLSM (Olympus, FV1000) with 633 nm excitation wavelength. The silica NPs were casted onto a glass slide and imaged using the CLSM. The experimental data of silica NPs (1) and silica NPs (2) are similar here so only data of silica NPs (2) are provided. The fluorescence image of the silica NPs (2) is shown in the inset of Fig. 1c, where strong fluorescence is observed under 633 nm excitation. The fluorescence spectrum of silica NPs (2) is presented in Fig. 1c. Obvious fluorescence emission is detected peaking at around 670 nm, which is typical fluorescence signals originated from A647 molecules.The extinction, excitation and emission spectra of silica NPs (1) and silica NPs (2) can better prove the successful attachment of A647. As indicated by the green shadows in Fig. 2a and b, a bump at around 650 nm is observed in both the extinction spectra of silica NPs (1) and silica NPs (2), which originates from the absorbance of A647 molecules. The emission and excitation spectra of A647, silica NPs (1) and silica NPs (2) are similar to each other (see Fig. 2c and d). The difference is that spectra of silica NPs (1) and silica NPs (2) exhibit a slight red shift as compared with those of A647. Besides, more red shift is observed for silica NPs (1) as compared with silica NPs (2). This is possibly caused by the fact that A647 in silica NPs (1) is surrounded by the silica matrix and thus experiences a higher refractive index. Fortunately, such a slight red shift would not affect fluorescence imaging in our experiments, because we use a 655 nm long pass filter to collect the fluorescence signals. These result well confirmed that A647 molecules have been successfully modified onto the silica NPs, because blank or empty silica NPs do not fluoresce or show obvious absorbance at around 650 nm. In addition, a photograph of A647 modified silica NPs and blank silica NPs is added to the inset of Fig. 1c, where a greenish color is observed in A647 modified silica NPs. Hence, we can conclude that modification of A647 is successful.
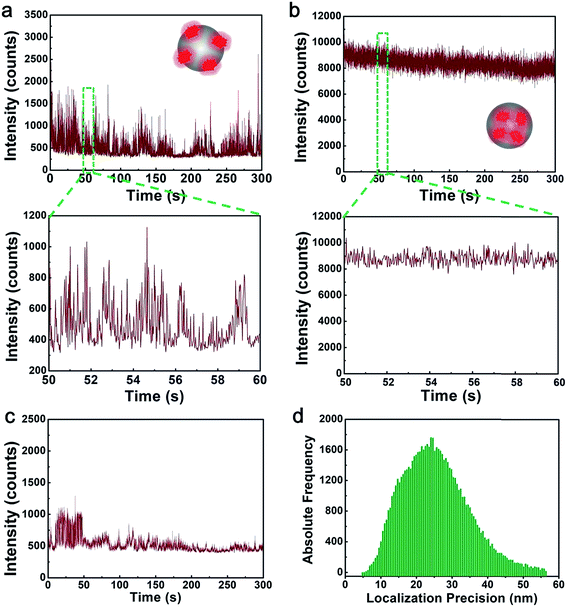
After confirming the successful fabrication of silica NPs (1) and silica NPs (2), the next step is to find out whether they are suitable for SMLM imaging. Hence, fluorescence blinking test is conducted to evaluate these two kinds of silica NPs using a Zeiss Elyra P.1 super resolution microscopy. Usually, repetitive fluorescence blinking of A647 molecules requires to lasers, one activation laser and one illumination laser.11 So in the experiment, the illumination wavelength is 642 nm and the activation wavelength is 405 nm. The exposure time is 20 ms and the filter is a 655 nm long pass filter which ensures that fluorescence from A647 is collected. Fig. 3 shows the representative time dependent fluorescence intensity profiles of individual silica NPs (1) and silica NPs (2). For silica NPs (2), dramatic blinking or fluctuation of the fluorescence signal is observed (Fig. 3a). While for silica NPs (1), the fluorescence signal intensity is quite stable and consecutive. The results in Fig. 3 clearly prove that silica NPs (2) are blinking and silica NPs (1) are non-blinking. Considering the requirements of SMLM, silica NPs (2) are excellent candidates for SMLM imaging while silica NPs (1) are not. The reason why silica NPs (1) are non-blinking might be that A647 molecules are encapsulated inside the silica NPs. The silica networks can well protect A647 from the outer environment (e.g. oxygen), which greatly prevents them from photobleaching and improves the photo stability of A647.23–25 Besides, researchers have reported that the transformation of many organic dyes (including A647) from the bright state to the dark state requires the assistance of a mercapto group containing buffer.26 A647 molecules encapsulated inside the silica NPs are isolated from the outer mercapto group containing buffer. Hence, they are not likely to get into the dark state. On the contrary, for silica NPs (2), since A647 molecules are decorated on the outmost surface of the silica NPs, they are exposed to the outer environment (e.g. the oxygen and mercapto groups), thus they will preserve their intrinsic fluorescence blinking property. As a result, the above experimental results show that to generate fluorescence blinking silica NPs for SMLM applications, the A647 molecules must be attached to the outer surface of silica NPs.
To find out the influence of dye contents on the blinking behavior, we fabricated silica NPs attached with less A647 as compared to the silica NPs (2). The fabrication is quite simple, we shortened the incubation time of APTES-A647 and silica NPs. For silica NPs (2), the incubation time is 12 h as shown in the experimental section. While for the silica NPs here, the incubation time is only 1 h. As a result, less A647 molecules are attached to these silica NPs. Blinking of these silica NPs with less A647 is tested using the same parameters (i.e. laser power, exposure time, etc.) as silica NPs (2). The results are shown in the Fig. 3c and d. As can be seen, although still blinking, the overall fluorescence intensity of these silica NPs is obviously weaker than silica NPs (2). This is rational since less A647 molecules are presented. The localization precision of these silica NPs is comparable to that of the silica NPs (2) (see Fig. 6e), which is also reasonable since the precision is mainly determined by the microscope system and the dye molecule itself. However, we prefer silica NPs (2) in practical applications since they can provide a intense fluorescence signal.
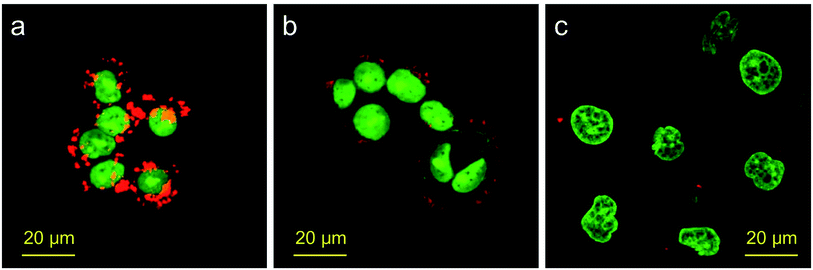
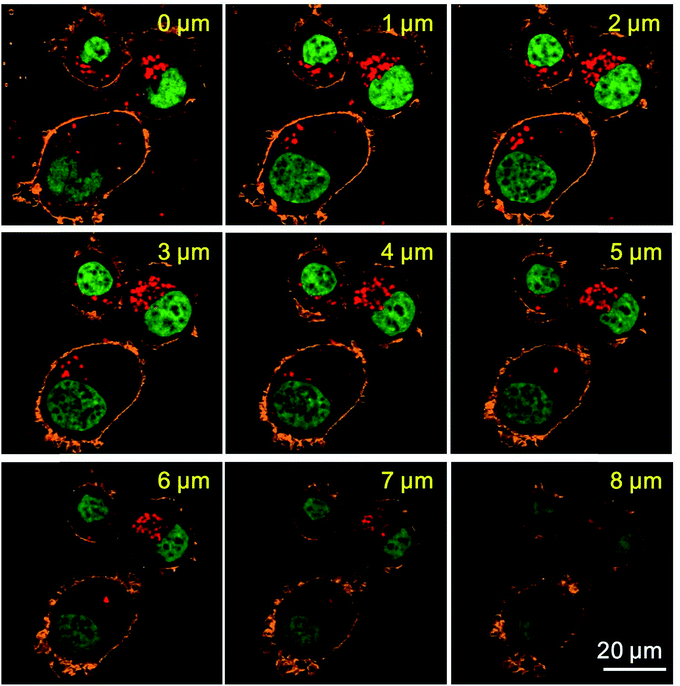
With the excellent fluorescence blinking behavior, silica NPs (2) can be used for cellular SMLM imaging. To facilitate cancer cell imaging, we also modified antibodies onto the surface of silica NPs (2). The cells are SKBR3 cells, which overexpress HER2 on their membranes. So rabbit anti-HER2 antibodies are attached to the surface of silica NPs (2). Before SMLM imaging, we first tested the cancer targeting ability of the antibody modified silica NPs (2). MDA-MB-231 cells with a low HER2 expression level and bare silica NPs (2) without antibodies are used as the controls. The silica NPs were incubated with cells and then these cells were imaged using CLSM. The results are shown in Fig. 4. It is obvious that SKBR3 cells take up more antibody modified silica NPs (2) as compared with the bare silica NPs (2) and MDA-MB-231 cells. This result confirms that anti-HER2 antibody modified silica NPs (2) can specifically recognize SKBR3 cells and increase the cellular uptake amount of the silica NPs (2). Moreover, we performed Z-slice imaging of SKBR3 cells incubated with antibody modified silica NPs (2) to check whether the nanoparticles are located inside the cells or not. The images were collected using the CLSM with a 60× oil objective, the stepsize is 1 μm. The results are shown in Fig. 5, which clearly proved that silica NPs (2) are located inside the cytoplasm of SKBR3 cells.
Finally, SMLM imaging experiments are conducted using SKBR3 cells incorporated with antibody modified silica NPs (2). Cells are immersed in imaging buffer prepared according to previously published literature. The imaging buffer can support the blinking of A647 molecules.11 The activation laser is 405 nm and illumination laser is 642 nm. The exposure time is 20 ms. SMLM image is reconstructed from 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 frames and analyzed using the Zeiss ZEN 2012 software. The results are illustrated in Fig. 6. Fig. 6a is conventional wide field total internal reflection (TIRF) microscopic image, Fig. 6b is SMLM image of the same cells and Fig. 6c is merged image of the TIRF and SMLM images. Basically, the SMLM image exhibits improved spatial resolution. For example, Fig. 6f shows the intensity profiles along the yellow dashed lines in the TIRF and SMLM images. The yellow dashed line indicates a cluster of silica NPs (2) in the cells. As can be seen, the intensity profile of TIRF image only shows one peak while that of the SMLM image shows two peaks. The reason is that TIRF microscopy holds a moderate spatial resolution and thus can not discriminate tiny structures of the clustered silica NPs (2). Contrarily, SMLM has a high spatial resolution and thus can unveil the subtle structures of the clustered silica NPs (2). The localization precision obtained in SMLM imaging experiments using silica NPs (2) is around 30 nm (Fig. 6e), which more vividly indicates a high spatial resolution. Consequently, the fabricated silica NPs (2) can indeed be used for SMLM imaging after taken up by cells.
000 frames and analyzed using the Zeiss ZEN 2012 software. The results are illustrated in Fig. 6. Fig. 6a is conventional wide field total internal reflection (TIRF) microscopic image, Fig. 6b is SMLM image of the same cells and Fig. 6c is merged image of the TIRF and SMLM images. Basically, the SMLM image exhibits improved spatial resolution. For example, Fig. 6f shows the intensity profiles along the yellow dashed lines in the TIRF and SMLM images. The yellow dashed line indicates a cluster of silica NPs (2) in the cells. As can be seen, the intensity profile of TIRF image only shows one peak while that of the SMLM image shows two peaks. The reason is that TIRF microscopy holds a moderate spatial resolution and thus can not discriminate tiny structures of the clustered silica NPs (2). Contrarily, SMLM has a high spatial resolution and thus can unveil the subtle structures of the clustered silica NPs (2). The localization precision obtained in SMLM imaging experiments using silica NPs (2) is around 30 nm (Fig. 6e), which more vividly indicates a high spatial resolution. Consequently, the fabricated silica NPs (2) can indeed be used for SMLM imaging after taken up by cells.
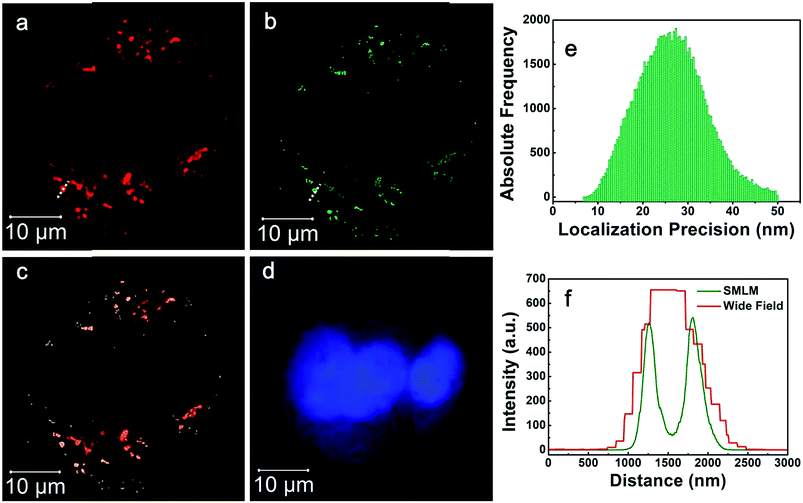 | ||
| Fig. 6 SMLM imaging results of SKBR3 cells incubated with antibody modified silica NPs (2). (a) Wide field TIRF image, (b) SMLM image, (c) merged image of (a) and (b), (d) cell nucleus. (e) Localization precision of silica NPs (2). (f) Intensity profiles along the yellow dashed lines as indicated in Fig. 4a and b. | ||
Conclusions
Fluorescent blinking silica NPs are successfully fabricated which are proven to be suitable for SMLM imaging applications. Fluorescence blinking behavior is obtained by attaching A647 molecules to the outer surface of silica NPs. Obvious signal fluctuation is observed with these blinking silica NPs. Besides, encapsulating A647 inside silica NPs produces non-blinking silica NPs, indicating that attaching A647 to the outer surface of silica NPs is a necessity in fabricating blinking silica NPs. By attaching cancer specific antibodies to the blinking silica NPs, cancer targeting is realized using the silica NPs. Moreover, SMLM imaging is achieved using cancer cells incorporated with the blinking silica NPs. A localization precision of about 30 nm is achieved in the experiment. We anticipate that the presented silica NPs with excellent fluorescence blinking behaviors and SMLM imaging abilities can have a fine potential in the investigation of the interaction between nanomaterials and cells.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Key Basic Research Program of China (2015CB352002) and the Natural Science Foundation of China (NSFC) (61535003, 61675042, 61505027).References
- B. Huang, H. Babcock and X. Zhuang, Cell, 2010, 143, 1047–1058 CrossRef CAS PubMed.
- L. Schermelleh, R. Heintzmann and H. Leonhardt, J. Cell Biol., 2010, 190, 165–175 CrossRef CAS PubMed.
- S. W. Hell, Science, 2007, 316, 1153–1158 CrossRef CAS PubMed.
- R. Heintzmann and M. G. L. Gustafsson, Nat. Photonics, 2009, 3, 362–364 CrossRef CAS.
- D. Li, L. Shao, B.-C. Chen, X. Zhang, M. Zhang, B. Moses, D. E. Milkie, J. R. Beach, J. A. Hammer III, M. Pasham, T. Kirchhausen, M. A. Baird, M. W. Davidson, P. Xu and E. Betzig, Science, 2015, 349, aab3500 CrossRef PubMed.
- M. J. Rust, M. Bates and X. Zhuang, Nat. Methods, 2006, 3, 793–795 CrossRef CAS PubMed.
- E. Betzig, G. H. Patterson, R. Sougrat, O. W. Lindwasser, S. Olenych, J. S. Bonifacino, M. W. Davidson, J. Lippincott-Schwartz and H. F. Hess, Science, 2006, 313, 1642–1645 CrossRef CAS PubMed.
- W. Wan, M.-Q. Zhu, Z. Tian and A. D. Q. Li, J. Am. Chem. Soc., 2015, 137, 4312–4315 CrossRef CAS PubMed.
- J. Yan, L.-X. Zhao, C. Li, Z. Hu, G.-F. Zhang, Z.-Q. Chen, T. Chen, Z.-L. Huang, J. Zhu and M.-Q. Zhu, J. Am. Chem. Soc., 2015, 137, 2436–2439 CrossRef CAS PubMed.
- M.-Q. Zhu, G.-F. Zhang, Z. Hu, M. P. Aldred, C. Li, W.-L. Gong, T. Chen, Z.-L. Huang and S. Liu, Macromolecules, 2014, 47, 1543–1552 CrossRef CAS.
- G. T. Dempsey, J. C. Vaughan, K. H. Chen, M. Bates and X. Zhuang, Nat. Methods, 2011, 8, 1027–1036 CrossRef CAS PubMed.
- M. Tan, N. Jatupaiboon, Y. Song, G. Sun and X. Ma, J. Controlled Release, 2013, 172, E57 CrossRef.
- H. Yamaguchi, M. Tsuchimochi, K. Hayama, T. Kawase and N. Tsubokawa, Int. J. Mol. Sci., 2016, 17, 1086 CrossRef PubMed.
- M. Liong, J. Lu, M. Kovochich, T. Xia, S. G. Ruehm, A. E. Nel, F. Tamanoi and J. I. Zink, ACS Nano, 2008, 2, 889–896 CrossRef CAS PubMed.
- L. Jiao, F. Song, B. Zhang, H. Ning, J. Cui and X. Peng, J. Mater. Chem. B, 2017, 5, 5278–5283 RSC.
- Y. Luo, W. Dou and G. Zhao, Anal. Bioanal. Chem., 2017, 409, 4139–4147 CrossRef CAS PubMed.
- Y. Wei, L. Gao, L. Wang, L. Shi, E. Wei, B. Zhou, L. Zhou and B. Ge, Drug Delivery, 2017, 24, 681–691 CrossRef CAS PubMed.
- J. Wang, J. Guo, J. Zhang, W. Zhang and Y. Zhang, Biosens. Bioelectron., 2017, 95, 100–105 CrossRef CAS PubMed.
- S. Zong, L. Wang, C. Chen, J. Lu, D. Zhu, Y. Zhang, Z. Wang and Y. Cui, Anal. Methods, 2016, 8, 5001–5008 RSC.
- Y. Zhang, Z. Wang, L. Wu, S. Zong, B. Yun and Y. Cui, RSC Adv., 2016, 6, 81046–81052 RSC.
- C. Wu, J. Hong, X. Guo, C. Huang, J. Lai, J. Zheng, J. Chen, X. Mu and Y. Zhao, Chem. Commun., 2008, 750–752, 10.1039/b717038f.
- J. Xue, C. Wang and Z. Ma, Mater. Chem. Phys., 2007, 105, 419–425 CrossRef CAS.
- G. Chen, F. Song, X. Wang, S. Sun, J. Fan and X. Peng, Dyes Pigm., 2012, 93, 1532–1537 CrossRef CAS.
- J. Liang, Z. Xue, J. Xu, J. Li, H. Zhang and W. Yang, Colloids Surf., A, 2013, 426, 33–38 CrossRef CAS.
- W.-H. Zhang, X.-X. Hu and X.-B. Zhang, Nanomaterials, 2016, 6, 81 CrossRef PubMed.
- G. T. Dempsey, M. Bates, W. E. Kowtoniuk, D. R. Liu, R. Y. Tsien and X. Zhuang, J. Am. Chem. Soc., 2009, 131, 18192–18193 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2017 |