Open Access Article
Open Access ArticleSerine promoted synthesis of peptide thioester-precursor on solid support for native chemical ligation†
Hader E.
Elashal
,
Yonnette E.
Sim
and
Monika
Raj
*
Department of Chemistry, Seton Hall University, 400 South Orange Ave, South Orange, NJ 07079, USA. E-mail: monika.raj@shu.edu
First published on 16th August 2016
Abstract
Fmoc solid phase peptide synthesis of thioesters for the chemical synthesis of proteins via native chemical ligation is a challenge. We have developed a versatile approach for direct synthesis of peptide thioesters from a solid support utilizing Fmoc chemistry. Peptide thioester synthesis is performed by the formation of a cyclic urethane moiety via a selective reaction of the backbone amide chain with the side group of serine. The activated cyclic urethane moiety undergoes displacement by a thiol to generate the thioester directly from the solid support. Importantly, the method activates the serine residue for the synthesis of peptide thioesters; thus it is fully automated and free of the types of resins, linkers, handles, and unnatural amino acids typically needed for the synthesis of peptide thioesters using Fmoc chemistry. The resulting thioester is free of epimerization and is successfully applied for the synthesis of longer peptides using NCL.
Introduction
Total chemical synthesis of proteins provides easy access to various modified proteins with high stability and improved biological activity which can be key in understanding the importance of various post translational modifications.1,2 Native Chemical Ligation (NCL) has revolutionized the field of chemical synthesis of proteins and relies on the reaction of a peptide thioester with a cysteinyl peptide.3 Thus, considerable effort has been applied to the synthesis of peptide thioesters using Fmoc or Boc solid-phase peptide synthesis (SPPS) approaches.The common method for generation of peptide thioesters by Boc-based SPPS utilizes special thioester linkers; nonetheless, Boc chemistry is unfavorable due to the use of harsh conditions, usually HF, for peptide cleavage, which makes this method incompatible for the synthesis of post translationally modified peptides such as glyco- or phospho-peptides.4 Fmoc SPPS is not applicable on thioester resins because of the instability of peptide thioesters towards the piperidine treatment required for the deprotection of the Fmoc group. Substantial efforts have been directed towards the synthesis of peptide thioesters by the Fmoc SPPS approach, such as the use of less basic Fmoc deprotection protocols,5 the use of sulfonamide safety catch linkers,6 activation of protected peptides in solution,7 O/N to S acyl shift methods,8–14 and peptide hydrazides.15–17 The activation of protected peptides in solution for the synthesis of thioesters is limited by the susceptibility of the C-terminal residue to racemization.7 Despite significant advances in O/N to S acyl shift methods, they are limited by the requirement for unnatural linkers.13,14
Another significant approach for the formation of thioesters is the activation of a relatively inert backbone amide bond followed by subsequent thiolysis. Examples include an activated N-acylurea linker18,19 or a backbone pyroglutamyl imide linker.20 The activated N-acylurea approach has been extensively applied to the synthesis of a variety of proteins18,19 but is limited by the use of unnatural amino acids, the formation of various side products,21,22 and the requirement of additional steps for the synthesis of N-methylated unnatural amino acid.19 On the other hand, backbone pyroglutamyl imide linkers are limited by the instability of the resin when stored for 24 h.20 Thus, a simple and versatile route to peptide thioesters via Fmoc SPPS is highly desirable.
Herein, we report an alternative approach for the Fmoc SPPS of peptide thioesters that overcomes some of the challenges associated with existing methods. The methodology is based on the activation of the amide backbone chain at the serine residue. The significance of this approach is that it utilizes natural serine residue and does not require special unnatural amino acids or loading procedures, which makes it distinct from the currently used methods.18,19 We believe that this accessible and robust Fmoc-based thioesterification technique provides a significant advance in chemical protein synthesis due to its uncomplicated nature, thereby eliminating the special precautions and additional steps typically needed for the synthesis of peptide thioesters.18,19
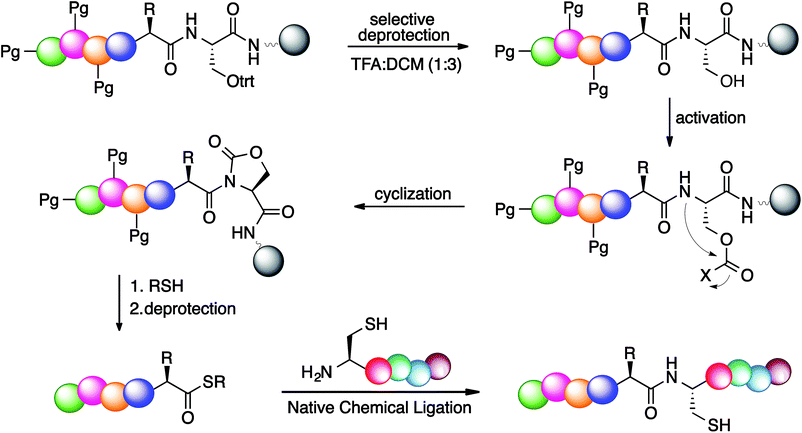
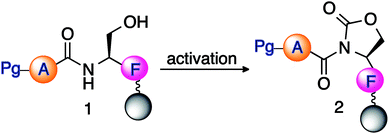
This approach relies on the selective activation of the backbone amide bond at the C-terminal serine residue for generation of the cyclic urethane moiety on a solid support. The synthesis of peptide thioesters by this strategy entails the anchoring of a C-terminal serine residue with a selectively removable side-chain protecting group to a solid support (Scheme 1). After chain assembly, the side-chain of serine is deprotected and activated by an electrophile, resulting in the formation of a cyclic urethane moiety on the resin (Scheme 1). Recently, we have reported a method for the selective cleavage of peptide bonds in solution for protein sequencing by employing the backbone amide activation strategy.23 Next, nucleophilic displacement of the cyclic urethane moiety by treatment with a thiol releases the peptide thioester from the solid support, which can then be deprotected in solution (Scheme 1). It is noteworthy that this approach utilizes serine during chain assembly followed by acylation of the backbone amide chain, and is compatible with various amino acids and protecting groups commonly utilized in Fmoc SPPS. Moreover, the thioester is directly obtained from the solid support, thus reducing the need for extra steps before proceeding with NCL, which is in contrast to traditional methods.
Results and discussion
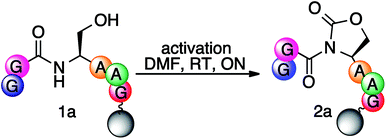
To test the proposed methodology for the synthesis of thioesters, the first step was the synthesis of the cyclic urethane moiety from the serine residue on solid support. To optimize the formation of the cyclic urethane moiety on solid support, a model hexapeptide Ac-GGSAAG was assembled on solid support and the trt group was selectively deprotected from C-terminal serine using TFA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DCM.
DCM.
For activation of the side chain of serine, various electrophilic reagents such as 4-nitrophenyl chloroformate, 1,1′-carbonyldiimidazole (CDI) and N,N′-disuccinimidyl carbonate (DSC) were screened (entries 1–4, Table 1). Quantitative conversion of peptide Ac-GGSAAG 1a to the cyclic urethane moiety Ac-GGOxdAAG 2a was observed by using DSC as an electrophile (entry 4, Table 1). The conversion yields were calculated from the HPLC data. The formation of the cyclic urethane moiety was confirmed by LCMS and NMR spectroscopy (ESI†). To further optimize the formation of the cyclic urethane moiety on solid support, various other reaction conditions such as solvent, time, temperature, and equivalents of reagents were explored for a model peptide, Fmoc-ASF-Rink AM 1b (entries 1–6, Table 2). The low conversion to activated peptide 2b (40%) in DCM is due to the poor solubility of DSC in DCM (entry 1, Table 2). When a mixture of DCM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMF (1
DMF (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was used as a solvent, the conversion to 2b increased from 40 to 60%, whereas when the reaction was performed in DMF, the conversion increased to 99% (entries 2–3, Table 2). This is due to the high solubility of DSC in DMF.
1) was used as a solvent, the conversion to 2b increased from 40 to 60%, whereas when the reaction was performed in DMF, the conversion increased to 99% (entries 2–3, Table 2). This is due to the high solubility of DSC in DMF.
| Entry | Reagent | Additive | Conv.b (%) |
|---|---|---|---|
| a Reaction conditions: to peptide Ac-GGSAAG 1a on solid support (25 mg, 0.52 mm g−1), a solution of 4-nitrophenylchloroformate/CDI/DSC (10 equiv.), DIEA (10 equiv.) and DMAP (catalytic amount) in DMF (3 mL) was added and the resin was left on a shaker overnight (ON) at room temperature (RT). b Conversion to Ac-GGOxdAAG 2a was calculated from the absorbance at 220 nm using HPLC. | |||
| 1 | 4-Nitrophenyl chloroformate | — | 0 |
| 2 | 4-Nitrophenyl chloroformate | DMAP | 20 |
| 3 | CDI | DMAP | 60 |
| 4 | DSC | DMAP | 99 |
| Entry | Pg | DSC (equiv.) | Solvent | Time (h) | Conv.b (%) |
|---|---|---|---|---|---|
| a Reaction conditions: peptide 1 (25 mg, 0.87 mm g−1) on solid support was reacted with DSC (5–10 equiv.), DIEA (5–10 equiv.), and a crystal of DMAP in different solvents (3 mL) at room temperature for 3–17 h. b Conversion to cyclic urethane moiety 2 was calculated from the absorbance at 220 nm using HPLC. | |||||
| 1 | 1b Fmoc | 10 | DCM | 17 | 40 |
| 2 | 1b Fmoc | 10 | DCM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMF DMF |
17 | 60 |
| 3 | 1b Fmoc | 10 | DMF | 17 | >99 |
| 4 | 1b Fmoc | 5 | DMF | 17 | 70 |
| 5 | 1b Fmoc | 10 | DMF | 3 | 30 |
| 6 | 1b Fmoc | 10 | DMF | 7 | 90 |
| 7 | 1c Ac | 10 | DMF | 17 | >99 |
| 8 | 1d tosyl | 10 | DMF | 17 | >99 |
| 9 | 1e Boc | 10 | DMF | 17 | >99 |
Next, to determine the role of the N-terminal protecting group (Pg) in the formation of the cyclic urethane moiety, a model peptide Pg-ASF-Rink AM with other N-terminal protecting groups such as acetyl, tosyl, and Boc was investigated (entries 7–9, Table 2). Contrary to earlier studies,24 the results indicated that the formation of the cyclic urethane moiety from serine by activating the backbone peptide chain is independent of the nature of the N-terminal protecting group.
Subsequently, the serine activation reaction was explored on solid support with various peptides containing different sequences (Table S1, ESI†). It is noteworthy that the formation of the activated cyclic urethane moiety is independent of the nature of the amino acid preceding serine, and high conversion to the cyclic urethane moiety was obtained even with bulky amino acid residues neighbouring serine (Table S1, ESI†). Moreover, the results in Tables 1, 2 and S1† showed that the activation of serine does not depend upon the relative position of serine and the resin.
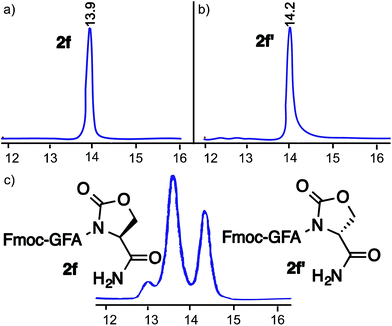
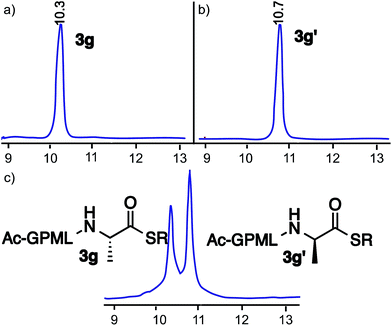
The stability of the cyclic urethane moiety on the resin-bound peptide was also evaluated. The resin-bound cyclic urethane moiety was found to be very stable in a desiccator for longer than a month, which is in contrast to the previously reported pyroglutamyl imide method.20 Moreover, the formation of the cyclic urethane moiety on peptide Fmoc-GFA(L)Oxd (2f) and corresponding diastereoisomer Fmoc-GFA(D)Oxd (2f′) showed no sign of epimerization as determined by HPLC (Fig. 1).
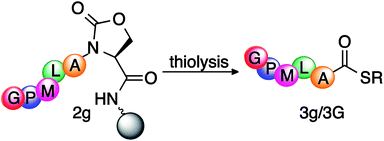
After activating the peptide backbone to form a cyclic urethane moiety on model peptide Ac-GPMLA-Oxd-Rink AM (2g) on solid support, the thiolysis reaction was performed (Table 3). The resin was treated with varying amounts of thiol/base mixtures and solvents at different temperatures, in order to release the peptide thioester Ac-GPMLA-COSR 3g into the solution. The resin was then filtered and the solvent was evaporated to obtain peptide thioester 3g, which was analyzed by HPLC and MS (ESI†).
| Entry | Base (equiv.) | Solvent | Temp. (°C) | Time (h) | Conv.b (%) | |
|---|---|---|---|---|---|---|
| 3g | 3G | |||||
| a Reaction conditions: cyclic urethane-activated peptide Ac-GPMLA-Oxd2g (25 mg, 0.7 mm g−1) on solid support was reacted with thiol (100 μL) and base (0.5–20 equiv.) in DMF (1 mL). b Conversion to peptide thioester Ac-GPMLA-COSR 3g or 3G was calculated from the absorbance at 220 nm using HPLC. c Activated peptide was completely released from the resin but hydrolysis product was observed along with the thioester 3g/3G. SR = S-(CH2)2-OH or S-(CH2)2-COOC2H5, RT = room temperature, entries in bold: optimized conditions at room temperature. Thioesterification at high temperature leads to 5% epimerization. | ||||||
| 1 | — | DMF | RT | 20 | 10 | 15 |
| 2 | DBU (5 equiv.) | DMF | RT | 20 | 40c | 60 |
| 3 | DIEA (20 equiv.) | DMF | RT | 20 | 60c | 65 |
| 4 | PhSNa (0.5 equiv.) | DMF | RT | 20 | 85 | 99 |
| 5 | PhSNa (0.5 equiv.) | ACN | RT | 20 | 15 | 25 |
| 6 | PhSNa (0.5 equiv.) | DMF | RT | 5 | 30 | 35 |
| 7 | PhSNa (0.5 equiv.) | DMF | 60 °C | 20 | 10c | 70c |
| 8 | PhSNa (0.5 equiv.) | DMF | 60 °C | 5 | 30c | 99 |
| 9 | PhSNa (0.5 equiv.) | DMF | 60 °C | 3 | 50c | 99 |
Initially, the reaction was performed at room temperature using DMF as solvent in the presence of a thiol and without the use of any base (entry 1, Table 3). This resulted in the release of 10% peptide thioester 3g from the resin after treatment for 20 h. To increase the conversion to peptide thioester 3g, various bases such as DIEA, DBU, and sodium thiolate were investigated (entries 2–4, Table 3). Addition of a catalytic amount of sodium thiolate (0.5 equiv. with respect to resin) as a base significantly increased the yield of peptide thioester 3g released from the resin (85%; entry 4, Table 3). Since it is difficult to remove DMF from the reaction mixture, a more volatile solvent, ACN, was explored, but it resulted in very low conversion to peptide thioester 3g (15%; entry 5, Table 3). The final yields in ACN were low due to the poor solubility of sodium thiolate in ACN. Temperatures higher than 60 °C resulted in significant hydrolysis of peptide thioester 3g into the corresponding acid (entries 7–9, Table 3). The formation of the hydrolyzed product was circumvented by the use of ethyl-3-mercaptopropionate, which gave stable peptide thioester 3G that is less susceptible to hydrolysis, as determined by NMR (ESI†). All reactions were clean at room temperature irrespective of the nature of the thiol used, and did not show significant amounts of hydrolysis.
Potential epimerization during the thioesterification process was rigorously examined by synthesizing peptide thioester Ac-GPMLA(L)-COS(CH2)2OH 3g and epimer Ac-GPMLA(D)-COS(CH2)2OH 3g′ under the optimized reaction conditions. No detectable levels of epimerization were observed by HPLC (Fig. 2). Next, we carried out epimerization studies on peptide thioester Ac-GVALF(L)-COS(CH2)2-COOC2H53j with a bulky Phe residue at the C-terminus by analyzing it with epimer Ac-GVALF(D)-COS(CH2)2-COOC2H53j′ (Fig. S1 and S2, ESI†). Less than 1% epimerization was observed by HPLC (Fig. S2, ESI†). Indeed, Phe is known as an amino acid relatively prone to epimerization, but these results indicated that our new methodology did not afford substantial epimerization.
Another significant feature of the cyclic urethane method is that it is independent of the nature of the resin and works equally well with Tentagel, Wang, and Chemmatrix resins.
Various peptides with different amino acids and protecting groups were evaluated for the synthesis of peptide thioesters on solid support (Table 4). Protecting groups were subsequently removed from the resulting peptide thioesters by using a TFA cleavage cocktail in solution. It was noted that the formation of peptide thioesters is independent of the nature of the amino acid residues preceding serine. For example, peptides with a bulky amino acid residue next to serine such as Glu(tBu), Phe, and Ser(tBu) and peptides with β-branched residues such as Val and Thr(tBu) generated the protected peptide thioesters in a clean manner and with high yields (entries 2–6, Table 4, ESI†). From these studies, we conclude that the cyclic urethane-activated peptide will be a versatile precursor for the synthesis of peptidyl thioesters.
| Entry | Substrate | Peptide | Conv.b (%) |
|---|---|---|---|
| a Reaction conditions: activated peptide 2 (25 mg, 0.7 mm g−1) on solid support was reacted with ethyl-3-mercaptopropionate (100 μL) and a catalytic amount of sodium thiolate (0.5 equiv.) in DMF (1 mL) at room temperature for 20 h. b Conversion to peptide thioester 3 was calculated from the absorbance at 220 nm using HPLC integration. SR = S-(CH2)2-COOC2H5. | |||
| 1 | 2h | Ac-AVGPPGVA-Oxd | 95 |
| 2 | 2i | Ac-R(Pbf)AFK(Boc)Y(tBu)GLE(tBu)-Oxd | 95 |
| 3 | 2j | Ac-GVALF-Oxd | 95 |
| 4 | 2k | Ac-Y(tBu)FD(tBu)IR(Pbf)AV-Oxd | 90 |
| 5 | 2l | Ac-S(tBu)GIS(tBu)GPLS(tBu)-Oxd | 95 |
| 6 | 2m | Ac-RFAT(tBu)-Oxd | 90 |
| 7 | 2n | Ac-GPMLA-MeOxd | 95 |
| 8 | 2o | Ac-GPMLA-Thz | 90 |
Next, the peptide backbone activation was investigated with threonine, whose side chain functionality exhibits strong similarity to serine. As expected, threonine underwent smooth cyclization under the optimized reaction conditions and generated activated peptide Ac-GPMLA-MeOxd 2n (entry 7, Table 4). In a similar manner, cysteine generated thiazolidinone-activated peptide Ac-GPMLA-Thz 2o under the reaction conditions (entry 8, Table 4). The activated peptides 2n and 2o generated peptide thioester 3G in high yields upon thiolysis.
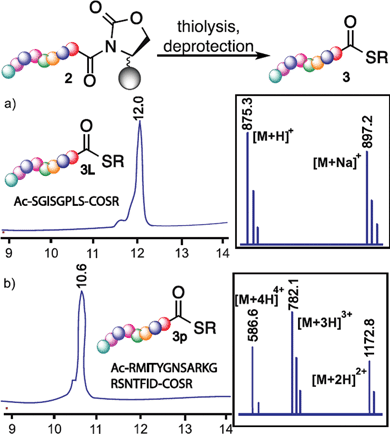
Importantly, our method was successfully applied for the synthesis of a multiserine containing bioactive peptide thioester Ac-SGISGPLS-COS(CH2)2COOC2H53L, a fragment of antimicrobial bovine beta-defensin 13,25 from cyclic urethane-activated peptide Ac-S(tBu)GIS(tBu)GPLS(tBu)-Oxd2l (a, Fig. 3).
The main role of peptide thioesters is to synthesize complex proteins and bioconjugates, therefore it is important that the cyclic urethane moiety is capable of making long peptide thioesters with diverse functional groups. To see the compatibility of the cyclic urethane technique, nineteen amino acid long peptide thioester Ac-RMITYGNSARKGRSNTFID-COS(CH2)2COOC2H53P, a fragment of rabies virus glycoprotein (Rvg),18,26 was successfully synthesized from corresponding cyclic urethane moiety 2p with a yield of 31% (b, Fig. 3, ESI†). These results (Fig. 3) and Table 4 demonstrated the versatility of the cyclic urethane technique in the synthesis of peptide thioesters.
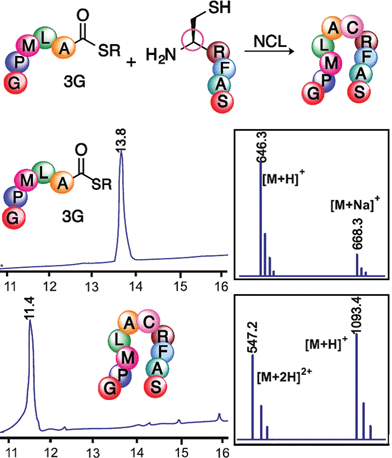
Next, we utilized peptide thioesters Ac-GPMLA-COS-(CH2)2-COOC2H53G and Ac-AVGPPGVA-COS-(CH2)2-COOC2H53h generated by the cyclic urethane technique in Native Chemical Ligation (NCL) with N-terminal cysteine containing peptide CRFAS-NH2 (Fig. 4 and 5). In addition, cyclic urethane technique-derived 14 amino acid long peptide thioester Ac-GNSARKGRSNTFID-COSR 3Q (39% yield) was ligated with N-terminal cysteine-containing peptide CPTGPRPNEPMWITY-NH2 to generate a 29 amino acid long peptide fragment of rabies virus glycoprotein (Rvg), an effective carrier for cargo delivery into cells and for crossing the blood–brain barrier (Fig. 6, ESI†).26
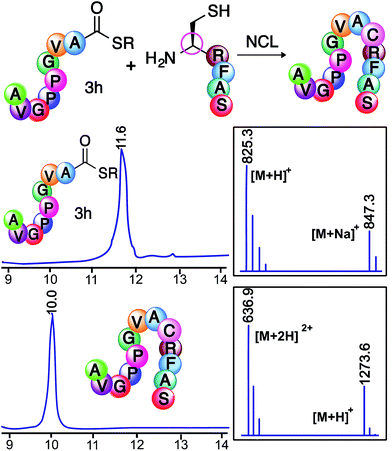 | ||
| Fig. 5 Native chemical ligation of peptide thioester Ac-AVGPPGVA-COSR 3h with N-terminal cysteine peptide CRFAS-NH2. HPLC/MS traces of peptide thioester 3h and ligated product. SR = S-(CH2)2-COOC2H5. | ||
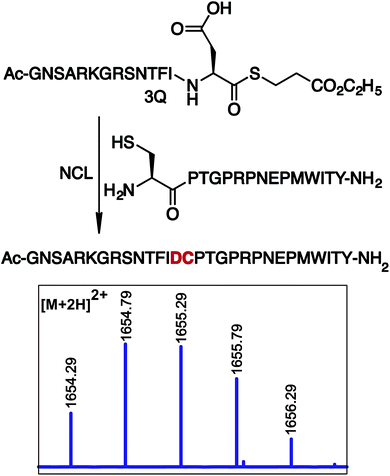 | ||
| Fig. 6 Ligation of peptide thioester Ac-GNSARKGRSNTFID-COSR 3Q with N-terminal cysteine peptide CPTGPRPNEPMWITY-NH2. MS of the ligated product. SR = S-(CH2)2-COOC2H5. | ||
It is noteworthy that relatively dilute conditions were used for the peptide reactants (1.2 mM) as well as low concentrations of the 4-mercaptophenylacetic acid catalyst (25 mM). The ligation was completed after 24 h at 37 °C, demonstrating the utility of the cyclic urethane technique in the synthesis of peptide thioesters for the formation of large peptides using NCL.
Conclusions
We have developed a novel cyclic urethane activation technique for the synthesis of peptide thioesters directly from a solid support. The cyclic urethane technique activates the peptide backbone, which undergoes nucleophilic displacement by a thiol to release the corresponding thioester into the solution. The significant advantage of this technique is that unnatural handles or linkers are not required, and the synthesis of peptides is carried out by Fmoc-SPPS without the need for any special reaction conditions and can be fully automated, thus constituting a major advance in the field. It is noteworthy that only those peptides that undergo complete activation are displaced by the thiol into the solution, thus delivering very pure thioesters. Moreover, activation of serine to form the cyclic urethane moiety and subsequent thiolysis is compatible with bulky amino acids including β-branched residues such as Val/Thr next to serine, and is resistant to epimerization. The results suggest that the cyclic urethane activation technique will be utilized in organic synthesis, including peptide and protein chemistry. Moreover, the cyclic urethane ring is stable and can be stored in a desiccator for long periods of time without any disintegration. The applicability of the strategy was demonstrated by the synthesis of a long peptide thioester, which was utilized for the synthesis of a 29 amino acid long peptide derived from rabies virus glycoprotein (Rvg) using NCL. As a result, access to C-terminally activated cyclic urethane peptides will be applicable for the synthesis of complex post-translationally modified peptides and other biomolecules. Further work is ongoing in our laboratory to apply the technique to other synthetically demanding targets.Experimental section
Peptide bond activation at serine on the solid support and synthesis of peptide thioesters by Fmoc SPPS approach
To a peptide on solid support (25 mg to 0.7 mm g−1), a solution of DSC (10 equiv.), DIEA (10 equiv.) and DMAP (catalytic amount) in DMF (3 mL) was added and the resin was left on shaker for 17 h. The solution was drained and the resin was washed with DMF. The activated resin was swelled in DMF (1 mL) followed by addition of a catalytic amount of sodium thiolate (0.5 equiv.) and thiol (100 μL). The resulting solution was left on a shaker at RT for 20 h. The resin was then filtered, followed by solvent removal under high vacuum. The resulting peptide thioester was then analyzed by HPLC, MS, and NMR. To determine the % release of the peptide thioester from the solid support, the resin was cleaved using a TFA cleavage cocktail and analyzed by HPLC and MS.Acknowledgements
We thank Ryan Cohen at Merck for structural analysis by NMR.Notes and references
- P. E. Dawson and S. B. Kent, Annu. Rev. Biochem., 2000, 69, 923–960 CrossRef CAS PubMed.
- C. Unverzagt and Y. Kajihara, Chem. Soc. Rev., 2013, 42, 4408–4420 RSC.
- P. E. Dawson, T. W. Muir, I. Clark-Lewis and S. B. Kent, Science, 1994, 266, 776–779 CAS.
- T. M. Hackeng, J. H. Griffin and P. E. Dawson, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 10068–10073 CrossRef CAS.
- X. Li, T. Kawakami and S. Aimoto, Tetrahedron Lett., 1998, 39, 8669–8672 CrossRef CAS.
- Y. Shin, K. A. Winans, B. J. Backes, S. B. H. Kent, J. A. Ellman and C. R. Bertozzi, J. Am. Chem. Soc., 1999, 121, 11684–11689 CrossRef CAS.
- A. R. Mezo, R. P. Cheng and B. Imperiali, J. Am. Chem. Soc., 2001, 123, 3885–3891 CrossRef CAS PubMed.
- T. Kawakami, M. Sumida, K. I. Nakamura, T. Vorherr and S. Aimoto, Tetrahedron Lett., 2005, 46, 8805–8807 CrossRef CAS.
- S. Tsuda, A. Shigenaga, K. Bando and A. Otaka, Org. Lett., 2009, 11, 823–826 CrossRef CAS PubMed.
- J. Kang, J. P. Richardson and D. Macmillan, Chem. Commun., 2009, 407–409 RSC.
- N. Ollivier, J. Dheur, R. Mhidia, A. Blanpain and O. Melnyk, Org. Lett., 2010, 12, 5238–5241 CrossRef CAS PubMed.
- J.-S. Zheng, H.-N. Chang, F.-L. Wang and L. Liu, J. Am. Chem. Soc., 2011, 133, 11080–11083 CrossRef CAS PubMed.
- F. Burlina, G. Papageorgiou, C. Morris, P. D. White and J. Offer, Chem. Sci., 2014, 5, 766–770 RSC.
- V. P. Terrier, H. Adihou, M. Arnould, A. F. Delmas and V. Aucagne, Chem. Sci., 2016, 7, 339–345 RSC.
- J. A. Camarero, B. J. Hackel, J. J. de Yoreo and A. R. Mitchell, J. Org. Chem., 2004, 69, 4145–4151 CrossRef CAS PubMed.
- J. S. Zheng, M. Yu, Y. K. Qi, S. Tang, F. Shen, Z. P. Wang, L. Xiao, L. Zhang, C. L. Tian and L. Liu, J. Am. Chem. Soc., 2014, 136, 3695–3704 CrossRef CAS PubMed.
- Y.-M. Li, Y.-T. Li, M. Pan, X.-Q. Kong, Y.-C. Huang, Z.-Y. Hong and L. Liu, Angew. Chem., Int. Ed., 2014, 53, 2198–2202 CrossRef CAS PubMed.
- J. B. Blanco-Canosa and P. E. Dawson, Angew. Chem., Int. Ed., 2008, 47, 6851–6855 CrossRef CAS PubMed.
- J. B. Blanco-Canosa, B. Nardone, F. Albericio and P. E. Dawson, J. Am. Chem. Soc., 2015, 137, 7197–7209 CrossRef CAS PubMed.
- A. P. Tofteng, K. K. Sørensen, K. W. Conde-Frieboes, T. Hoeg-Jensen and K. J. Jensen, Angew. Chem., Int. Ed., 2009, 48, 7411–7414 CrossRef CAS PubMed.
- S. K. Mahto, C. J. Howard, J. C. Shimko and J. J. Ottesen, ChemBioChem, 2011, 12, 2488–2494 CrossRef CAS PubMed.
- S. K. Mong, A. A. Vinogradov, M. D. Simon and B. L. Pentelute, ChemBioChem, 2014, 15, 721–733 CrossRef CAS PubMed.
- H. E. Elashal and M. Raj, Chem. Commun., 2016, 52, 6304–6307 RSC.
- R. De Marco, A. Tolomelli, M. Campitiello, P. Rubini and L. Gentilucci, Org. Biomol. Chem., 2012, 10, 2307–2317 CAS.
- M. E. Selsted, Y. Q. Tang, W. L. Morris, P. A. McGuire, M. J. Novotny, W. Smith, A. H. Henschen and J. S. Cullor, J. Biol. Chem., 1993, 268, 6641–6648 CAS.
- P. Kumar, H. Wu, J. L. McBride, K.-E. Jung, M. Hee Kim, B. L. Davidson, S. Kyung Lee, P. Shankar and N. Manjunath, Nature, 2007, 448, 39–43 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental procedures and analytical data for new compounds. See DOI: 10.1039/c6sc02162j |
| This journal is © The Royal Society of Chemistry 2017 |