Open Access Article
Open Access ArticleSpecific protein labeling with caged fluorophores for dual-color imaging and super-resolution microscopy in living cells†
Sebastian
Hauke‡
 a,
Alexander
von Appen‡
a,
Tooba
Quidwai
b,
Jonas
Ries
b and
Richard
Wombacher
*a
a,
Alexander
von Appen‡
a,
Tooba
Quidwai
b,
Jonas
Ries
b and
Richard
Wombacher
*a
aInstitute of Pharmacy and Molecular Biotechnology, Ruprecht-Karls-University Heidelberg, Im Neuenheimer Feld 364, 69120 Heidelberg, Germany. E-mail: wombacher@uni-heidelberg.de; Fax: +49 6221 546439; Tel: +49 6221 544879
bEuropean Molecular Biology Laboratory, Meyerhofstraße 1, 69117 Heidelberg, Germany
First published on 5th September 2016
Abstract
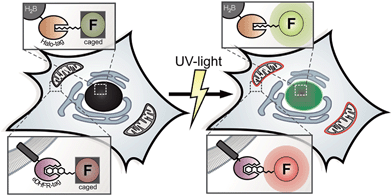
We present new fluorophore-conjugates for dual-color photoactivation and super-resolution imaging inside live mammalian cells. These custom-designed, photo-caged Q-rhodamines and fluoresceins are cell-permeable, bright and localize specifically to intracellular targets. We utilized established orthogonal protein labeling strategies to precisely attach the photoactivatable fluorophores to proteins with subsequent activation of fluorescence by irradiation with UV light. That way, diffusive cytosolic proteins, histone proteins as well as filigree mitochondrial networks and focal adhesion proteins were visualized inside living cells. We applied the new photoactivatable probes in inverse fluorescence recovery after photo-bleaching (iFRAP) experiments, gaining real-time access to protein dynamics from live biological settings with resolution in space and time. Finally, we used the caged Q-rhodamine for photo-activated localization microscopy (PALM) on both fixed and live mammalian cells, where the superior molecular brightness and photo-stability directly resulted in improved localization precisions for different protein targets.
Introduction
For decades, photoactivation experiments, based on small caged fluorophores inside live cells have provided access to decipher multi-parametric dynamic processes and fates of cellular proteins.1,2 However, attachment of caged fluorophores to specific proteins in live cells remained challenging and microinjection of purified and labeled proteins has been the common strategy to perform live cell studies. Since the report of caged coumarin, fluorescein and rhodamine, the palette of synthetic bright and photo-stable, caged fluorophores is steadily expanding.3–9 New, facile synthesis routes and commercial availability provide the scientific community easy access to caged synthetic fluorophores.10 Despite this progress in synthesis of caged fluorophores, live cell studies mainly suffered from insufficient cell permeabilities of applied caged dyes or were hampered by precisely localizing photoactivatable fluorophores to proteins of interest (POIs) within the particular living specimen under investigation.1,8,9 Therefore, multi-color photoactivation experiments inside living cells have been mainly restricted to the application of photo-modulatable FPs so far.11,12 Antibody-labeling using caged chemical fluorophores is limited to applications in fixed cells.13,14 Previously reported benzyl-guanine (BG) conjugates bearing caged fluorophores, as applied in SNAP-protein labeling technology possess poor cell permeabilities and have mainly been restricted to either fixed cells, to the outside of live cells or relied on harsh loading techniques.8,9 Recently, novel super-resolution microscopy (SRM) techniques such as photo-activated localization microscopy (PALM) were developed to push the resolution of light microscopy images beyond the diffraction limit and to allow imaging of cellular components at nanometer resolution.15,16 In general, PALM requires specifically designed and targetable fluorochromes that can be selectively switched from dark ‘off’ to bright ‘on’ states.17 Caged fluorophores carry photo-labile groups to be maintained in non-fluorescent dark states. Upon irradiation with light of appropriate wavelength, the photo-labile cages fall off and the (prior dark) probe is converted into a bright fluorophore. In PALM, only a sparse subset of fluorophores is stochastically activated, recorded and bleached over the course of thousands of raw frames. Here, the precision of localizing individual emitters depends inversely on the square root of the number of collected photons.18 Before going to the dark due to photo-bleaching, even the most photo-stable fluorescent proteins (FPs) on average emit 10-fold fewer photons compared to organic fluorophores with good photophysical properties.19,20 In addition, only a sparse repertory of green photoactivatable FPs (PA-FPs) is currently available with sufficient good photo-physical properties for single-molecule-based imaging.12,21 Moreover, PA-FPs come with different limitations like poor photophysical properties such as low photostabilities, poor activation properties,22 high background, low contrast ratios and photon yields,11 slow maturation behaviors, sizes or oligomeric states.23 Therefore, bright and stable synthetic fluorophores with high photon output offer a significant advantage over conventional FPs in this respect, even though previous SR experiments predominantly relied on photo-controllable FPs.24–28 Compared to conventional FPs, the color palette of photosensitive representatives is limited. Even a smaller number of FPs such as photoactivatable GFP (PA-GFP) and photoactivatable mCherry (PA-mCherry1) are spectroscopically distinguishable.28,29 In contrast, small organic fluorophores allow for the synthetic tuning of spectral properties, solubility and target specificity.10,30,31 As chemical fluorophores can be conjugated to proteins utilizing a variety of protein tagging strategies in living cells,32 they have the potential to become valuable tools for SRM.33 Lee et al. reported SRM of live bacteria, based on photoactivatable fluorophores that were localized to POIs via HaloTagging.7 However, the visualization of multi-parametric dynamic processes inside live cells on the basis of controlled activation of specifically localized, caged chemical fluorophores with good photophysical properties and extendable applicability for SRM inside live mammalian cells has not been realized so far. These approaches require cell-permeable, photo-modulatable, spectrally separable, bright and photostable small-molecule fluorescent probes to be simultaneously and specifically introduced into particular genetically encoded, protein-specific attachment sites (tags) in living cells.34,35 The eDHFR-tag and the HaloTag are well known for live cell protein labeling and both provide substrates with superior cell permeability. The eDHFR-tag is based on the high affinity interaction of the antibiotic 5-(3,4,5-trimethoxybenzyl)pyrimidine-2,4-diamine (trimethoprim, TMP) to the protein dihydrofolate-reductase (eDHFR) from Escherichia coli (E. coli).36 Fluorophores conjugated to TMP have been shown to possess excellent cell permeability37 and recently found application in live cell direct stochastic optical reconstruction microscopy (dSTORM).38 The ligand-reactive haloalkane dehalogenase tag (HaloTag)39 is based on the presence of a haloalkane attached to the probe (Scheme 1).This renders the fluorophore ligand small, which is likewise beneficial to pass cell membranes. Therefore, the combination of both aforementioned labeling strategies helps us to overcome persisting limitations concerning cell permeability for probe delivery and protein labeling inside live mammalian cells.
Here we present the combined application of previously reported orthogonal protein tagging strategies (eDHFR-tag36 and Halo-tag39) for the installation of custom designed, spectrally separable and photoactivatable fluorescein and Q-rhodamine to various protein targets for advanced diffraction-limited fluorescence microscopy and super-resolution imaging within live cell contexts.
Results and discussion
Synthesis and photo-spectrometric analysis and characterization of photoactivatable probes
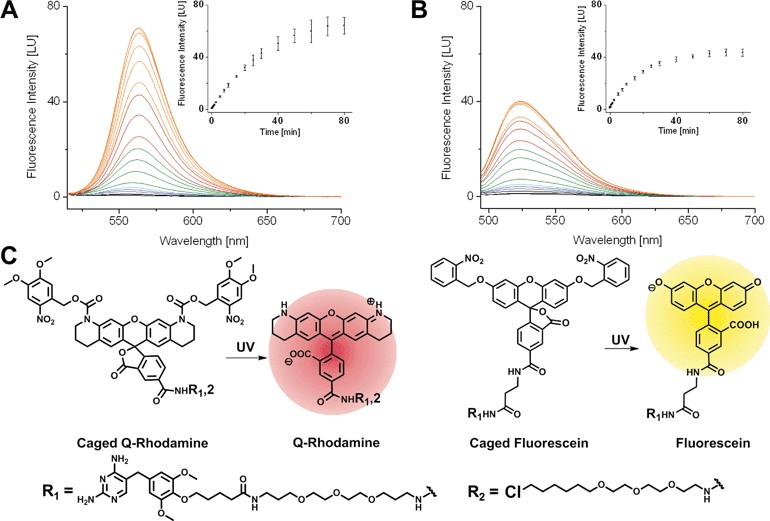
First, we synthesized 5-carboxyfluorescein, bearing photo-removable ortho-nitrobenzyl groups on the hydroxyl function of the xanthone according to literature protocols.1ortho-Nitroveratryloxycarbonyl-5-carboxy-Q-rhodamine (PA-Q-Rh) was commercially available.3 TMP was modified with an amine bearing linker as described previously37 and finally coupled to the caged Fl and caged Q-Rh via peptide bond formation to yield the TMP–PA-Fl respective TMP–PA-Q-Rh conjugates. Halo–PA-Q-Rh was synthesized accordingly. Structures of synthesized dyes are shown in Fig. 1. As non-photoactivatable controls we also synthesized fluorescent TMP- and Halo-conjugates with di-acetyl-fluorescein (diAcFl) and tetramethyl-rhodamine (TMR). These conjugates also found application as complementary fluorophore in dual-color imaging when only one photoactivatable fluorophore was used. Prior to live cell applications, caged conjugates were tested for activation by light in photo-spectrometric in vitro measurements (Fig. 1). Conjugates bearing the caged Q-rhodamine showed 46-fold increase of fluorescence upon UV-irradiation (Fig. 1A). The caged fluorescein containing conjugates showed 23-fold increase in fluorescence at the emission maximum (Fig. 1B).Live cell mono- and dual-color photoactivation of fluorophore-conjugates, localized to target proteins in nuclei, mitochondria and focal adhesions
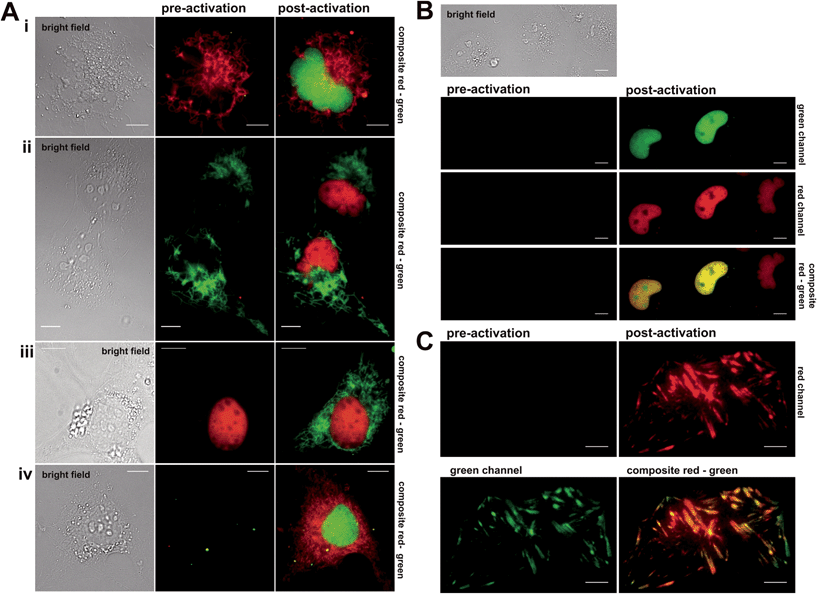
Next, the synthesized conjugates were evaluated in protein labelling experiments inside different live mammalian cellular regimes for their general labelling properties (see (ESI), Fig. S1 and S2†). Within these experiments we used epifluorescence microscopy to test for photoactivation by using a standard DAPI (4′,6-diamidin-2-phenylindol) filter. We prepared various plasmids that express protein constructs containing either the Halo- or eDHFR-tag. To demonstrate the orthogonality in labelling, Halo- and eDHFR-fusions of the histone proteins H2A and H2B were chosen, as well as the mitochondrial import receptor subunit Tom20 (translocase of outer mitochondrial membrane). Live COS-7 cells were transiently transfected and incubated with spectrally separable photoactivatable TMP- and Halo-fluorophore conjugates. That way, the synthesized probes were evaluated for membrane permeability, cell toxicity, photoactivation properties and selectivity for target proteins. Dual-color imaging of histones H2A and H2B in live cells was successfully tested using non-activatable dyes Halo–TMR and TMP–diAcFl first (see Fig. S3 in the ESI†). Photoactivation in epifluorescence mode was conducted by exposure of transfected and stained cells to UV light of the DAPI channel, which exhibited spectral ranges that were ideal for the cleavage of the applied photolabile caging groups (352–402 nm, 14.5 mW cm−2). First, mono-color photoactivation was established on histone proteins H2A and H2B, using Halo–PA-Q-Rh or Halo–TMR and TMP–PA-Fl. We observed increasing fluorescence intensity within nuclei over exposure time to light from the DAPI channel (see Fig. S4–S6 in the ESI†). Fig. 2 shows the experimental results from mono- and dual-color activation experiments of photoactivatable TMP and Halo-fluorophores inside different mammalian cell lines. For mono-color photoactivation experiments, we transfected COS-7 cells with H2B–eDHFR and Halo–Tom20 and incubated with either TMP–PA-Fl (5 μM) or Halo–TMR (1 μM) (Fig. 2A, upper panel, i) or TMP–PA-Q-Rh (5 μM) and Halo–diAcFl (1 μM) (Fig. 2A, second panel, ii). In both experiments we observed strong fluorescence signals from photoactivated fluorophores, localized to nuclei upon 60 s of irradiation with UV-light (DAPI-filtered) (Fig. 2i and ii, post activation images). Next, Tom20–eDHFR and Halo–H2B were transiently co-overexpressed in live COS-7 cells which were stained with TMP–PA-Fl (5 μM) and Halo–TMR (1 μM) (Fig. 2A, third panel, iii). Subsequent photoactivation revealed the filigree structure of the mitochondrial network in the green fluorescent channel. In general, no fluorescence signal emerged from the caged dyes in pre-activation recordings (middle column), while localized, non-activatable fluorophore conjugates revealed constant signal pre- and post-activation (Fig. 2A, panels i–iii). These initial mono-color activation experiments clearly demonstrate that the synthesized caged fluorophore conjugates are well suited for live cell photoactivation experiments. Both TMP- and Halo-photoactivatable fluorophore conjugates readily penetrate live cell membranes with incubation times no longer than 30 min. Cell viability was not affected (Fig. S15†). Visible signs of cell toxicity of synthesized dye conjugates could not be observed even after 12 h of incubation. Very low cellular autofluorescence and negligible background signal of the (caged) fluorescent probes were determined in non-transfected cells (see Fig. S7 in the ESI†). Dual-color photoactivation was approached on nuclear and cytosolic target proteins. For this, we transiently over-expressed nuclear H2B–eDHFR and diffusive EGFP–Halo in COS-7 cells which were stained with TMP–PA-Fl (5 μM) and Halo–PA-Q-Rh (5 μM). Exposure to light from the DAPI channel for 120 s was enough for strong signal in both fluorescent channels (see Fig. S8 in the ESI†). Based on these results, we simultaneously localized two spectrally separable caged fluorescent dyes to nuclear and mitochondrial target proteins for live cell dual-color photoactivation. For this, NIH 3T3 fibroblasts were transiently transfected with H2B–eDHFR and Halo–Tom20 and stained with both TMP–PA-Fl and Halo–PA-Q-Rh (5 μM, each). As expected, we did not observe any fluorescence signal prior to activation (Fig. 2A, lower panel iv, middle column). Upon irradiation with DAPI-filtered UV light, the fluorescence signal significantly increased in the green channel from the photoactivated fluorescein at the nucleus as well as at mitochondrial structures from the photoactivated Q-Rh (Fig. 2A, lower panel iv, right column). Further, we demonstrate dual-color photoactivation of TMP–PA-Fl and Halo–PA-Q-Rh, localized to histones H2A–Halo and H2B–eDHFR in COS-7 cells. Strong, nucleus-localized fluorescent signal in both red and green channels was detected, following UV irradiation (Fig. 2B).Having successfully shown live cell dual-color photoactivation of localized fluorophores for mitochondrial and nuclear targets in epifluorescence mode, we wanted to demonstrate the application of the herein presented, combined protein labeling strategies for photoactivatable fluorophores within more complex structural cellular regimes. For this, we selected the focal adhesion protein vinculin (Vcl) to be visualized by the herein presented localizable photoactivable fluorophore conjugates. Focal adhesions are macromolecular protein assemblies that link the extracellular matrix (ECM) to the individual cell. The so-called adhesion plaques are located close to the cell membrane which is in contact with the substrate surface at the water–glass interface of the cover slip. Therefore, focal adhesions and the proteins integrated into this protein assembly can be ideally monitored via total internal reflection microscopy (TIRFM). We prepared Halo–EGFP-Vcl and eDHFR–EGFP–Halo constructs that were expressed in mouse embryonic fibroblasts (MEF) Vcl-knock-out (Vcl−/−) cells, which were then incubated with Halo–PA-Q-Rh respective TMP–PA-Q-Rh (5 μM, each). The signal from the EGFP-tag in the green channel was used to identify the focal plane prior to activation. Fig. 2C depicts the photoactivation experiment, with PA-Q-Rh localized to Halo–EGFP–Vcl (or eDHFR–EGFP–Vcl, see Fig. S10 and S11 in the ESI†). Photoactivation was achieved upon illumination with a 405 nm laser (60 mW cm−2, 50% laser power). Optimal activation intervals were determined by repetitively exposing a selected ROI (region of interest) (405 nm, 5 s-intervals). For maximum activation of localized caged Q-rhodamine conjugates, optimal exposure times were determined to 5–10 s (50% laser power) or 50–60 s (10% laser power). Prior to photoactivation, no rhodamine fluorescence signal was observed in the red channel (Fig. 2C, upper left) whereas localization of Vcl was constantly monitored by EGFP fluorescence (Fig. 2C, green channel, bottom left image). After photoactivation, the bright fluorescence signal of the activated rhodamine was detectable in the red channel (Fig. 2C, upper right image). Excellent co-localization to the EGFP fluorescence signal was observed, as indicated by the merged images of the red and green channel (Fig. 2C, lower right image and Fig. S11†).
In all experiments, photoactivation was either performed by a laser at 405 nm or via DAPI-filtered UV light. Most epifluorescence or confocal microscopes are equipped with such light sources, which makes the implementation of the presented approach straightforward at low costs.
Photoactivated fluorophores allow dynamic studies of proteins inside live cells by inverse fluorescence recovery after photobleaching (iFRAP)
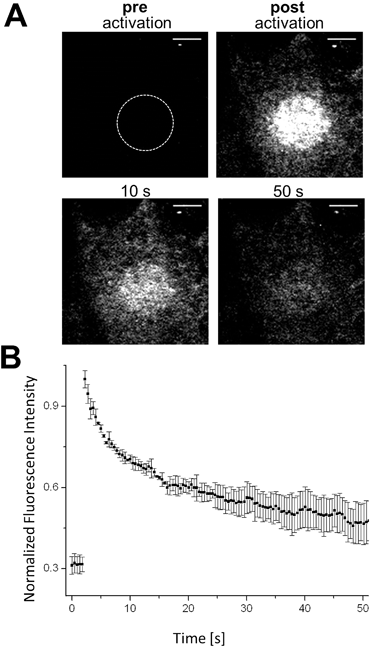
The controlled change of a non-fluorescent ‘off’-state to a fluorescent ‘on’-state (and vice versa) in a defined region can be used to retrieve information about dynamic processes in living biological systems. Fluorescence recovery after photobleaching (FRAP) is a common technology to investigate diffusion processes of proteins in live cells.40,41 Whereas FRAP is based on the bleaching of fluorophores (i.e. the irreversible change from the bright ‘on’- to dark ‘off’-state), the light-mediated activation of a fluorophore (i.e. conversion from the ‘off’- to the ‘on’-state) allows the reversed approach, termed ‘inverse-‘FRAP (iFRAP). iFRAP represents an excellent alternative to conventional FRAP experiments. As the recovery of fluorescence emerges from the mobility of molecules within the surroundings of a bleached spot, classical FRAP is often referred to as an imprecise method for the determination of biomolecular dynamics. Monitoring membrane dynamics via FRAP, fluorescence recovery might emerge from insertion of non-bleached fluorescently tagged proteins into the membrane, as well as due to the lateral mobility of surrounding molecules.9Photoactivation of chemically labeled cell surface proteins permits to overcome this problem as only photoactivated molecules originating from the illuminated region of interest (ROI) at the membrane are tracked. As we have shown that our photoactivatable fluorophores can be selectively localized to individual POIs, we utilize our Halo–PA-Q-Rh to gain information about the dynamic behaviour of a protein inside a living cell by means of an iFRAP experiment. Q-Rh is a very photostable dye which is ideally suited for the long-term tracking of molecule dynamics. To track the dynamics of proteins at the plasma membrane (PM), live HeLa cells were transfected with Halo-tagged lymphocyte-specific protein tyrosine kinase (Halo-Lck), with Lck localizing the Halo-tag at the inside of the PM. Transfected cells were stained with Halo–PA-Q-Rh (5 μM). Using this PM construct, we observed the expected fast diffusion dynamics with a half-residence time of 7.8 s and an equilibration time of 47.4 s. Activation was conducted in pre-defined ROIs (diameter: 10 μm) at 405 nm (60 mW cm−2), which resulted in an immediate stepped increase of the fluorescence signal (Fig. 3B). The fluorescence signal exponentially decreased as expected to reach a plateau, slightly above the starting value after 40–50 s of observation (Fig. 3B). To further show the suitability of photoactivatable fluorophore conjugates to follow processes with slow dynamics, we chose rather slowly rearranging chromatin as a suitable target structure (Fig. S12†). COS-7 cells were transiently transfected with H2B–Halo and stained with Halo–PA-Q-Rh. Circular ROIs (4 μm diameter) for photoactivation were pre-defined within live cell nuclei. Halo–PA-Q-Rh, localized to H2B was activated by illumination at λ = 405 nm with 60 mW cm−2 laser power. As expected, the observed dynamics of the tagged H2B are much slower.42 The experiments demonstrate that the photoactivatable fluorophore conjugates can be used to study protein dynamics of different rates in live cells with iFRAP. Nevertheless, it has to be noted that background signal from unbound photoactivatable fluorophores can falsify the results as they have significant faster dynamics than the tagged protein and can result in a multi exponential decay of the signal.
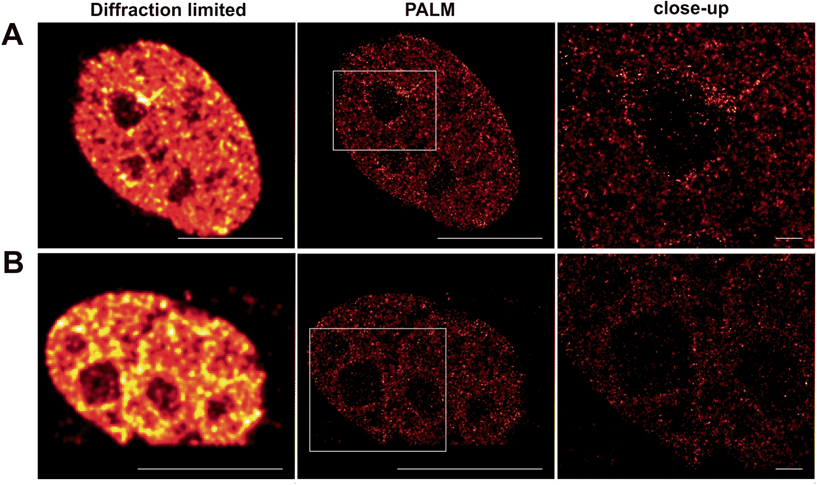
Localizable photoactivatable Q-rhodamine in super-resolution microscopy
The molecular characteristic of photoactivatable fluorophores to be actively switched from a fluorescent dark ‘off-state’ to a bright fluorescent ‘on-state' is a fundamental requirement to perform pointillistic super-resolution fluorescence microscopy.Therefore, we intended to further apply the herein presented localizable photoactivatable fluorophores in combination with the super-resolution microscopy technology PALM. The original publication inventing PALM in 2006 (ref. 15) utilized PA-GFP. However, we would like to point out that the authors already noted that photoactivatable small organic fluorophores like PA-Q-Rh would be highly beneficial for PALM.15
Whereas dSTORM entirely relies on the reversible photoswitching of fluorochromes, herein presented caged Q-rhodamine-conjugates are activatable only once and allow for their tracking of molecular targets over many frames from then on. The obvious reason is the high number of photons that can be collected from such small organic fluorophores before going to the dark. Such high photon yields offer significant improvement in resolution, as the precision of localizing individual emitters depends inversely on the square root of the number of collected photons.18 In particular live-cell PALM critically depends on the technical limitation of collecting enough photons from single molecule emitters within very short time periods, to counter the mobility of the living specimen. As SRM is still commonly performed on fixed cellular targets, even the herein presented caged fluorescent conjugates were first tested for their precise labeling properties of cellular POIs inside fixed cells for PALM. For this, Tom20–Halo was transiently overexpressed in NIH 3T3 cells which were then stained with Halo–PA-Q-Rh and subsequently fixed, using paraformaldehyde. PALM revealed the mitochondrial network in high contrast (Fig. S13 in the ESI†). Encouraged by the successful application on fixed targets, we decided to use the nuclear target H2B to demonstrate the applicability of our probes for live cell PALM. For this, NIH 3T3 cells were transiently transfected with H2B–Halo, respective H2B–eDHFR to be stained with Halo–PA-Q-Rh or TMP–PA-Q-Rh and imaged by PALM (Fig. 4). Information extracted from the raw data was used to calculate the statistics on molecular photon yields and localization precision. The PA-Q-Rh conjugates showed excellent photophysical properties after photoactivation, with much higher mean photon yields than photoactivatable-mCherry (PA-mCherry) (Halo–Q-PA-Rh and TMP–Q-PA-Rh 410% respectively 312% of the mean photon yield determined for PA-mCherry). The increased molecular brightness is translated into an improvement in localization, demonstrated by the median value of uncertainty in position, which is ∼10 nm for Halo–Q-PA-Rh and TMP–Q-PA-Rh, a factor of about 2.5 better than for PA-mCherry (∼25 nm) (Fig. S14†). The results demonstrate that the herein present PA-Q-Rh conjugates are excellent candidates for PALM on live cell targets.
Conclusions
In summary, we herein presented caged, spectrally separable fluorophore conjugates for dual-color imaging and SRM inside living cells. Custom-designed caged conjugates with excellent cell permeabilities selectively label intracellular tagged POIs. We successfully demonstrated the labeling and photoactivation of caged fluorescein and caged Q-rhodamine within different cellular regimes such as nuclear-, mitochondrial-, cytosolic-, membrane- and focal adhesion localized proteins. To our knowledge, the presented work is the first example of well cell-permeable photoactivatable fluorescent probes for multi-color protein labeling in live mammalian cells. We further demonstrate the application of our probes for analyzing protein dynamics in live cell membranes, using iFRAP. We finally exploit the superior molecular brightness and photostability of the herein applied Q-rhodamine for advanced live cell super-resolution imaging, using PALM. The presented conjugates represent attractive experimental alternatives to PA-FPs, especially when high photo-stability and brightness is required. The herein demonstrated combination of the photoactivatable fluorophore conjugates with protein labeling in live cells unleash the enormous potential of photoactivatable small organic fluorophores for live cell super-resolution microscopy.Acknowledgements
R. W. acknowledges funding from the Deutsche Forschungsgemeinschaft DFG (SPP1623, WO 1888/1-2). S. H. was supported by a stipend of the Studienstiftung des Deutschen Volkes. The authors thank Christian Tischer and Stefan Terjung from the Advanced Light Microscopy Facility at the European Molecular Biology Laboratory for support with iFRAP and TIRFMicroscopy as well as image analysis. We also thank Ulrike Müller (IPMB, Ruprecht-Karls-Universität) for access to microscopes. We further would like to acknowledge Marcel Best, Dirk Ollech and Jessica Schwab for technical support and thank Wolfgang H. Goldmann (Universität Erlangen-Nürnberg) for providing MEF Vinc−/− cells and Felix Hövelmann (EMBL) for the Halo-Lck construct. We gratefully acknowledge Virginia W. Cornish (Columbia University, NY) for advice and generous support.Notes and references
- T. J. Mitchison, J. Cell Biol., 1989, 109, 637–652 CrossRef CAS PubMed.
- J. C. Politz, Trends Cell Biol., 1999, 9, 284–287 CrossRef CAS PubMed.
- K. R. Gee, E. S. Weinberg and D. J. Kozlowski, Bioorg. Med. Chem. Lett., 2001, 11, 2181–2183 CrossRef CAS PubMed.
- T. Kobayashi, Y. Urano, M. Kamiya, T. Ueno, H. Kojima and T. Nagano, J. Am. Chem. Soc., 2007, 129, 6696–6697 CrossRef CAS PubMed.
- J. Ottl, D. Gabriel and G. Marriott, Bioconjugate Chem., 1998, 9, 143–151 CrossRef CAS PubMed.
- Y. Zhao, Q. Zheng, K. Dakin, K. Xu, M. L. Martinez and W.-H. Li, J. Am. Chem. Soc., 2004, 126, 4653–4663 CrossRef CAS PubMed.
- H. D. Lee, S. J. Lord, S. Iwanaga, K. Zhan, H. Xie, J. C. Williams, H. Wang, G. R. Bowman, E. D. Goley, L. Shapiro, R. J. Twieg, J. Rao and W. E. Moerner, J. Am. Chem. Soc., 2010, 132, 15099–15101 CrossRef CAS PubMed.
- S. Banala, D. Maurel, S. Manley and K. Johnsson, ACS Chem. Biol., 2012, 7, 289–293 CrossRef CAS PubMed.
- D. Maurel, S. Banala, T. Laroche and K. Johnsson, ACS Chem. Biol., 2010, 5, 507–516 CrossRef CAS PubMed.
- L. M. Wysocki, J. B. Grimm, A. N. Tkachuk, T. A. Brown, E. Betzig and L. D. Lavis, Angew. Chem., Int. Ed. Engl., 2011, 50, 11206–11209 CrossRef CAS PubMed.
- J. Lippincott-Schwartz and G. H. Patterson, Trends Cell Biol., 2009, 19, 555–565 CrossRef CAS PubMed.
- D. M. Shcherbakova, P. Sengupta, J. Lippincott-Schwartz and V. V. Verkhusha, Annu. Rev. Biophys., 2014, 43, 303–329 CrossRef CAS PubMed.
- M. Heilemann, S. van de Linde, A. Mukherjee and M. Sauer, Angew. Chem., 2009, 121, 7036–7041 CrossRef.
- J. Fölling, M. Bossi, H. Bock, R. Medda, C. A. Wurm, B. Hein, S. Jakobs, C. Eggeling and S. W. Hell, Nat. Methods, 2008, 5, 943–945 CrossRef PubMed.
- E. Betzig, G. H. Patterson, R. Sougrat, O. W. Lindwasser, S. Olenych, J. S. Bonifacino, M. W. Davidson, J. Lippincott-Schwartz and H. F. Hess, Science, 2006, 313, 1642–1645 CrossRef CAS PubMed.
- D. Toomre and J. Bewersdorf, Annu. Rev. Cell Dev. Biol., 2010, 26, 285–314 CrossRef CAS PubMed.
- M. Fernández-Suárez and A. Y. Ting, Nat. Rev. Mol. Cell Biol., 2008, 9, 929–943 CrossRef PubMed.
- R. E. Thompson, D. R. Larson and W. W. Webb, Biophys. J., 2002, 82, 2775–2783 CrossRef CAS PubMed.
- M. A. Thompson, J. S. Biteen, S. J. Lord, N. R. Conley and W. E. Moerner, Methods Enzymol., 2010, 475, 27–59 CAS.
- T. Schmidt, U. Kubitscheck, D. Rohler and U. Nienhaus, Single Mol., 2002, 3, 327 CrossRef CAS.
- K. A. Lukyanov, D. M. Chudakov, S. Lukyanov and V. V. Verkhusha, Nat. Rev. Mol. Cell Biol., 2005, 6, 885–891 CrossRef CAS PubMed.
- J. S. Marchant, G. E. Stutzmann, M. A. Leissring, F. M. LaFerla and I. Parker, Nat. Biotechnol., 2001, 19, 645–649 CrossRef CAS PubMed.
- J. Wiedenmann, F. Oswald and G. U. Nienhaus, IUBMB Life, 2009, 61, 1029–1042 CrossRef CAS PubMed.
- R. Ando, H. Mizuno and A. Miyawaki, Science, 2004, 306, 1370–1373 CrossRef CAS PubMed.
- S. T. Hess, T. J. Gould, M. V Gudheti, S. A. Maas, K. D. Mills and J. Zimmerberg, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 17370–17375 CrossRef CAS PubMed.
- Z. Liu, L. D. Lavis and E. Betzig, Mol. Cell, 2015, 58, 644–659 CrossRef CAS PubMed.
- R. M. Dickson, D. J. Norris, Y. L. Tzeng and W. E. Moerner, Science, 1996, 274, 966–969 CrossRef CAS PubMed.
- G. H. Patterson and J. Lippincott-Schwartz, Science, 2002, 297, 1873–1877 CrossRef CAS PubMed.
- F. V. Subach, G. H. Patterson, S. Manley, J. M. Gillette, J. Lippincott-Schwartz and V. V. Verkhusha, Nat. Methods, 2009, 6, 153–159 CrossRef CAS PubMed.
- V. N. Belov, C. A. Wurm, V. P. Boyarskiy, S. Jakobs and S. W. Hell, Angew. Chem., Int. Ed. Engl., 2010, 49, 3520–3523 CrossRef CAS PubMed.
- L. D. Lavis, J. B. Grimm, L. M. Wysocki, A. N. Tkachuk, T. A. Brown and E. Betzig, Microsc. Microanal., 2012, 18, 668–669 CrossRef.
- R. Wombacher and V. W. Cornish, J. Biophotonics, 2011, 4, 391–402 CrossRef CAS PubMed.
- F. M. Raymo, J. Phys. Chem. Lett., 2012, 3, 2379–2385 CrossRef CAS PubMed.
- R. Wieneke, A. Raulf, A. Kollmannsperger, M. Heilemann and R. Tampé, Angew. Chem., Int. Ed., 2015, 54, 10216–10219 CrossRef CAS PubMed.
- A. Kollmannsperger, A. Sharei, A. Raulf, M. Heilemann, R. Langer, K. F. Jensen, R. Wieneke and R. Tampé, Nat. Commun., 2016, 7, 10372 CrossRef CAS PubMed.
- L. W. Miller, Y. Cai, M. P. Sheetz and V. W. Cornish, Nat. Methods, 2005, 2, 255–257 CrossRef CAS PubMed.
- N. T. Calloway, M. Choob, A. Sanz, M. P. Sheetz, L. W. Miller and V. W. Cornish, ChemBioChem, 2007, 8, 767–774 CrossRef CAS PubMed.
- R. Wombacher, M. Heidbreder, S. van de Linde, M. P. Sheetz, M. Heilemann, V. W. Cornish and M. Sauer, Nat. Methods, 2010, 7, 717–719 CrossRef CAS PubMed.
- G. V. Los, L. P. Encell, M. G. McDougall, D. D. Hartzell, N. Karassina, C. Zimprich, M. G. Wood, R. Learish, R. F. Ohana, M. Urh, D. Simpson, J. Mendez, K. Zimmerman, P. Otto, G. Vidugiris, J. Zhu, A. Darzins, D. H. Klaubert, R. F. Bulleit and K. V. Wood, ACS Chem. Biol., 2008, 3, 373–382 CrossRef CAS PubMed.
- D. Axelrod, D. E. Koppel, J. Schlessinger, E. Elson and W. W. Webb, Biophys. J., 1976, 16, 1055–1069 CrossRef CAS PubMed.
- J. Lippincott-Schwartz, N. Altan-Bonnet and G. H. Patterson, Nat. Cell Biol., 2003, S7–S14 CAS.
- H. Kimura and P. R. Cook, J. Cell Biol., 2001, 153, 1341–1354 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6sc02088g |
| ‡ Current address: European Molecular Biology Laboratory, Meyerhofstraße 1, 69117 Heidelberg, Germany. |
| This journal is © The Royal Society of Chemistry 2017 |