Open Access Article
Open Access ArticleCopper-catalyzed click reaction on/in live cells†
Siheng
Li
 a,
Lin
Wang
ab,
Fei
Yu
a,
Zhiling
Zhu
a,
Dema
Shobaki
a,
Haoqing
Chen
a,
Mu
Wang
a,
Jun
Wang
a,
Guoting
Qin
a,
Uriel J.
Erasquin
a,
Li
Ren
b,
Yingjun
Wang
b and
Chengzhi
Cai
*a
a,
Lin
Wang
ab,
Fei
Yu
a,
Zhiling
Zhu
a,
Dema
Shobaki
a,
Haoqing
Chen
a,
Mu
Wang
a,
Jun
Wang
a,
Guoting
Qin
a,
Uriel J.
Erasquin
a,
Li
Ren
b,
Yingjun
Wang
b and
Chengzhi
Cai
*a
aDepartment of Chemistry, University of Houston, 4800 Calhoun Rd., Houston, TX 77204, USA. E-mail: cai@uh.edu
bCollege of Materials Science and Engineering, South China University of Technology, Guangzhou, 510640, China
First published on 25th November 2016
Abstract
We demonstrated that the copper-catalyzed azide–alkyne cycloaddition (CuAAC) reaction could be performed inside live mammalian cells without using a chelating azide. Under optimized conditions, the reaction was performed in human ovary cancer cell line OVCAR5 in which newly synthesized proteins were metabolically modified with homopropargylglycine (HPG). This model system allowed us to estimate the efficiency of the reaction on the cell membranes and in the cytosol using mass spectrometry. We found that the reaction was greatly promoted by a tris(triazolylmethyl)amine CuI ligand tethering a cell-penetrating peptide. Uptake of the ligand, copper, and a biotin-tagged azide in the cells was determined to be 69 ± 2, 163 ± 3 and 1.3 ± 0.1 μM, respectively. After 10 minutes of reaction, the product yields on the membrane and cytosolic proteins were higher than 18% and 0.8%, respectively, while 75% of cells remained viable. By reducing the biothiols in the system by scraping or treatment with N-ethylmalemide, the reaction yield on the cytosolic proteins was greatly improved to ∼9% and ∼14%, respectively, while the yield on the membrane proteins remained unchanged. The results indicate that out of many possibilities, deactivation of the current copper catalysts by biothiols is the major reason for the low yield of the CuAAC reaction in the cytosol. Overall, we have improved the efficiency for the CuAAC reaction in live cells by 3-fold. Despite the low yield inside live cells, products that strongly bind to the intracellular targets can be detected by mass spectrometry. Hence, the in situ CuAAC reaction can be potentially used for screening of cell-specific enzyme inhibitors or biomarkers containing 1,4-substituted 1,2,3-triazoles.
Introduction
Bioconjugation of biologically active or proactive molecules in living organisms using bioorthogonal click reactions represents a great challenge and opportunity in the fields of chemical biology, drug discovery and biomaterials.1–8 A few such reactions, particularly strain-promoted azide–alkyne cycloaddition (SPAAC)2,5,9 and tetrazine ligation,10 exhibit a high reaction rate and excellent biocompatibility, yield and selectivity, without a catalyst. Hence, they have been widely used for labelling of biomolecules inside living cells and even in animals.11 In comparison, the well-known copper-catalyzed azide–alkyne cycloaddition (CuAAC)12,13 uses small handles (azide/ethynyl) to afford the smallest linkage (triazole) that resembles an amide linkage and possibly imposes the least perturbation on the biological function of the conjugates.14 Also, the ethynyl and azido handles are readily incorporated to synthetic- and bio-molecules. Hence, the CuAAC reaction has been widely used for bioconjugation ex situ and in fixed cells, and for screening for enzyme inhibitors.6,15–17 However, its use in living systems has been limited by the cytotoxicity of excess CuI that mediates the generation of reactive oxygen species (ROS).18–20Under optimized conditions,21,22 the use of certain CuI ligands, especially tris(triazolylmethyl)amine derivatives,23 such as the ligand 1 (BTTAA, Fig. 1), greatly accelerates the CuAAC reaction and reduces oxidative damage, thus allowing the reaction to be performed on the surfaces of live cells.22,24–27 Performing the reaction inside live cells proved to be much more challenging.1,28 To date, few examples were reported for bacterial cells,29,30 and only one for mammalian cells31 in which copper-chelating azide was used to enhance the CuAAC reactivity but at the expense of enlarging the resultant triazole linkage. Besides the copper toxicity, other obstacles for the intracellular CuAAC reaction may include (1) the presence of a variety of endogenous copper-binding ligands that may deactivate the catalyst, (2) the low concentration of reagents limited by their uptake and cytotoxicity, and (3) high biomacromolecular crowding and heterogeneity within the cytoplasm.
Another challenge for studying CuAAC reactions on/in live cells is the quantitation of reagents and products confined in a small cellular volume containing numerous biomolecules. To date, fluorescence imaging is the most common technique to monitor bioconjugation reactions on/in cells, which requires a fluorophore and is difficult to quantify due to the high heterogeneity of the intracellular environment.
Herein, we report our study of the CuAAC reaction in live cells without using a chelating azide. The reaction was performed on cellular proteins incorporating homopropargylglycine (HPG). This model system allowed us to estimate the reaction yields on the cell membranes and in the cytosol of live cells using mass spectrometry.
Results and discussion
Cell-penetrating peptide-conjugated CuI ligand
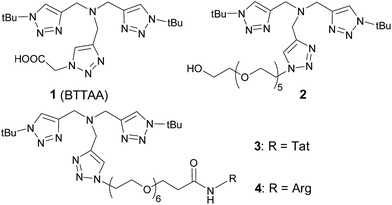
We began by identifying a suitable catalyst and conditions for the intracellular CuAAC reaction. We first evaluated the most efficient reported CuI ligand 1,22 and its analogue 2. To maximize the cellular uptake of the ligand, we replaced the carboxylic acid in 1 with an arginine-rich, cell-penetrating peptide (RKKRRQRRR, referred to as Tat peptide)32,33 to form 3. For the subsequent LC-MS/MS analysis, 3 was hydrolyzed to 4 as will be described later.Evaluation of in vitro CuAAC reactivity
The CuAAC reactivity of ligands 1–3 was evaluated with a fluorogenic reaction assay (Fig. 2).34 In this assay, the CuAAC reaction of the non-fluorescent azide 5 and L-homopropargylglycine (HPG, 6) generates the fluorescent product (7) that can be conveniently quantified. The reaction rate is substantially affected by the solvents. While superior reactivity was reported for 1 in 5/95 DMSO/PBS,22 we found that the reaction mixture in this medium was cytotoxic. The cytotoxicity could be reduced using 10% Dulbecco's modified eagle medium (DMEM, Fig. S1†) which slightly reduced the reaction rate and yield (Fig. S2†). In this medium, the Tat-conjugated ligand 3 exhibited a two-fold increase in yield compared to 1 and 2 (Fig. 2). The enhanced reactivity of 3 was presumably originated from the sacrificial oxidation of arginine and lysine residues in the Tat peptide which protected the active CuI species, similar to the use of excess reducing agent as the sacrificial reagent.21 Indeed, as the concentration of the reducing agent (sodium ascorbate) increased by 10-fold to 5 mM, the rate enhancement of 3 became less significant (Fig. S3†). Note that, however, an ascorbate concentration higher than 0.5 mM in the CuAAC reaction mixture exhibited increasing cytotoxicity (Fig. S1b†).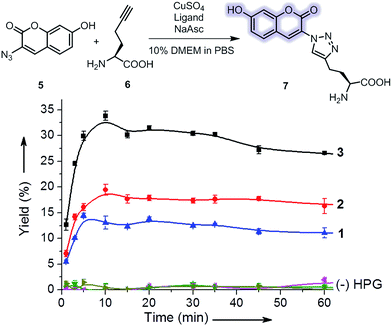 | ||
| Fig. 2 Evaluation of the CuAAC reactivity of ligands 1–3 using a fluorogenic CuAAC reaction. Reaction conditions: 100 μM 5, 50 μM 6, 100 μM CuSO4, 200 μM ligand, 500 μM sodium ascorbate (NaAsc) in 10/90 (v/v) methionine-free DMEM/PBS, room temperature. Yield was derived from the measured mean fluorescence intensity of 7 (Fig. S4†). Controls were performed without HPG. Error bars represent the standard deviation of data from three samples. | ||
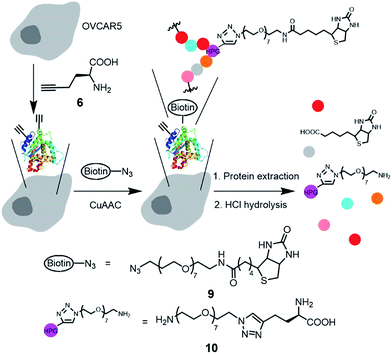
For the CuAAC reaction on live cells, we used the biotin-coumarin-azide 8 (Fig. 3a) as a probe, attempting to use the coumarin moiety as a fluorogenic indicator, and the biotin moiety as a handle to attach an additional functional moiety (e.g. avidin–FITC) via biotin–avidin interaction. To introduce ethynyl groups to the cellular proteins, HPG (6) as a surrogate of methionine35,36 was metabolically incorporated into the newly synthesized proteins of human umbilical vein endothelial cells (HUVECs) and human ovarian cancer cells (OVCAR5). The effective HPG concentration in the culture medium was optimized with a western blot (Fig. S6†). The CuAAC reaction was performed by treatment of the adhered cells with a mixture of 8, CuSO4, ligand and sodium ascorbate in 10% methionine-free DMEM/PBS.
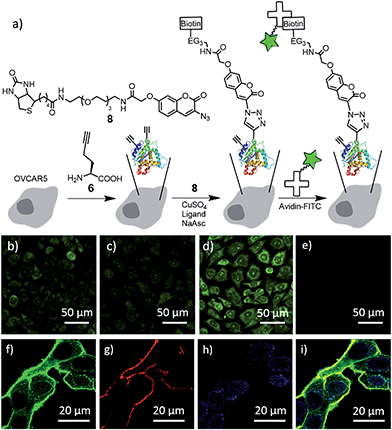 | ||
| Fig. 3 Reactivity of ligands 1–3 for CuAAC reaction in live cells. (a) Schematic of metabolic incorporation of HPG (6) to the newly synthesized proteins, CuAAC reaction and fluorescence labeling with avidin–FITC. (b)–(h) Representative wide field (b–e) and confocal (f–h) fluorescence images of OVCAR5 cells after treatment with a mixture of 8 (100 μM), CuSO4 (100 μM), 200 μM of the ligand 1 (b), 2 (c), 3 (d and f–i), and sodium ascorbate (500 μM) in 10/90 (v/v) methionine-free DMEM/PBS for 10 minutes, followed by fixing and labeling with avidin–FITC (b–f), a plasma membrane dye (g), and DAPI (h). (e) Control without ligand and CuSO4. (i) Overlay of (f)–(h). For the control without avidin–FITC, see Fig. S9.† | ||
We initially attempted to observe the triazole-coumarin product with fluorescence emitted at 388–521 nm,34 but the signal was barely distinguishable from the background, presumably due to the intrinsic low absorption coefficient and relatively low quantum yield for the triazole-coumarin dyes,37 especially for the ether derivative (8vs.5, Fig. S7†). We then visualized the biotin moieties by staining with avidin–FITC after cell fixation (Fig. 3b–e). The fluorescence images in Fig. 3b–d clearly show that the cells were labelled with avidin–FITC after being subjected to the above CuAAC conditions for conjugating the biotin-azide 8 to the HPG-incorporated cells, while no labelling was observed in the absence of the copper catalyst under identical conditions (Fig. 3e and S8b†). In addition, ligand 3 largely improved the labelling efficiency as compared to ligands 1 and 2 (Fig. 3dvs.b and c). Confocal microscopy imaging was used to indicate the location of biotinylated proteins, with the plasma membrane stained with wheat germ agglutinin, Alexa Fluor 555 conjugate (Fig. 3g) and the nucleus stained with DAPI (Fig. 3h). Without the use of avidin–FITC, no fluorescence signal in the FITC channel was observed, eliminating the possibility of autofluorescence interference from cellular components (Fig. S9†). After staining with avidin–FITC, strong FITC fluorescence was observed, mostly colocalized with the plasma membrane dye, indicating the CuAAC reaction occurred more efficiently on the cell surface (Fig. 3i and S8a†). Significantly, the FITC signal from the cytoplasm was clearly observed in the magnified image (Fig. 3f). Although the signal in the cytosol was much weaker than that on the cell surface, it should be noted that the cytosol has a larger volume than the plasma membrane and the extracellular matrix. This result shows the presence of the biotin–protein conjugate product inside the cells. Due to the intrinsic heterogeneity of these cellular samples, a quantitative comparison of the CuAAC reaction with different catalysts, as well as the CuAAC reaction in different cellular compartments is difficult. To verify CuAAC reaction product formation and to quantify the reaction mixture in the complex cellular matrix, we developed the following analytical method based on LC-ESI-MS/MS.
Quantification of the cellular uptake of CuAAC catalysts
We first used LC-ESI-MS/MS to quantify the cellular uptake of ligands 1–3. Specifically, both HUVEC and OVCAR5 cells were treated with the above CuAAC reaction conditions for 10 min, and immediately washed 4 times with PBS to remove the extracellular ligand as confirmed by LC-ESI-MS/MS analysis (Fig. S10†). The cells were then lysed with trypsin–EDTA solution. Trypsin efficiently hydrolyzes amides from the C-terminus of lysine and arginine residues, including those in the Tat peptide-conjugated ligand 3. The complete hydrolysis of 3 afforded 4 with only the N-terminus arginine residue tethering to the OEG chain. The amounts of ligands 1–3 in the sample were then estimated by the intensity of the most abundant secondary ions in the MS/MS spectra of 1, 2 and 4 using the corresponding calibration curves (Fig. S11–S13†). The results summarized in Table 1 represent the sum of the ligand/Cu cellular uptake and ligand/Cu strongly associated to the cell membrane, which is hereafter referred to as uptake for the sake of simplicity.| HUVEC | OVCAR5 | |||
|---|---|---|---|---|
| [Ligand] | [Cu] | [Ligand] | [Cu] | |
| a Derived from the total uptake (measured by LC-MS/MS) divided by the number of cells and the cell volume assuming to be 1.7 picoliters.38 b After incubation in 100 μM CuSO4, 200 μM ligand, 500 μM NaAsc in 10/90 (v/v) methionine-free DMEM/PBS for 10 minutes, the same conditions used for the intracellular CuAAC reaction but without the azide. c Control 1 was performed without both ligand and CuSO4. d Control 2 was performed without the ligand. For deriving the error propagated from the measured amounts of ligand/copper and cell counting, see ESI†. | ||||
| Control 1c | 22 ± 2 | 23 ± 1 | ||
| Control 2d | 46 ± 3 | 95 ± 2 | ||
| 1/CuSO4 | 1.0 ± 0.1 | 52 ± 3 | 4.3 ± 0.2 | 104 ± 2 |
| 2/CuSO4 | 10.8 ± 0.3 | 69 ± 4 | 8.6 ± 0.2 | 139 ± 3 |
| 3/CuSO4 | 32.9 ± 0.4 | 86 ± 3 | 68.8 ± 1.8 | 163 ± 3 |
The result in Table 1 shows that tethering a cell-penetrating Tat peptide to the ligand as in 3 largely enhanced its uptake in both cell lines as compared to 1 and 2. The low uptake of 1 is presumably due to its negatively charged carboxylate group that blocks the transportation through the plasma membrane.22
Copper uptake after 10 min treatment with various ligand/copper mixtures was measured by inductively coupled plasma mass spectrometry (ICP-MS). In general, the presence of the ligand facilitated copper uptake (Table 1). However, the enhancement was relatively small for 1, presumably due to the low internalization of the ligand itself. The uptake was mostly enhanced in the presence of the Tat-conjugated ligand 3, indicating that the formation of the ligand 3–CuI complex improved copper uptake. Notably, a higher copper uptake was observed for OVCAR5 cells compared to HUVECs, which is consistent with the higher metabolic activity of the cancer cell line.
Evaluation of cytotoxicity
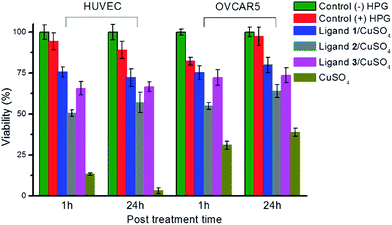
With the high uptake of copper aided by the ligands, cytotoxicity becomes a major concern. The cytotoxicity of the CuAAC reaction mixtures were evaluated with the MTS assay on both HPG-incorporated HUVECs and OVCAR5 cell lines (Fig. 4). After the reaction, the cells were incubated in their respective media (EGM™-2 for HUVECs, RPMI-1640 for OVCAR5) for an additional 24 hours. Slower proliferation for cells incorporated with HPG was observed, probably due to retardation of protein biosynthesis.39 As showed in Fig. 4, both cell lines treated with ligand 1 or 3 proliferated in a slightly slower rate compared to the control without the catalyst. Remarkably, even though the intracellular copper concentration was significantly higher in the presence of ligand 3 as compared to 1, the cell proliferation was only slightly slower. Meanwhile, the use of ligand 2 decreased the viability for both cell lines even with a lower copper uptake. Without ligand, 100 μM of copper caused significantly higher cytotoxicity, particularly for HUVECs.Quantification of intracellular CuAAC reaction yields
After confirming that the Tat-conjugated ligand 3 enhanced copper/ligand uptake while not significantly affecting cell viability, we then performed a quantitative analysis of the CuAAC products in live, HPG-incorporated OVCAR5 cells. We used the biotin-azide 9 to perform the CuAAC reaction as outlined in Fig. 5. The uptake of 9 was determined to be 1.3 ± 0.1 μM. After the CuAAC reaction, the remaining azide was completely removed by washing the adhered cells 4 times with PBS, as confirmed by LC-MS/MS analysis (ESI†). We then determined the yields for the reaction on the proteins in the cell membrane and cytosol, respectively. Thus, the adherent cells were detached by trypsin–EDTA solution, counted with a cell counter, and pelleted by centrifugation. This step removed the extracellular matrix proteins not bound to the membrane. The cells were then lysed and fractionated into membrane proteins and cytosolic proteins using a Mem-PER™ plus membrane protein extraction kit.40 To measure the contamination of the cytosolic protein fraction by membrane proteins, we used the same method to fractionate 5-tagged membrane proteins from the native cytosolic proteins. Indeed, the cytosolic protein fraction was found to contain less than 2% fluorescent membrane proteins (ESI†). The extracted proteins were subsequently hydrolyzed into amino acids using standard conditions (6 M HCl, 110 °C, 24 hours). It is worth noting that the amide linkage between the biotin moiety and the OEG chain was also cleaved to yield 10 as the final product that was quantified by LC-ESI-MS/MS (Fig. S19–S21†).The complete hydrolysis of the biotinylated proteins to yield the triazole 10 enabled us to estimate the yield of the CuAAC reaction on the proteins in the cell membrane and cytosol (Table 2). To confirm that product 10 did not come from the free HPG that was not completely removed after the metabolic incorporation, OVCAR5 cells were incubated in methionine-abundant (0.2 mM) medium containing 4 mM HPG. In this negative control (Table 2, control 1), HPG was outcompeted by methionine for protein synthesis.41 Indeed, the subsequent CuAAC reaction and protein hydrolysis did not yield a detectable amount (<0.10 pg per μg proteins as the lower limit of quantification) of 10, confirming that 10 was originated from the biotinylated proteins. To prepare the standard representing 100% yield, we performed the CuAAC reaction on the total extracted HPG-containing proteins from OVCAR5 cell lysate. The reaction was performed under nitrogen atmosphere and in CuAAC compatible lysis buffer (0.2% SDS in PBS) for 5 hours. These conditions largely enhanced the yield of the fluorogenic reaction assay (Fig. 2) from 30% to ∼100% (Fig. S3b†). The detergent SDS was needed to dissolve the membrane proteins.
| Pre-treatment | Protein | Intracellular GSHf (μM) | CuAAC with 1 | CuAAC with 3 | ||
|---|---|---|---|---|---|---|
| Amount of 10 (pg) per μg proteins | Yieldh (%) | Amount of 10 (pg) per μg proteins | Yieldh (%) | |||
| a Control 1 (negative control), the CuAAC reaction was performed with the adhered cells without HPG incorporation into cellular proteins. b Control 2 (positive control), the CuAAC reaction was performed with the total extracted HPG-containing proteins from cell lysate under the optimal conditions (see text). c Detached the cells with a cell scraper in cold PBS after HPG incorporation. d Adherent OVCAR5 cells were incubated in 100 μM NEM in PBS for 10 minutes after HPG incorporation. e Adherent OVCAR5 cells were incubated in RPMI-1640 medium containing 20 μM BSO for 24 hours before HPG incorporation. f Determined by LC-ESI-MS/MS upon alkylation of the cellular GSH with NEM (ESI). g Not measurable with this method. h Yield (%) was determined by the amount of CuAAC reaction product 10 (pg) per protein (μg) divided by the result obtained in control 2 (ESI†). All measured yields of cytosolic proteins were deducted by 0.2% due to the 2% contamination of the cytosolic proteins by the membrane proteins that contain 18% of the product 10. | ||||||
| Control 1a | Whole-cell | <0.10 | <0.2 | |||
| Control 2b | Whole-cell | 49.18 ± 0.95 | 100 | |||
| (—) | Membrane | 3.03 ± 0.25 | 6.2 | 8.83 ± 0.82 | 18.0 | |
| (—) | Cytosolic | 1585 ± 29 | 0.34 ± 0.02 | 0.5 | 0.48 ± 0.05 | 0.8 |
| Scrapingc | Cytosolic | 427 ± 17 | 1.09 ± 0.05 | 2.0 | 4.28 ± 0.28 | 8.5 |
| NEMd | Cytosolic | N.D.g | 2.08 ± 0.29 | 4.0 | 7.02 ± 0.42 | 14.1 |
| BSOe | Cytosolic | 4.9 ± 0.4 | 0.49 ± 0.05 | 0.8 | 1.04 ± 0.06 | 1.9 |
It should be noted that SDS denatures and linearizes the proteins. Hence, nearly all ethynyl groups in the proteins were converted to 10 under these conditions to provide the standard for 100% yield of the reaction (Table 2, control 2), whereas only the HPG groups on the surface of the native proteins were available to the reaction in live cells. Therefore, the yields in Table 2 represent an underestimate of the yields for the reaction on the proteins in the membranes and cytosols of the live cells.
Significantly, we confirmed that the CuAAC reaction in the presence of ligand 1 or 3 and CuI did occurred on/inside live OVCAR5 cells. On the membrane proteins, the presence of ligand 3 resulted in a 3-fold increase of product formation than ligand 1 (∼18% vs. ∼6%). We initially performed the CuAAC reaction on a freshly prepared cell suspension (∼2 × 107 cells per mL). In suspended cells, the yield of the intracellular CuAAC reaction was surprisingly high, reaching 8.5% when the Tat-conjugated ligand 3 was used. However, when we performed the reaction later under identical conditions on the adherent cells, we were disappointed to find that the yield was largely decreased to ∼0.8% (Table 2).
To account for this result, we suspected that the cell membrane might be compromised to some extent during the preparation of the suspended cells. Specifically, the suspended cells were prepared by detaching the cells from the culture dish in ice-cold PBS with a cell scraper, rather than using trypsin solution that promotes hydrolysis of the surface proteins. Indeed, we found later that about half of the cells detached by this method were positive in the trypan blue staining test, suggesting the compromised cellular membrane. The higher uptake of ligand 3/CuIvs.1/CuI did not apparently enhance the CuAAC reaction yield for the adhered cells (0.8% vs. 0.5%, Table 2). The result might be due to the deactivation of the CuAAC catalyst in the intracellular environment. Specifically, in the membrane compromised OVCAR5 cells, the rapid efflux of GSH42 as the CuAAC reaction inhibitor (see below) in the cytoplasm might lead to the great enhancement of the reaction yield in the cytosolic proteins. Indeed, the use of a cell scraper for cell detachment resulted in reduction of cellular GSH by 73% (from 1585.3 ± 28.6 μM to 427.1 ± 17.2 μM) as determined by LC-ESI-MS/MS (ESI†).43
Main inhibitor of the intracellular CuAAC reaction
Although extra copper is cytotoxic, most organisms require a trace amount of copper, predominantly CuI, for metabolism and other functions. Cellular copper uptake, trafficking and metabolism are mainly regulated by coordination with a series of metalloproteins and endogenous small molecule ligands, and are directed by their CuI affinity.44–46 Once the copper ligands 1–3 enter such an intracellular environment, a new equilibrium for the distribution of CuI will be established, depending on the concentration and CuI affinity of these ligands and the endogenous ligands. However, only limited reports to date discussed the effect of endogenous CuI ligands, including histidine19 and free thiols,21 on CuAAC reaction.Although many metalloproteins have a high affinity (KD < 10−14 M) to CuI, they are generally expressed at a low micromolar or nanomolar level. Relatively low affinity CuI ligands, mainly reduced glutathione (GSH), are abundant in the intracellular environment,47 and may compete with the exogenous CuAAC ligands for binding with copper. Indeed, the dissociation constants (KD) of CuI–GSH and CuI–3 were similar (3.10 × 10−11 M vs. 3.52 × 10−11 M), as determined using a competition assay with bicinchoninic acid (BCA) (ESI†).48 Also, the concentration of GSH was ∼23 times higher than that of the ligand 3 (∼1.6 mM vs. 69 μM) in the cytoplasm. The abundant GSH may deactivate the CuI catalyst. Indeed, in the presence of 1 mM GSH in the reaction mixture, only ∼1% yield was obtained in the fluorogenic reaction assay (Fig. S22†). Under conditions similar to the intracellular CuAAC reaction conditions (69 μM 3, 163 μM CuI, 20 μM HPG and 1.3 μM azide in PBS for 10 minutes), the reaction yield dropped from 25% to 2% in the presence of 500 μM GSH (Table S5†).
To deactivate the intracellular GSH and other biothiols from complexation with CuI, we treated the OVCAR5 cells with N-ethylmaleimide (NEM, 100 μM) in HBS for 10 minutes (ESI†) prior to the CuAAC reaction. As expected, the yield of the intracellular CuAAC reaction increased significantly from 0.5% to 4.0% for ligand 1 and from 0.8% to 13.2–14.9% (results from three experiments) for ligand 3. After the 10 minute reaction, the cells were immediately treated with the culture medium supplemented with glutathione reduced ethyl ester (GSH-OEt, a membrane permeable GSH derivative) to restore the intracellular GSH level against oxidative damage. This treatment increased the cell viability from ∼15% to ∼50% (Fig. S26b†).
In searching for a method to deplete intracellular GSH without inducing cytotoxicity, we found the work by De Nicola and Ghibelli who showed that cells can survive slow depletion of GSH by using buthionine sulfoximine (BSO) which inhibits GSH biosynthesis.49 Indeed, incubating OVCAR5 cells with 20 μM BSO in the culture medium for 24 hours resulted in a reduction of the cellular GSH level by 99.7% (from 1585.3 ± 28.6 μM to 4.9 ± 0.4 μM, Table 2). No cytotoxicity was observed for using the low concentration of BSO to deplete cellular GSH (Fig. S25†). The cells were subsequently incorporated with HPG and incubated with the CuAAC reagents (100 μM 9, 100 μM CuSO4, 200 μM ligand, 500 μM sodium ascorbate in 10/90 (v/v) methionine-free DMEM/PBS) for 10 minutes. However, the yield for the intracellular reaction was only slightly increased to 0.8% for ligand 1 and 1.9% for ligand 3. This result was in contrast to our expectation that the yield for cells losing 99.7% GSH should be higher than 8.5% for scraped cells (that rapidly lost 73% of the cellular GSH, Table 2). A possible explanation is that instead of undergoing the apoptosis pathway leading to rapid depletion of GSH for membrane-compromised (scraped) cells, slow depletion of cellular GSH by low concentration of BSO allowed the cells to overexpress other biothiols as antioxidants (and strong inhibitors of the copper catalyst) to maintain the reducing environment against oxidative damage.50
Conclusions
In summary, we have demonstrated the first CuAAC reaction inside live cells without using a chelating azide, and estimated the yields for the reaction occurring at the cell membrane and cytosol using mass spectrometry. The reaction was performed on cellular HPG-containing proteins to minimize the loss of products due to diffusion, and to allow for determination of the CuAAC reaction yields on the membrane and cytosolic proteins, respectively. These yields reflect the efficiencies of the reaction occurring on the cell surface and in the cytosol. Due to the high heterogeneity of the cellular microenvironment, the reaction was not homogenous in different cellular compartments, as shown by the fluorescence image in Fig. 3f. We show that the conjugation of ligand 1 with a Tat-peptide resulted in a 3-fold enhancement of the yield at the cell membranes to ∼18%. We have established that out of many possible factors in the complex intracellular environment, the presence of a millimolar level of GSH in the cytoplasm is the predominant attribute to the low yield (∼0.8%) for the intracellular reaction.Experimental section
CuAAC reaction in live cells
The adhered HUVEC or OVCAR5 cells were cultured to ∼80% confluence, and the medium was changed to HBS. After 30 minutes of incubation to exhaust methionine, the medium was then changed again to methionine-free DMEM supplemented with 4 mM HPG. After incubation for 4 h to incorporate HPG to the newly synthesized proteins, the cells were washed 3 times with PBS, and treated with a solution of CuAAC reagents (100 μM the azide, 100 μM CuSO4, 200 μM ligand) in 10% methionine-free DMEM/PBS; the sodium ascorbate stock solution was prepared freshly and added at a final concentration of 500 μM to initiate the CuAAC reaction. After incubation on an orbital shaker for 10 minutes at room temperature, the solution was carefully removed by suction with a vacuum, and the cells were immediately washed 4 times with PBS. For the reaction in suspended cells detached from the surface using a cell scraper, see Section 12 in ESI.†Fluorescent microscopy imaging
After the CuAAC reaction, the cells were fixed with 2% formaldehyde in PBS for 30 min at room temperature. After fixation, all samples were incubated in 20 μg mL−1 avidin–FITC in PBS at room temperature for 30 minutes. Samples were washed with PBS for six times and their wide-field fluorescence images were captured with a Nikon Eclipse 80i microscope in bright field and fluorescence modes using a 10× objective, a FITC emission filter (Nikon C-FL B-2E/C, 465–495 nm) and 400 ms of exposure time. For confocal imaging, the fixed cells were first incubated in 3 μg mL−1 wheat germ agglutinin, Alexa Fluor 555 conjugate (Life Technologies) in PBS for 10 minutes for staining the plasma membrane. Samples were washed with PBS for 3 times and incubated in 20 μg mL−1 avidin–FITC in PBS at room temperature for 30 minutes. Samples were washed with PBS six times and mounted on glass slide with DAPI containing mounting medium (VECTOR Laboratories). The samples were imaged with a Leica SP8 upright confocal microscope using a 63× objective and the following excitation wavelengths: 405 nm (DAPI), 496 nm (FITC), 561 nm (Alexa 555).LC-ESI MS/MS analysis of azide and ligand uptakes and reaction yields
Liquid chromatography was performed on a Kinetex XB-C18 column (Phenomenex, 50 × 2.1 mm, 5 μm) eluted with a linear gradient of MeCN and water containing 0.1% formic acid at a flow rate of 200 μL min−1. A Thermo LCQ Deca XP Plus mass spectrometer was set at positive electrospray ionization. For the preparation of the standard calibration curve, a stock solution containing 1 ng μL−1 of analyte was diluted to 1, 2, 5, 10, 20, 50, 100, 200, 500 pg μL−1, respectively. All samples were spiked with an internal standard (ISTD, see ESI†). The standard samples were analyzed with five replicate 5 μL injections at each concentration level and a blank (water) injection between each level. The chromatogram was generated by summing the most intense product ions observed in the full scan MS/MS spectra.To determine the intracellular ligand and azide concentrations, the cells were lysed with cyropulverization, and incubated at 37 °C for 1 day with trypsin to hydrolyze the Tat peptide. The solution was dried in vacuum, and the residue was dissolved in 150 μL water containing 0.1% TFA, and centrifuged (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g, 20 min). Samples in the supernatant were extracted with a C18 ziptip. The internal standard (ESI,† final concentration: 100 pg μL−1) was added to the sample solution and the mixture was subjected to LC-ESI-MS/MS analysis.
000 × g, 20 min). Samples in the supernatant were extracted with a C18 ziptip. The internal standard (ESI,† final concentration: 100 pg μL−1) was added to the sample solution and the mixture was subjected to LC-ESI-MS/MS analysis.
To determine the reaction yield, the cells were counted, and lysed with a Mem-PER™ plus Membrane protein extraction kit (Life Technologies) to separate the membrane proteins and cytosolic proteins. The amounts of proteins in both fractions were measured by Micro BCA™ protein assay kit (Life Technologies). The protein solutions were freeze-dried. As described in ESI,† the proteins were completely hydrolyzed to amino acids including the triazole-amino acid 10 that was quantified by LC-ESI-MS/MS. For preparing and analyzing protein samples representing 100% yield for the CuAAC reaction, see the ESI.†
ICP-MS analysis of the intracellular copper
Detached cells (∼107 cells determined by a hemocytometer) were pelleted by centrifugation at 1000 rpm for 5 minutes. The cell pellet was dried at 60 °C in reduced pressure, and treated with 70% HNO3 (100 μL, Sigma, trace metal grade). The mixture was incubated at 40 °C overnight, diluted with Millipore water to 2% (v/v) HNO3, and centrifuged at 5500 rpm for 5 minutes to precipitate the insoluble cell debris. The supernatant was collected for ICP-MS analysis of copper content using a Varian 810 ICP-MS. The measured c/s (count/second) was converted to the copper concentration using the standard calibration curve.Acknowledgements
This work was supported by the National Institute of Health grants (R21HD058985 and 5R01EY013175), the National Science Foundation grant (DMR-1207583), and the GEAR grant from the University of Houston. Lin Wang thanks the support by The China Scholarship Council. We thank Dr Shadi Moghaddas for allowing use of the Western Blotting apparatus.Notes and references
- C. S. McKay and M. G. Finn, Chem. Biol., 2014, 21, 1075–1101 CrossRef CAS PubMed.
- J. A. Prescher and C. R. Bertozzi, Nat. Chem. Biol., 2005, 1, 13–21 CrossRef CAS PubMed.
- Z. Hao, S. Hong, X. Chen and P. R. Chen, Acc. Chem. Res., 2011, 44, 742–751 CrossRef CAS PubMed.
- K. Lang and J. W. Chin, Chem. Rev., 2014, 114, 4764–4806 CrossRef CAS PubMed.
- E. M. Sletten and C. R. Bertozzi, Angew. Chem., Int. Ed., 2009, 48, 6974–6998 CrossRef CAS PubMed.
- P. Thirumurugan, D. Matosiuk and K. Jozwiak, Chem. Rev., 2013, 113, 4905–4979 CrossRef CAS PubMed.
- C. A. DeForest, B. D. Polizzotti and K. S. Anseth, Nat. Mater., 2009, 8, 659–664 CrossRef CAS PubMed.
- L. Wang, M. Zhao, S. Li, U. J. Erasquin, H. Wang, L. Ren, C. Chen, Y. Wang and C. Cai, ACS Appl. Mater. Interfaces, 2014, 6, 8401–8406 CAS.
- N. J. Agard, J. A. Prescher and C. R. Bertozzi, J. Am. Chem. Soc., 2004, 126, 15046–15047 CrossRef CAS PubMed.
- N. K. Devaraj, R. Upadhyay, J. B. Haun, S. A. Hilderbrand and R. Weissleder, Angew. Chem., Int. Ed., 2009, 48, 7013–7016 CrossRef CAS PubMed.
- P. Agarwal, B. J. Beahm, P. Shieh and C. R. Bertozzi, Angew. Chem., Int. Ed., 2015, 54, 11504–11510 CrossRef CAS PubMed.
- V. V. Rostovtsev, L. G. Green, V. V. Fokin and K. B. Sharpless, Angew. Chem., Int. Ed., 2002, 41, 2596–2599 CrossRef CAS.
- C. W. Tornoe, C. Christensen and M. Meldal, J. Org. Chem., 2002, 67, 3057–3064 CrossRef CAS PubMed.
- I. E. Valverde, A. Bauman, C. A. Kluba, S. Vomstein, M. A. Walter and T. L. Mindt, Angew. Chem., Int. Ed., 2013, 52, 8957–8960 CrossRef CAS PubMed.
- A. Salic and T. J. Mitchison, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 2415–2420 CrossRef CAS PubMed.
- R. Manetsch, A. Krasiński, Z. Radić, J. Raushel, P. Taylor, K. B. Sharpless and H. C. Kolb, J. Am. Chem. Soc., 2004, 126, 12809–12818 CrossRef CAS PubMed.
- V. Hong, N. F. Steinmetz, M. Manchester and M. G. Finn, Bioconjugate Chem., 2010, 21, 1912–1916 CrossRef CAS PubMed.
- E. Lallana, R. Riguera and E. Fernandez-Megia, Angew. Chem., Int. Ed., 2011, 50, 8794–8804 CrossRef CAS PubMed.
- D. C. Kennedy, C. S. McKay, M. C. B. Legault, D. C. Danielson, J. A. Blake, A. F. Pegoraro, A. Stolow, Z. Mester and J. P. Pezacki, J. Am. Chem. Soc., 2011, 133, 17993–18001 CrossRef CAS PubMed.
- S. Li, H. Cai, J. He, H. Chen, S. Lam, T. Cai, Z. Zhu, S. J. Bark and C. Cai, Bioconjugate Chem., 2016, 27, 2315–2322 CrossRef CAS PubMed.
- V. Hong, S. I. Presolski, C. Ma and M. G. Finn, Angew. Chem., Int. Ed., 2009, 48, 9879–9883 CrossRef CAS PubMed.
- C. Besanceney-Webler, H. Jiang, T. Zheng, L. Feng, D. Soriano del Amo, W. Wang, L. M. Klivansky, F. L. Marlow, Y. Liu and P. Wu, Angew. Chem., Int. Ed., 2011, 50, 8051–8056 CrossRef CAS PubMed.
- T. R. Chan, R. Hilgraf, K. B. Sharpless and V. V. Fokin, Org. Lett., 2004, 6, 2853–2855 CrossRef CAS PubMed.
- D. Soriano del Amo, W. Wang, H. Jiang, C. Besanceney, A. C. Yan, M. Levy, Y. Liu, F. L. Marlow and P. Wu, J. Am. Chem. Soc., 2010, 132, 16893–16899 CrossRef CAS PubMed.
- C. Uttamapinant, A. Tangpeerachaikul, S. Grecian, S. Clarke, U. Singh, P. Slade, K. R. Gee and A. Y. Ting, Angew. Chem., Int. Ed., 2012, 51, 5852–5856 CrossRef CAS PubMed.
- H. Jiang, B. P. English, R. B. Hazan, P. Wu and B. Ovryn, Angew. Chem., Int. Ed., 2015, 54, 1765–1769 CrossRef CAS PubMed.
- Y. Yuan, S. Xu, X. Cheng, X. Cai and B. Liu, Angew. Chem., Int. Ed., 2016, 55, 6457–6461 CrossRef CAS PubMed.
- M. Yang, J. Li and P. R. Chen, Chem. Soc. Rev., 2014, 43, 6511–6526 RSC.
- M. Yang, Y. Song, M. Zhang, S. Lin, Z. Hao, Y. Liang, D. Zhang and P. R. Chen, Angew. Chem., Int. Ed., 2012, 51, 7674–7679 CrossRef CAS PubMed.
- M. Yang, A. S. Jalloh, W. Wei, J. Zhao, P. Wu and P. R. Chen, Nat. Commun., 2014, 5, 4981–7679 CrossRef CAS PubMed.
- V. Bevilacqua, M. King, M. Chaumontet, M. Nothisen, S. Gabillet, D. Buisson, C. Puente, A. Wagner and F. Taran, Angew. Chem., Int. Ed., 2014, 53, 5872–5876 CrossRef CAS PubMed.
- E. Vivès, P. Brodin and B. Lebleu, J. Biol. Chem., 1997, 272, 16010–16017 CrossRef.
- E. G. Stanzl, B. M. Trantow, J. R. Vargas and P. A. Wender, Acc. Chem. Res., 2013, 46, 2944–2954 CrossRef CAS PubMed.
- K. Sivakumar, F. Xie, B. M. Cash, S. Long, H. N. Barnhill and Q. Wang, Org. Lett., 2004, 6, 4603–4606 CrossRef CAS PubMed.
- J. C. M. van Hest, K. L. Kiick and D. A. Tirrell, J. Am. Chem. Soc., 2000, 122, 1282–1288 CrossRef CAS.
- J. A. Johnson, Y. Y. Lu, J. A. Van Deventer and D. A. Tirrell, Curr. Opin. Chem. Biol., 2010, 14, 774–780 CrossRef CAS PubMed.
- J. A. Key, S. Koh, Q. K. Timerghazin, A. Brown and C. W. Cairo, Dyes Pigm., 2009, 82, 196–203 CrossRef CAS.
- H. Oberleithner, Kidney Int., 2005, 67, 1680–1682 CrossRef CAS PubMed.
- J. T. Ngo and D. A. Tirrell, Acc. Chem. Res., 2011, 44, 677–685 CrossRef CAS PubMed.
- B. Zhao, P. Shah, A. V. Budanov, L. Qiang, M. Ming, A. Aplin, D. M. Sims and Y.-Y. He, J. Biol. Chem., 2014, 289, 35806–35814 CrossRef CAS PubMed.
- K. L. Kiick, E. Saxon, D. A. Tirrell and C. R. Bertozzi, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 19–24 CrossRef CAS PubMed.
- R. Franco, M. I. Panayiotidis and J. A. Cidlowski, J. Biol. Chem., 2007, 282, 30452–30465 CrossRef CAS PubMed.
- D. Giustarini, I. Dalle-Donne, A. Milzani, P. Fanti and R. Rossi, Nat. Protoc., 2013, 8, 1660–1669 CrossRef CAS PubMed.
- L. Banci, I. Bertini, S. Ciofi-Baffoni, T. Kozyreva, K. Zovo and P. Palumaa, Nature, 2010, 465, 645–648 CrossRef CAS PubMed.
- L. A. Finney and T. V. O'Halloran, Science, 2003, 300, 931–936 CrossRef CAS PubMed.
- B. E. Kim, T. Nevitt and D. J. Thiele, Nat. Chem. Biol., 2008, 4, 176–185 CrossRef CAS PubMed.
- H. Ostergaard, C. Tachibana and J. R. Winther, J. Cell Biol., 2004, 166, 337–345 CrossRef CAS PubMed.
- Z. G. Xiao, P. S. Donnelly, M. Zimmermann and A. G. Wedd, Inorg. Chem., 2008, 47, 4338–4347 CrossRef CAS PubMed.
- H. P. Broquist, Nutr. Rev., 1992, 50, 110–111 CrossRef CAS PubMed.
- M. De Nicola and L. Ghibelli, Front. Pharmacol., 2014, 5, 267 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Detailed experimental procedures, characterizations and supporting data. See DOI: 10.1039/c6sc02297a |
| This journal is © The Royal Society of Chemistry 2017 |