Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
“Turn-on” fluorescence sensing of cytosine: development of a chemosensor for quantification of cytosine in human cancer cells†
Himadri Sekhar Sarkar a,
Sujoy Das
a,
Sujoy Das a,
Debasish Mandalb,
Md Raihan Uddinc,
Sukhendu Mandal
a,
Debasish Mandalb,
Md Raihan Uddinc,
Sukhendu Mandal c and
Prithidipa Sahoo
c and
Prithidipa Sahoo *a
*a
aDepartment of Chemistry, Visva-Bharati University, Santiniketan, 731235, India. E-mail: prithidipa@hotmail.com
bInstitute of Chemistry, The Hebrew University of Jerusalem, 91904 Jerusalem, Israel
cDepartment of Microbiology, University of Calcutta, Kolkata-700019, India
First published on 23rd November 2017
Abstract
A simple new chemosensor, pyrene-appended 5-hydroxyisophthalic acid derivative (PIA), was developed and characterized for selective detection and quantification of cytosine in different human cancer cells. PIA shows “turn-on” fluorescence in presence of cytosine through intermolecular hydrogen bonding with a detection limit of 32 nM at neutral pH.
Introduction
Purine and pyrimidine bases present in two nucleic acids DNA/RNA pair among themselves through hydrogen bonding in different patterns. This phenomenon has great influence in living systems as it precedes genetic codons of essential amino acids.1–3 Cytosine, a base of DNA, plays an important role in this base pairing through forming large organic assemblies with appropriate receptors. The molecular recognition among cytosine and organic hetero synthons needs attention to properly develop the fields of structure–function and structure–stability relationships between DNA sequences, structure, and flexibility because of the interaction of DNA with both small molecules and proteins along with the function of resulting molecular assemblies.4–8The concept of molecular recognition9–12 is fundamentally important both in chemistry and biology. The specific recognition process is responsible for many chemical and biological properties of big and small molecules. Researchers have extensively studied to learn complexation and behaviour of neutral biomolecules in complex biological systems.13 Complexation is stabilized through hydrogen bonding14–16 with host–guest molecules. The biomolecular recognition system, such as enzyme/protein–substrate recognition, replication of nucleic acids, and structural maintenance of proteins, mostly is based on hydrogen bonding via complementary base pairing.17 In molecular recognition research, model receptors have been established to mimic biological events and to recognize the biochemical significance of substrates.18–20
Furthermore, there has been substantial interest in the development of facile and reliable methods to detect nucleotides and nucleic acids owing to their potential applications in bio-analytical, clinical, and forensic analysis. Numerous analytical methods are available for detecting different bioactive molecules. In contrast, methods based on fluorescent sensors/chemosensors21,22 propose distinct advantages with desirable properties like high sensitivity, selectivity, convenience, cost effectiveness, fast response times, easy implementation, and local observation (e.g., by fluorescence imaging spectroscopy). High sensitivity and selectivity is potentially able to detect physiological and pathological consequences of biomolecules in a cell with spatial and chronological reliability. Detection and monitoring of biomolecules for live cell imaging studies23–25 using these sensors becomes critically important for biological, medical/pharmaceutical, analytical, and environmental applications.
As part of our ongoing research towards developing fluorophores for purine and pyrimidine derivatives,26,27 here, we prepared a new and simple chemosensor, a pyrene-appended 5-hydroxyisophthalic acid derivative (PIA), which showed efficient fluorescence signal upon selective binding with cytosine in aqueous medium (Scheme 1). This receptor contains hydrogen-bond donor–acceptor groups (carboxylic acid groups) for favourable complexation with cytosine.
So far, published articles have not highlighted any chemosensor which explains the detection and direct quantification of cytosine in live cells, although there are a few published HPLC-based,28,29 electrochemical-based,30 and nanopore-based31 cytosine detection techniques. Previously, a chemosensor32 has been reported with big limitations which cannot detect or quantify free cytosine present in a cell. The superiority of our chemosensor over others is that it can work on biotic and abiotic samples, in simple and in a complex matrix, with much less time and cost, through a simple one-step, direct detection method. Moreover, our aim is to detect free cytosine as it remains with higher concentration in human cancer cells. We can easily detect and quantify free cytosine in different human cancer cells by using our chemosensor. The above chemosensor is thus far better than other biosensors or detection techniques and easily detects cytosine in biological samples and in-solution, in a single experiment or in high-throughput screening with a selective manner (performance comparison is shown in Table S1†).
PIA exhibits remarkable change in fluorescence intensity in the presence of cytosine while the same remain unchanged for other structurally similar pyrimidine/purine bases. To the best of our knowledge, this is the first report, where we explored the prospect of selective recognition of cytosine executing “turn-on” fluorescence sensing in human cancer cells via confocal imaging experiments by a pyrene-based fluorescent receptor in aqueous medium.
The “turn-on” fluorescence is most likely due to photoinduced electron transfer (PET). Before interaction with the cytosine, the fluorescence of pyrene is quenched by the benzoic acid moiety, but with cytosine added, the pyrene recovers fluorescence. The electron-rich isophthalic acid can form a stronger hydrogen bond with cytosine to make PIA highly fluorescent. The design and procedure of developing such a chemosensor could also be an initial step in order to develop better and next generation molecules with better selectivity.
Experimental
PIA was synthesized from a simple reaction of 1-pyrenemethylamine hydrochloride and bromoacetyl chloride, followed by reaction with 5-hydroxyisophthalic acid resulting in 86% overall yield (Scheme S1†). The structure of PIA was confirmed by 1H NMR, 13C NMR, and MS analysis (Fig. S1–S5†).Spectrophotometric and spectrofluorimetric titrations (details in ESI†) were carried out to understand the PIA–cytosine interaction upon adding varying concentrations of cytosine to a fixed concentration of PIA (1 μM) in aqueous medium at neutral pH.
Results and discussion
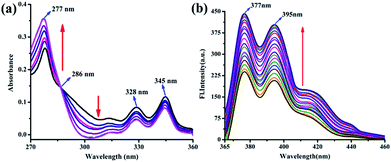
The UV-vis absorption spectra of PIA showed absorption maxima at 277 nm, which gradually increased upon addition of increasing amounts of cytosine (Fig. 1a). Furthermore, these spectra also showed a clear isosbestic point at 286 nm which indicates possible stronger interaction of PIA with cytosine.PIA shows very weak fluorescence with a low quantum yield (Φfs = 0.008)33 at room temperature (ESI†). A remarkable enhancement in fluorescence intensity at 377 nm and 395 nm (λex = 345 nm) was achieved upon binding of cytosine with PIA (Fig. 1b). The corresponding binding constant (Ka) from the fluorescence titration is 3.48 × 105 M−1 (Fig. S6†). The association constant of the complex was determined by non-linear fitting analyses of the titration curves following published reports.34,35 To understand the nature of the PIA–cytosine interaction, the continuous variation method was applied, which indicated a typical 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry (Fig. S7†). This implies that PIA could be utilized as a selective sensor to measure cytosine through a “turn-on” fluorescence sensing mechanism with a detection limit of 32 nM at neutral pH (Fig. S8†).
1 stoichiometry (Fig. S7†). This implies that PIA could be utilized as a selective sensor to measure cytosine through a “turn-on” fluorescence sensing mechanism with a detection limit of 32 nM at neutral pH (Fig. S8†).
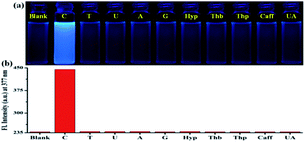
In order to establish the sensing selectivity and binding efficacy of the probe PIA, parallel experimentations were carried out with a variety of biologically relevant pyrimidine/purine derivatives such as thymine, uracil, adenine, guanine, hypoxanthine, theobromine, theophylline, caffeine, and uric acid. The change in fluorescence of PIA upon addition of various pyrimidine/purine derivatives was observed under a UV lamp (Fig. 2a). As in the presence of cytosine, a change of discernible fluorescence intensity of PIA was observed while other derivatives showed negligible effects (Fig. 2b). The above observation shows consistency with the fluorescence titration experiments where no such binding of PIA with other nucleobases was found (Fig. S9†). This result demonstrates that PIA exhibits high selectivity and sensitivity towards cytosine over other pyrimidine/purine bases in aqueous medium.
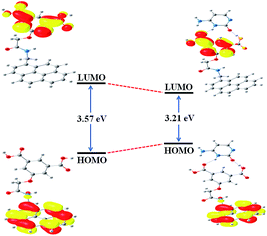
Significant binding of cytosine with PIA has also been examined by quantum chemical calculations at the TDDFT level using the B3LYP/6-31+G(d,p) method basis set in presence of water as a solvent. The COSMO solvent model,36 implemented in ORCA,37,38 incorporates the solvent effects. The HOMO of PIA was primarily localized on the pyrene moiety while the LUMO chiefly resides along the isophthalic acid, though these were extended with the amide chain. Geometry optimization resulted in conformational changes at the two different positions of PIA, hence two carboxylic acid groups take part to accommodate cytosine efficiently (Fig. S10†). Conformational changes can be estimated through the torsional angles of the two sides (Tables S2 and S3†) and the structures are given in Fig. 3. The π-electrons on the HOMO of PIA–cytosine complex are mainly located on the whole π-conjugated –NH–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O framework including the pyrene moiety but the LUMO is mostly positioned at the interior of the isophthalic acid. Moreover, the HOMO–LUMO energy gap of the PIA–cytosine complex (3.21 eV) becomes much smaller relative to that of PIA (3.57 eV) (Fig. 3).
O framework including the pyrene moiety but the LUMO is mostly positioned at the interior of the isophthalic acid. Moreover, the HOMO–LUMO energy gap of the PIA–cytosine complex (3.21 eV) becomes much smaller relative to that of PIA (3.57 eV) (Fig. 3).
In order to know more about interactions of PIA with cytosine, 1H NMR titration was also executed in DMSO-d6 (Fig. S11†). Both the carboxylic acid protons from PIA became abolished, indicating the presence of stronger hydrogen bonding interactions of the carboxylic acids with the amide moiety of cytosine. In contrast, the aromatic protons of PIA shifted downfield and became broader upon addition of cytosine in the solution. This may be due to the decrease in electron density of the pyrene ring of PIA because of the drifting of electrons from pyrene toward cytosine.
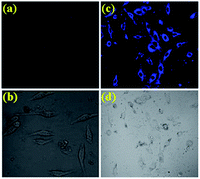
The desirable features of PIA such as high sensitivity and high selectivity with “turn-on” fluorescence at physiological pH encouraged us to further evaluate the potential of the sensor for imaging cytosine in live cells. A549 cells (Human cell A549, ATCC no. CCL-185) treated with PIA (1 μM) exhibited very weak fluorescence (probably for the endogenous cytosine), whereas a clear brilliant blue fluorescence signal was observed in the cells stained with PIA (1 μM) and cytosine (10 μM), which is in good agreement with the fluorescence “turn-on” profile of the chemosensor PIA in the presence of cytosine, thus corroborating the in-solution observation. As far as we are aware, this is the first report where we are executing the possible use of the pyrene-based amide of 5-hydroxyisophthalic acid via fluorescence change for the selective recognition of cytosine in living cells (Fig. 4). The confocal microscopic analysis also suggested that PIA could readily cross the membrane barrier of A549 cells, and rapidly sense intracellular cytosine as well. These findings open an avenue for future biomedical applications of the chemosensor to recognize cytosine. Cells were intact and showed a healthy spread and adherent morphology during and after the labeling process with PIA and PIA–cytosine complex indicating absence of cytotoxic effects (Fig. S12†).39 These results clearly indicate that PIA is effective for intracellular cytosine imaging agents having significant cell permeability.
The concentration of cytosine was also quantified from A549 human cancer cells (details in ESI†). Lysate of 107 A549 cells was added to 1 μM of PIA and the fluorescence signal was recorded. Cytosine presence in this cancer cell was detected with the help of a PIA–cytosine standard fluorescence curve (Fig. 5) using the selective detection ability of the chemosensor PIA.
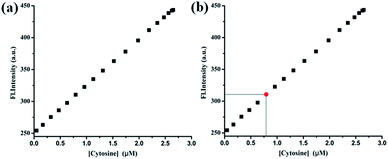 | ||
| Fig. 5 (a) Calibration curve obtained for the estimation of cytosine. (b) Estimation of the concentration of cytosine (red point) from the calibration curve. | ||
From the standard curve it was found that the concentration of cytosine in the tested sample was 0.82 μM present in 16.7 mm3 A549 cell volume. The above result was authenticated by spiking a real sample with a known concentration of cytosine and different fluorescence signals were observed by adding known volumes from each sample to the PIA. The recovery of the spiked samples was estimated to be in the range of 96–98% (Table 1).
| Sample | PIA used (μM) | Initial cytosine used | Addition of exogenous cytosine (μM) | Amount of cytosine derived from fluorescence signal (μM) | Fluorescence signal recovery (%) |
|---|---|---|---|---|---|
| 1 | 1 | Cytosine present in 16.7 mm3 A549 cell volume | 0 | 0.82 | — |
| 2 | 1 | 1 | 1.79 | 98.3 | |
| 3 | 1 | 3 | 4.67 | 96.8 | |
| 4 | 1 | 5 | 9.46 | 96.3 |
We further validated assay of cytosine from multiple samples of different human cancer cells (A549, HeLa, and Hep-2) using PIA (see ESI†). Each sample was assayed in triplicate. The signal to noise ratio of each independent reading was between 1.88 and 2.03 (Table S4†). The fold increase of fluorescence signals also were statistically validated after calculating the Z′ value for each set of measurements.41 It was found that in all tested samples the Z′ score was more than 0.9, indicating an optimized and validated assay of cytosine.
Conclusions
In summary, the present study reveals a new chemosensor (PIA) for detecting cytosine expressed in cells through a “turn-on” fluorescence sensing mechanism. The driving force for the binding to the host (PIA) with guest (cytosine) was found to be directed during H-bonding. The binding interaction was demonstrated on the basis of absorption, fluorescence, 1H NMR, TDDFT calculations, and live cell imaging studies. PIA exhibits a highly sensitive and selective fluorescence enhancement towards cytosine over other biologically important pyrimidine and purine derivatives at neutral pH. The “turn-on” fluorescence of PIA for the selective detection and quantification of cytosine was successfully demonstrated in different human cancer cells with a detection limit of 32 nM. Thus, this sensor has high potential for usage to investigate cytosine in chemical, biological, and medical research. Furthermore, this chemosensor can be marketed as a unique cytosine detector for its application in single or high throughput screening in a simple or complex matrix.Conflicts of interest
There are no conflicts to declare.Acknowledgements
PS acknowledges SERB-DST, Govt. of India for awarding her the young scientist grant [Project file no. SB/FT/CS-021/2014]. HSS is sincerely thankful to UGC, Delhi for the junior research fellowship. Authors are also grateful to DBT-IPLS facility, CU.Notes and references
- W. Xu, J.-g. Wang, M. F. Jacobsen, M. Mura, M. Yu, R. E. A. Kelly, Q.-q. Meng, E. Lægsgaard, I. Stensgaard, T. R. Linderoth, J. Kjems, L. N. Kantorovich, K. V. Gothelf and F. Besenbacher, Angew. Chem., 2010, 122, 9563–9567 CrossRef.
- G. Portalone and M. Colapietro, J. Chem. Crystallogr., 2009, 39, 193–200 CrossRef CAS.
- G. Portalone, Chem. Cent. J., 2011, 5, 51 CrossRef CAS PubMed.
- S. S. Bag, Y. Saito, K. Hanawa, S. Kodate, I. Suzuka and I. Saito, Bioorg. Med. Chem. Lett., 2006, 16, 6338–6341 CrossRef CAS PubMed.
- Y. Sato, Y. Toriyabe, S. Nishizawa and N. Teramae, Chem. Commun., 2013, 49, 9983–9985 RSC.
- T. Lee and P. Y. Wang, Cryst. Growth Des., 2010, 10, 1419–1434 CAS.
- C. Vilaivan, W. Srinarang, N. Yotapan, W. Mansawat, C. Boonlua, J. Kawakami, Y. Yamaguchi, Y. Tanaka and T. Vilaivan, Org. Biomol. Chem., 2013, 11, 2310–2317 CAS.
- S. R. Perumalla, E. Suresh and V. R. Pedireddi, Angew. Chem., 2005, 117, 7930–7935 CrossRef.
- J. M. Lehn, in Supramolecular Chemistry: Concepts and Perspectives, VCH, Weinheim, 1995 Search PubMed.
- S. Dey, D. Saina and S. P. Goswami, RSC Adv., 2014, 4, 428–433 RSC.
- P. Sahoo, Bioorg. Chem., 2015, 58, 26–47 CrossRef CAS PubMed.
- S. Dey, D. Sain and S. P. Goswami, RSC Adv., 2014, 4, 428 RSC.
- W. Saenger, Principles of Nucleic Acid Structure, Springer-Verlag, New York, 1988 Search PubMed.
- C. O. Hugo, J. F. Arambula, S. R. Ramisetty, A. M. Baranger and S. C. Zimmerman, Chem. Commun., 2009, 668–670 Search PubMed.
- J. S. Lindsey and P. C. Kearney, J. Am. Chem. Soc., 1988, 110, 6575–6577 CrossRef CAS.
- Papers in the special issue on molecular recognition of Chem. Rev., 1997, 97, 1231 Search PubMed.
- B. Alerts, D. Bray, J. Lewis, M. Raff, K. Roberts and J. D. Watson, Molecular Biology of the Cell, Garland Publishing, New York, 2nd edn, 1989 Search PubMed.
- J. Rebek, Chem. Commun., 2000, 637–643 RSC.
- J. Rebek, Heterocycles, 2000, 52, 493 CrossRef CAS.
- V. M. Rotello, E. A. Viani, G. Deslongchamps, B. A. Murray and J. Rebek Jr, J. Am. Chem. Soc., 1993, 115, 797–798 CrossRef CAS.
- W. Jorgensen and J. Pranata, J. Am. Chem. Soc., 1990, 112, 2008 CrossRef CAS.
- T. Tjivikua, G. Deslongchamps and J. Rebek Jr, J. Am. Chem. Soc., 1990, 112, 8408 CrossRef CAS.
- D. T. Quang and J. S. Kim, Chem. Rev., 2010, 110, 6280–6303 CrossRef CAS PubMed.
- H. S. Sarkar, S. Das, M. R. Uddin, S. Mandal and P. Sahoo, Asian J. Org. Chem., 2017, 6, 71 CrossRef CAS.
- S. Das, H. S. Sarkar, M. R. Uddin, K. Rissanen, S. Mandal and P. Sahoo, Chem. Commun., 2017, 53, 7600–7603 RSC.
- P. Sahoo, H. S. Sarkar, S. Das, K. Maiti, M. R. Uddin and S. Mandal, RSC Adv., 2016, 6, 66774–66778 RSC.
- P. Sahoo, S. Das, H. S. Sarkar, K. Maiti, M. R. Uddin and S. Mandal, Bioorg. Chem., 2017, 71, 315–324 CrossRef CAS PubMed.
- J. L. Gaoa, K. S. Y. Leung, Y. T. Wang, C. M. Lai, S. P. Li, L. F. Hu, G. H. Lu, Z. H. Jiang and Z. L. Yu, J. Pharm. Biomed. Anal., 2007, 44, 807–811 CrossRef PubMed.
- H. Fan, S. P. Li, J. J. Xiang, C. M. Lai, F. Q. Yang, J. L. Gao and Y. T. Wang, Anal. Chim. Acta, 2006, 567, 218–228 CrossRef CAS.
- H. Li, X. Wang, Z. Wang and W. Zhao, J. Solid State Electrochem., 2016, 20, 2223 CrossRef CAS.
- B. M. Venkatesan and R. Bashir, Nat. Nanotechnol., 2011, 6, 615–624 CrossRef CAS PubMed and references therein.
- R. K. Verma, F. Takei and K. Nakatani, Org. Lett., 2016, 18, 3170–3173 CrossRef PubMed.
- A. T. R. Williams, S. A. Winfield and J. N. Miller, Analyst, 1983, 108, 1067–1071 RSC.
- P. T. Chou, G. R. We, C. Y. Wei, C. C. Cheng, C. P. Chang and F. T. Hung, J. Phys. Chem. B, 2000, 104, 7818–7829 CrossRef CAS.
- J. H. Liao, C.-T. Chen, H.-C. Chou, C.-C. Cheng, P.-T. Chou, J.-M. Fang, Z. Slanina and T. J. Chow, Org. Lett., 2002, 4, 3107–3110 CrossRef CAS PubMed.
- F. Neese, Wiley Interdiscip. Rev.: Comput. Mol. Sci., 2012, 2, 73–78 CrossRef CAS.
- S. Grimme, J. Antony, S. Ehrlich and H. J. Krieg, Chem. Phys., 2010, 132, 154104 Search PubMed.
- S. Grimme, S. Ehrlich and L. Goerigk, J. Comput. Chem., 2011, 32, 1456 CrossRef CAS PubMed.
- J. Ratha, K. A. Majumdar, S. K. Mandal, R. Bera, C. Sarkar, B. Saha, C. Mandal, K. D. Saha and R. Bhadra, Mol. Cell. Biochem., 2006, 290, 113 CrossRef CAS PubMed.
- T. W. Traut, Mol. Cell. Biochem., 1994, 140, 1–22 CrossRef CAS PubMed.
- J. H. Zhang, T. D. Y. Chung and K. R. Oldenburg, J. Biomol. Screening, 1999, 4, 67–73 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra11096k |
| This journal is © The Royal Society of Chemistry 2017 |