Open Access Article
Open Access ArticleSupramolecular assembly affording a ratiometric two-photon fluorescent nanoprobe for quantitative detection and bioimaging†
Peng
Wang
a,
Cheng
Zhang
a,
Hong-Wen
Liu
 a,
Mengyi
Xiong
a,
Sheng-Yan
Yin
a,
Yue
Yang
a,
Xiao-Xiao
Hu
ab,
Xia
Yin
*a,
Xiao-Bing
Zhang
a,
Mengyi
Xiong
a,
Sheng-Yan
Yin
a,
Yue
Yang
a,
Xiao-Xiao
Hu
ab,
Xia
Yin
*a,
Xiao-Bing
Zhang
 *a and
Weihong
Tan
*a and
Weihong
Tan
 a
a
aMolecular Science and Biomedicine Laboratory, State Key Laboratory of Chemo/Biosensing and Chemometrics, College of Chemistry and Chemical Engineering, Collaborative Innovation Center for Chemistry and Molecular Medicine, Hunan University, Changsha, Hunan 410082, China. E-mail: xbzhang@hnu.edu.cn
bMolecular Science and Biomedicine Laboratory, State Key Laboratory of Chemo/Biosensing and Chemometrics, College of Life Sciences, Hunan University, Changsha, Hunan 410082, China
First published on 9th October 2017
Abstract
Fluorescence quantitative analyses for vital biomolecules are in great demand in biomedical science owing to their unique detection advantages with rapid, sensitive, non-damaging and specific identification. However, available fluorescence strategies for quantitative detection are usually hard to design and achieve. Inspired by supramolecular chemistry, a two-photon-excited fluorescent supramolecular nanoplatform (TPSNP) was designed for quantitative analysis with three parts: host molecules (β-CD polymers), a guest fluorophore of sensing probes (Np–Ad) and a guest internal reference (NpRh–Ad). In this strategy, the TPSNP possesses the merits of (i) improved water-solubility and biocompatibility; (ii) increased tissue penetration depth for bioimaging by two-photon excitation; (iii) quantitative and tunable assembly of functional guest molecules to obtain optimized detection conditions; (iv) a common approach to avoid the limitation of complicated design by adjustment of sensing probes; and (v) accurate quantitative analysis by virtue of reference molecules. As a proof-of-concept, we utilized the two-photon fluorescent probe NHS–Ad-based TPSNP-1 to realize accurate quantitative analysis of hydrogen sulfide (H2S), with high sensitivity and good selectivity in live cells, deep tissues and ex vivo-dissected organs, suggesting that the TPSNP is an ideal quantitative indicator for clinical samples. What’s more, TPSNP will pave the way for designing and preparing advanced supramolecular sensors for biosensing and biomedicine.
Introduction
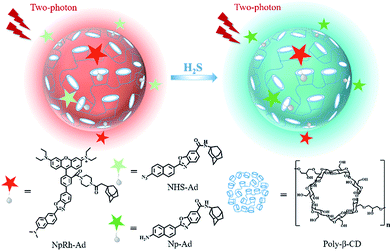
Optical visualization of vital biomolecules including gasotransmitters, enzymes, and nucleic acids holds a particularly prominent position owing to its high sensitivity, rapid response, non-damaging detection and real-time imaging.1–3 To date, as powerful tools, small molecular fluorescent probes have realized biosensing for various biomolecules, offering simplicity in operation and satisfactory performance.4–7 However, it should be noted that very little optimization work has been carried out on quantitative analysis for monitoring biomolecules in living cells, deep tissues, and organs. Current small molecular fluorescent probes have generally failed to achieve this goal due to their intrinsic limitations, such as short excitation wavelengths, shallow tissue penetration depths, environmentally sensitive fluorescence-intensity-based read-out, poor water-solubility and undesirable biocompatibility. Among them, ratiometric fluorescent probes are regarded as an alternative strategy for quantitative detection with self-calibration ability.8–11 Nevertheless, complicated energy transfer strategies and unmanageable ratiometric read-out signals delay the extensive and efficient quantitative analysis of ratiometric probes in biosamples. Therefore, there is an urgent need to design a fluorescent detection platform to realize efficient quantitative bioanalysis, but it is still a significant challenge.Since the Nobel Prize was awarded to host–guest and supramolecular chemistry, the field of supramolecular chemistry has attracted much attention from scientists, and numerous supramolecular systems have been developed to investigate their bioapplications.12–16 Utilizing host–guest self-assembled supramolecular polymers between macrocyclic molecules and functionalized guest molecules has already become a robust and flexible method to afford biosensing nanoprobes, which show tunable or stimuli-responsive physicochemical signals, prominent physiochemical shielding, improved water-solubility, and enhanced biocompatibility as well as avoiding the cumbersome synthesis provided by macrocyclic host molecules.17–19 Inspired by supramolecular chemistry, we intend to build a fluorescent supramolecular quantitative analysis nanoplatform with three parts: host molecules, guest probe molecules and guest internal reference molecules.
Within the common supramolecular self-assembly systems, cyclodextrins (CDs) are a popular class of macrocyclic ring as host molecules by virtue of their ready availability from their starch precursors, functionalization of a variety of guests, satisfactory water solubility and low toxicity toward biological systems.20–22 β-CD polymers (poly-β-CD) have been gradually used for molecular recognition, drug delivery and biosensing because of their large loading and tunable guest self-assembly.23–26 Thus, poly-β-CD are the optimal choice for our nanoplatform strategy. To achieve deep tissue penetration, bioimaging with near-infrared (NIR) photons as the excitation source is expected. Two-photon microscopy (TPM), which excites a two-photon (TP) fluorophore with NIR laser pulses, has emerged as a novel and indispensable imaging tool for biosensing.27–30 Compared with one-photon microscopy, TPM opens the opportunity for biosensing in living cells and deep tissues with the advantages of minimized fluorescence background, reduced photodamage, better three-dimensional spatial localization and increased penetration depth (>500 μm). Hence, based on our previous works, we chose a naphthalene-based TP fluorescent dye combined with adamantane (Np–Ad) as a fluorophore of the potential guest probes and TP through-bond energy transfer (TBET)-based rhodamine–adamantine (NpRh–Ad) as a TP guest internal reference with host nanoparticle poly-β-CD, to build a TP fluorescent supramolecular nanoplatform (TPSNP) for quantitative analysis. In this strategy, TPSNP would possess numerous merits from the host nanoparticles and functionalized guest molecules. First, poly-β-CD could improve the water-solubility and biocompatibility of small molecule fluorescent probes. Besides, as the host molecules, poly-β-CD could implement the quantitative and tunable assembly of functional guest molecules to obtain optimized conditions. Secondly, TP fluorescent guest molecules would increase the tissue penetration depth for bioimaging. Furthermore, this internal reference-doped supramolecular assembly strategy avoids the limitation of a complicated ratiometric design. Owing to the facile modification of Np–Ad, vast Np–Ad derivative probes could be utilized for various targets for physiological evaluation and disease diagnosis. To demonstrate the feasibility of the TPSNP design, hydrogen sulfide (H2S) was chosen due to its essential role as a gasotransmitter in the human body as the model target for the TPSNP-1, which was assembled with the Np–Ad derivative H2S sensing probe NHS–Ad, NpRh–Ad and poly-β-CD (Scheme 1).
Results and discussion
Preparation and characterization of the TPSNP
Poly-β-CD were synthesized according to previous literature.311H NMR spectra showed that most of the bands of β-CD at 4.2–3.4 ppm were broadened in the spectrum of the poly-β-CD, which confirmed the successful synthesis of poly-β-CD (Fig. S1†). The morphology and size of the poly-β-CD were investigated. Field emission scanning electron microscope (SEM) images showed that the poly-β-CD were spherical in shape and the size distribution was about 90 nm (Fig. S2†).TBET is a flexible strategy to design ratiometric probes without the need for spectral overlap between the donor emission and the acceptor absorption.32–35 Our group previously proposed a TBET system (Np–Rh), with a naphthalene derivative and rhodamine spirolactam to achieve deep-tissue imaging for Cu2+.36 We introduced adamantine, a common guest molecule for β-CD, into the rhodamine portion of Np–Rh by amide linkage with piperazine (termed NpRh–Ad) to make rhodamine always keep its ring-opening form and emit red fluorescence. Meanwhile, we utilized the naphthalene-based TP fluorescent probe with green fluorescence to functionalize the poly-β-CD. Owing to the TBET strategy, two emission bands from the naphthalene-based fluorophore and the rhodamine moiety displayed weak interplay when the sensing probe was activated. NpRh–Ad, Np–Ad and NHS–Ad were efficiently synthesized by following the synthetic route shown in Scheme S1† and all of the reaction products were fully characterized by 1H NMR, 13C NMR and mass spectrometry (see ESI, Fig. S16–S32†).
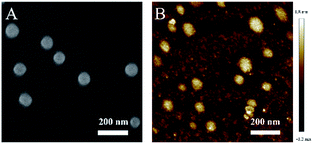
As a proof-of-concept, the poly-β-CD were incubated with NpRh–Ad (10 μM) and Np–Ad (5 μM) in a 10 mM phosphate buffered solution (PBS, pH = 7.4) at the same time and subjected to vibration treatment for 4 h at 300 rpm (25 °C). The morphology and size of the TPSNP were investigated using SEM, atomic force microscopy (AFM) and dynamic light scattering (DLS). The SEM and AFM images show that the self-assembled supramolecular nanoparticles were spherical in shape with a size distribution of ∼90 nm (Fig. 1), and its average hydrodynamic diameter in PBS was ∼140 nm by DLS analysis (Fig. S3†). There was no obvious change in the morphology and size between the TPSNP and poly-β-CD from the SEM images. To confirm the combination of the NpRh–Ad and/or Np–Ad with the poly-β-CD by supramolecular self-assembly, agarose gel electrophoresis (AGE) experiments were carried out. As can be seen in Fig. S4b,† under the effect of an electric field, NpRh–Ad and/or Np–Ad in the presence of poly-β-CD (channel 2, 4 and 5) remained owing to the high-molecular-weight, while the free dyes (channel 1 and 3) migrated. Furthermore, compared with the free NpRh–Ad and Np–Ad, the fluorescence emissions of NpRh–Ad/poly-β-CD and Np–Ad/poly-β-CD were both dramatically enhanced with 365 nm UV illumination (Fig. S4a†), which indicates the formation of NpRh–Ad/poly-β-CD and Np–Ad/poly-β-CD to improve the solubility of the corresponding dyes in buffer solution. These results are consistent with the previous literature that direct macrocyclic dye molecule host–guest supramolecular self-assembly enhanced the fluorescence intensity in aqueous solution.37–39 Obviously, supramolecular polymers have an advantage for detection over our previously proposed probe, Np–Rh, in biosamples, which requires 10% ethyl alcohol in buffer solution to work rather than an absolute water solution.
Spectroscopic studies of the TPSNPin vitro
The spectroscopic properties of the TPSNP were studied in PBS (pH = 7.4). It should be pointed out that, compared with the single dye-doped TPSNP, the two dye-doped TPSNP showed satisfactory overlay of the absorption spectra and distinction of the fluorescence emission spectra (Fig. S5†). What’s more, the red fluorescence region maintained no obvious difference after co-assembling (Fig. S5b†), suggesting that NpRh–Ad should be a competent TP internal reference. As shown in Fig. S6,† 1/(F − F0) vs. 1/[β-CD] was a linear relationship, indicating the formation of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
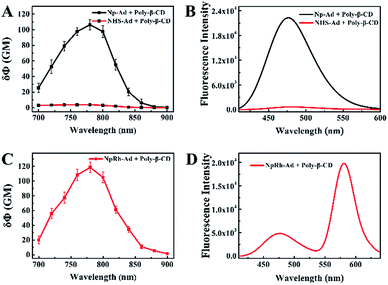
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 host–guest supramolecular polymer between the dyes with β-CD. Analyzed by the Benesi–Hildebrand plot, the binding constant (K) of NpRh–Ad with poly-β-CD in PBS (pH = 7.4) was 2.5 × 102 M−1, and that of Np–Ad with poly-β-CD was 2.5 × 103 M−1. At the same time, more than 96% of the dyes combined with β-CD cavities of the poly-β-CD. To achieve the capability of imaging deep inside a tissue and at a higher spacial resolution, the TP active absorption cross-section of host–guest supramolecular polymer was investigated. As shown in Fig. 2, the TP active absorption cross-section of NpRh–Ad with the poly-β-CD was 118.55 GM (1 GM = 10−50 (cm4 s) per photon) at 780 nm, and that of Np–Ad with the poly-β-CD was 106.38 GM. According to our previous reports, the TP properties of the supramolecular polymer were similar to those of the free dyes, which certifies that the assembled nanoprobes keep the TP properties of dyes. We also studied the effect of pH on the TPSNP and Np–Ad/poly-β-CD. Compared with Np–Ad/poly-β-CD (Fig. S7b†), pH changes (from 3.0 to 13.0) had no obvious effect on the performance of the TPSNP (Fig. S7a†), indicating that the fluorescent internal reference system can avoid interferences from the surroundings (such as pH value, viscosity and polarity) and have a predictable performance in its self-calibration ability. The stability of the TPSNP in the PBS (pH = 7.4) and cell culture medium was investigated by monitoring its fluorescence ratio (F475/F575) changes (Fig. S8†). The results showed that the prepared host–guest supramolecular polymer had good stability in detecting conditions and complex environments for long periods of time.
1 host–guest supramolecular polymer between the dyes with β-CD. Analyzed by the Benesi–Hildebrand plot, the binding constant (K) of NpRh–Ad with poly-β-CD in PBS (pH = 7.4) was 2.5 × 102 M−1, and that of Np–Ad with poly-β-CD was 2.5 × 103 M−1. At the same time, more than 96% of the dyes combined with β-CD cavities of the poly-β-CD. To achieve the capability of imaging deep inside a tissue and at a higher spacial resolution, the TP active absorption cross-section of host–guest supramolecular polymer was investigated. As shown in Fig. 2, the TP active absorption cross-section of NpRh–Ad with the poly-β-CD was 118.55 GM (1 GM = 10−50 (cm4 s) per photon) at 780 nm, and that of Np–Ad with the poly-β-CD was 106.38 GM. According to our previous reports, the TP properties of the supramolecular polymer were similar to those of the free dyes, which certifies that the assembled nanoprobes keep the TP properties of dyes. We also studied the effect of pH on the TPSNP and Np–Ad/poly-β-CD. Compared with Np–Ad/poly-β-CD (Fig. S7b†), pH changes (from 3.0 to 13.0) had no obvious effect on the performance of the TPSNP (Fig. S7a†), indicating that the fluorescent internal reference system can avoid interferences from the surroundings (such as pH value, viscosity and polarity) and have a predictable performance in its self-calibration ability. The stability of the TPSNP in the PBS (pH = 7.4) and cell culture medium was investigated by monitoring its fluorescence ratio (F475/F575) changes (Fig. S8†). The results showed that the prepared host–guest supramolecular polymer had good stability in detecting conditions and complex environments for long periods of time.
Cytotoxicity and intracellular uptake of the TPSNP
To elucidate its further bioapplication, the cytotoxicity of the TPSNP was first tested by MTS assays. As shown in Fig. S9,† the cells maintained high viability after incubation with the TPSNP for 24 h even at high concentrations, clearly indicating the low cytotoxicity of the TPSNP and suggesting its suitable application in intracellular sensing. To demonstrate the cellular uptake of this nanoprobe, imaging of HeLa cells was carried out after incubation with the TPSNP for different periods of time. As shown in Fig. S10,† the TPSNP were well internalized after 2 h of incubation. These results are consistent with vesicular uptake mechanisms for nanoparticles. The intracellular distribution of the TPSNP was also investigated by co-localization with a commercial lysosome-specific fluorescent dye, LysoTracker Red (Fig. S11†). The red fluorescence from the LysoTracker Red and the green fluorescence from the Np–Ad/poly-β-CD overlapped well with a Pearson’s correlation factor of 0.9295, which indicated that the nanoprobe was localized in the lysosomes. Therefore, we speculated that the TPSNP could be internalized by cells and used for biosensing.H2S sensing of the TPSNP-1in vitro
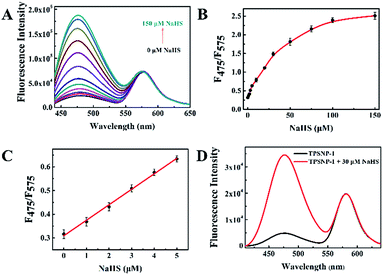
To exhibit the outstanding performance of quantitative determination of biomolecules in living cells and deep tissues by using TPSNP, H2S was chosen as the model target for further study. As an important member of the gasotransmitter family, H2S has been identified in mediating many physiological systems, such as the central nervous system, immune system, and blood circulation system.40–44 It is generally accepted that abnormal H2S level in live cells would result in a variety of human diseases, such as Alzheimer’s disease and Down’s syndrome.45,46 Therefore, it is of scientific interest to develop effective methods for the quantitative analysis of H2S in biosystems. We previously proposed a naphthalene derivative-based TP fluorescent probe, termed NHS1, which could be used for rapid detection of endogenous H2S in live cells.47 Based on this research, we designed a TP fluorescent probe, NHS–Ad, and combined it with NpRh–Ad and poly-β-CD to construct a TP fluorescent supramolecular nanoprobe (TPSNP-1) for H2S detection. TPSNP-1 operated through reaction between the H2S and azide group of NHS–Ad, leading to reduction of the azide to an amino and fluorescence enhancement of the NHS–Ad.The fluorescence spectra changes of the TPSNP-1 upon the gradual addition of H2S were investigated. As shown in Fig. 3A, the addition of NaHS triggered a large fluorescence enhancement of the TPSNP-1 at 475 nm due to the production of Np–Ad. Furthermore, accompanying the fluorescence enhancement at 475 nm, the fluorescence intensity at 575 nm remain unchanged, which proved the outstanding performance of the TP internal reference. To verify the advantage of poly-β-CD in the quantitative and tunable assembly of guests, different molar ratios of NHS–Ad and NpRh–Ad were combined with the poly-β-CD for H2S sensing. As shown in Fig. 3A and S12,† the ratios of the two dopants ranging from 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 1
1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 made the H2S-responsive sensitivity of the TPSNP-1 different. To obtain the optimal sensitivity and effectiveness of the internal reference, we chose TPSNP-1 prepared with a molar ratio of 2
2 made the H2S-responsive sensitivity of the TPSNP-1 different. To obtain the optimal sensitivity and effectiveness of the internal reference, we chose TPSNP-1 prepared with a molar ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (NHS–Ad to NpRh–Ad) for further study. As can be seen from Fig. 3C, the F475/F575 was linearly proportional to NaHS in the concentration range of 0–5 μM. The detection limit (S/N = 3) for NaHS was calculated to be 86 nM, which is favorable for direct imaging of intracellular H2S, the concentrations of which are usually in a submicromolar range. Notably, the detection sensitivity of this nanoprobe is similar to that of the reported probe NHS1, which certifies that the predominant biosensing capacity of the small molecule probe is kept after being combined with the host–guest supramolecular system. The selectivity of the TPSNP-1 for H2S was also investigated. As shown in Fig. S13,† the F475/F575 of the TPSNP-1 exhibited no obvious changes in the presence of these potential intracellular coexisting species, which demonstrates the excellent selectivity of the TPSNP-1 for H2S. The time-dependent fluorescence response was investigated. As shown in Fig. S14,† the fluorescence signal reached a plateau after about 40 min. The TP properties of NHS–Ad/poly-β-CD were investigated. As shown in Fig. 2A, the TP active absorption cross-section of NHS–Ad with poly-β-CD was 3.85 GM at 780 nm. The TP excited fluorescence spectra of the TPSNP-1 upon addition of H2S are shown in Fig. 3D. A distinct TP excited fluorescence enhancement at 475 nm was observed upon excitation by 780 nm femtosecond pulses. The drastic TP excited fluorescence enhancement is favorable for intracellular H2S imaging with high resolution and sensitivity by TPM.
1 (NHS–Ad to NpRh–Ad) for further study. As can be seen from Fig. 3C, the F475/F575 was linearly proportional to NaHS in the concentration range of 0–5 μM. The detection limit (S/N = 3) for NaHS was calculated to be 86 nM, which is favorable for direct imaging of intracellular H2S, the concentrations of which are usually in a submicromolar range. Notably, the detection sensitivity of this nanoprobe is similar to that of the reported probe NHS1, which certifies that the predominant biosensing capacity of the small molecule probe is kept after being combined with the host–guest supramolecular system. The selectivity of the TPSNP-1 for H2S was also investigated. As shown in Fig. S13,† the F475/F575 of the TPSNP-1 exhibited no obvious changes in the presence of these potential intracellular coexisting species, which demonstrates the excellent selectivity of the TPSNP-1 for H2S. The time-dependent fluorescence response was investigated. As shown in Fig. S14,† the fluorescence signal reached a plateau after about 40 min. The TP properties of NHS–Ad/poly-β-CD were investigated. As shown in Fig. 2A, the TP active absorption cross-section of NHS–Ad with poly-β-CD was 3.85 GM at 780 nm. The TP excited fluorescence spectra of the TPSNP-1 upon addition of H2S are shown in Fig. 3D. A distinct TP excited fluorescence enhancement at 475 nm was observed upon excitation by 780 nm femtosecond pulses. The drastic TP excited fluorescence enhancement is favorable for intracellular H2S imaging with high resolution and sensitivity by TPM.
TP fluorescence imaging of H2S in living cells and deep tissues
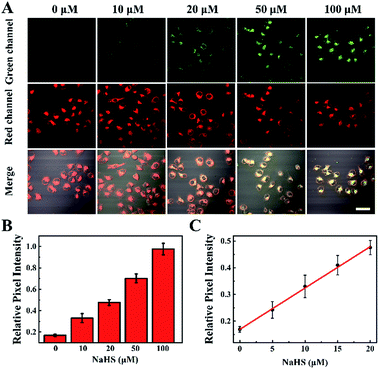
Owing to the highly sensitive and selective response to H2S and its remarkable TP properties, TP excited confocal fluorescence imaging experiments of the TPSNP-1 for H2S in living HeLa cells were then carried out. As shown in Fig. 4A, incubation of the HeLa cells with the TPSNP-1 afforded a gradually enhanced green fluorescence emission and steady red fluorescence emission when co-incubated with additional NaHS upon excitation at 780 nm. Notably, due to the presence of a red fluorescent internal reference, the green fluorescence emission enhancement could veritably reflect the response between the TPSNP-1 and H2S, avoiding the interference of fluorescence intensity from the surroundings, such as pH value, viscosity and polarity. Therefore, we could calculate the relative pixel fluorescence intensity of the HeLa cells images (Fig. 4B) and obtain a standard curve of the H2S response (Fig. 4C).To further show the excellent performance of the TPSNP-1, TP imaging of H2S in rat normal liver tissue slices was carried out. The changes in fluorescence signal intensity with scan depth were recorded by TPM in the Z-scan mode. As can be seen from Fig. S15,† the TPSNP-1 was capable of tissue imaging at depths of 80–170 μm by TPM. In the absence of exogenous H2S, the green channel exhibited a weak TP fluorescence signal (Fig. 5A and S15A†), while the red channel had an intense signal (Fig. 5B and S15B†), indicating that the TPSNP-1 had excellent tissue penetration and staining capabilities as well as non-interfering two-color (green and red) imaging ability. In addition, endogenic H2S in the normal rat liver tissue was at a low level. When NaHS was added, a drastic TP fluorescence signal enhancement was observed in the green channel (Fig. 5C and S15C†) and the fluorescence signal intensity of the red channel almost remained the same (Fig. 5D and S15D†). These data showed that the TPSNP-1 not only achieved TP fluorescence imaging in deep tissues but also possessed the power of ratiometric TP imaging by virtue of the red fluorescent TP internal reference.
H2S quantitative analysis in organs by the TPSNP-1
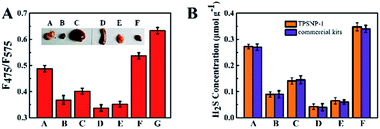
Since the TPSNP-1 demonstrated excellent performance in cells and deep tissues, we attempted to quantitatively evaluate the concentrations of H2S in ex vivo-dissected organs. As can be seen from the inset of Fig. 6, we dissected the living mice to get the visceral organs, including the brain, heart, liver, spleen, lung and kidney. Then fluorescence tests with the TPSNP-1 and commercial kits for endogenous H2S were carried out. We calculated the content of H2S in the aforementioned organs on the basis of the standard curve of the TPSNP-1, and made a comparison with that of H2S commercial kits (Fig. 6). The results demonstrated that the TPSNP-1 had a remarkable property for quantitative analysis of physiological factors. As shown in Fig. 6, the concentrations of H2S in the brain and kidney were much higher than that in the other organs. It is well known that H2S is a neuromodulator and neuroprotectant in the brain, and a wide range of H2S concentrations are reported from undetectable to micromolar, which are determined by brain activity.48 The fact that endogenous H2S related enzyme cystathionine γ-lyase (CSE) is at a high level may lead to a higher concentration of H2S in the kidney.49 Hence, the TPSNP-1 offered exciting opportunities for quantitative analysis in various organs and was promising for clinical samples to diagnose disease.Conclusions
In summary, we developed a novel two-photon-excited supramolecular nanoplatform TPSNP based on the host–guest supramolecular recognition between the TP fluorophore of a potential guest probe (Np–Ad), a TP guest internal reference (NpRh–Ad) and host nanoparticles (poly-β-CD). The TPSNP exhibited outstanding physicochemical properties (supramolecular self-assembly, spectroscopic properties, TP property and stability), feasible cell uptake and good biocompatibility. Thanks to facile modification of Np–Ad, the TPSNP has the most significant potential in quantitative analysis for biomolecules for physiological evaluation and disease diagnosis. We employed the TPSNP-1 to realize the quantitative analysis of gasotransmitter H2S in living cells, deep tissues and ex vivo-dissected organs with satisfactory performance (high sensitivity, good selectivity, deep tissue imaging and accurate quantitative detection). What’s more, the TPSNP is conducive to widening the functions and applications of supramolecular chemistry. We believe that the TP fluorescent supramolecular nanoplatform, TPSNP, will pave the way for designing and preparing advanced supramolecular sensor materials for biosensing and biomedicine.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grants 21325520, 21327009 and J1210040), the Foundation for Innovative Research Groups of NSFC (Grant 21521063), and the Science and Technology Project of Hunan Province (2016RS2009 and 2016WK2002).Notes and references
- X. Li, X. Gao, W. Shi and H. Ma, Chem. Rev., 2014, 114, 590–659 CrossRef CAS PubMed.
- J. A. Thomas, Chem. Soc. Rev., 2015, 44, 4494–4500 RSC.
- W. Xu, Z. Zeng, J. H. Jiang, Y. T. Chang and L. Yuan, Angew. Chem., Int. Ed., 2016, 55, 13658–13699 CrossRef CAS PubMed.
- A. R. Lippert, E. J. New and C. J. Chang, J. Am. Chem. Soc., 2011, 133, 10078–10080 CrossRef CAS PubMed.
- Y. Gabe, Y. Urano, K. Kikuchi, H. Kojima and T. Nagano, J. Am. Chem. Soc., 2004, 126, 3357–3367 CrossRef CAS PubMed.
- D. Asanuma, M. Sakabe, M. Kamiya, K. Yamamoto, J. Hiratake, M. Ogawa, N. Kosaka, P. L. Choyke, T. Nagano, H. Kobayashi and Y. Urano, Nat. Commun., 2015, 6, 6463 CrossRef CAS PubMed.
- X. Wu, L. Li, W. Shi, Q. Gong and H. Ma, Angew. Chem., Int. Ed., 2016, 55, 14728–14732 CrossRef CAS PubMed.
- J. Fan, M. Hu, P. Zhan and X. Peng, Chem. Soc. Rev., 2013, 42, 29–43 RSC.
- K. Gu, Y. Liu, Z. Guo, C. Lian, C. Yan, P. Shi, H. Tian and W. H. Zhu, ACS Appl. Mater. Interfaces, 2016, 26622–26629 CAS.
- Z. Dai, L. Tian, B. Song, Z. Ye, X. Liu and J. Yuan, Anal. Chem., 2014, 86, 11883–11889 CrossRef CAS PubMed.
- C. Huang, T. Jia, M. Tang, Q. Yin, W. Zhu, C. Zhang, Y. Yang, N. Jia, Y. Xu and X. Qian, J. Am. Chem. Soc., 2014, 136, 14237–14244 CrossRef CAS PubMed.
- B. Shi, K. Jie, Y. Zhou, J. Zhou, D. Xia and F. Huang, J. Am. Chem. Soc., 2016, 138, 80–83 CrossRef CAS PubMed.
- P. Xing and Y. Zhao, Adv. Mater., 2016, 28, 7304–7339 CrossRef CAS PubMed.
- Y. Zhou, W. Huang, J. Liu, X. Zhu and D. Yan, Adv. Mater., 2010, 22, 4567–4590 CrossRef CAS PubMed.
- R. Dong, Y. Zhou, X. Huang, X. Zhu, Y. Lu and J. Shen, Adv. Mater., 2015, 27, 498–526 CrossRef CAS PubMed.
- J. Murray, K. Kim, T. Ogoshi, W. Yao and B. C. Gibb, Chem. Soc. Rev., 2017, 46, 2479–2496 RSC.
- X. Ma and Y. Zhao, Chem. Rev., 2015, 115, 7794–7839 CrossRef CAS PubMed.
- G. Yu, K. Jie and F. Huang, Chem. Rev., 2015, 115, 7240–7303 CrossRef CAS PubMed.
- J. Hu and S. Liu, Acc. Chem. Res., 2014, 47, 2084–2095 CrossRef CAS PubMed.
- S. Li and W. C. Purdy, Chem. Rev., 1992, 92, 1457–1470 CrossRef CAS.
- T. Irie and K. Uekama, J. Pharm. Sci., 1997, 86, 147–162 CrossRef CAS PubMed.
- M. E. Brewster and T. Loftsson, Adv. Drug Delivery Rev., 2007, 59, 645–666 CrossRef CAS PubMed.
- R. Liu, Y. Zhang and P. Feng, J. Am. Chem. Soc., 2009, 131, 15128–15129 CrossRef CAS PubMed.
- L. He, X. Yang, F. Zhao, K. Wang, Q. Wang, J. Liu, J. Huang, W. Li and M. Yang, Anal. Chem., 2015, 87, 2459–2465 CrossRef CAS PubMed.
- H. Yan, L. He, C. Ma, J. Li, J. Yang, R. Yang and W. Tan, Chem. Commun., 2014, 50, 8398–8401 RSC.
- H. Yan, L. He, W. Zhao, J. Li, Y. Xiao, R. Yang and W. Tan, Anal. Chem., 2014, 86, 11440–11450 CrossRef CAS PubMed.
- H. M. Kim and B. R. Cho, Chem. Rev., 2015, 115, 5014–5055 CrossRef CAS PubMed.
- G. S. He, L.-S. Tan, Q. Zheng and P. N. Prasad, Chem. Rev., 2008, 108, 1245–1330 CrossRef CAS PubMed.
- H. M. Kim and B. R. Cho, Acc. Chem. Res., 2009, 42, 863–872 CrossRef CAS PubMed.
- H. W. Liu, Y. Liu, P. Wang and X. B. Zhang, Methods Appl. Fluoresc., 2017, 5, 012003 CrossRef PubMed.
- C. Koopmans and H. Ritter, Macromolecules, 2008, 41, 7418–7422 CrossRef CAS.
- G. S. Jiao, L. H. Thoresen and K. Burgess, J. Am. Chem. Soc., 2003, 125, 14668–14669 CrossRef CAS PubMed.
- W. Lin, L. Yuan, Z. Cao, Y. Feng and J. Song, Angew. Chem., Int. Ed., 2010, 49, 375–379 CrossRef CAS PubMed.
- J. Fan, P. Zhan, M. Hu, W. Sun, J. Tang, J. Wang, S. Sun, F. Song and X. Peng, Org. Lett., 2013, 15, 492–495 CrossRef CAS PubMed.
- H. W. Liu, X. B. Zhang, J. Zhang, Q. Q. Wang, X. X. Hu, P. Wang and W. Tan, Anal. Chem., 2015, 87, 8896–8903 CrossRef CAS PubMed.
- L. Zhou, X. Zhang, Q. Wang, Y. Lv, G. Mao, A. Luo, Y. Wu, Y. Wu, J. Zhang and W. Tan, J. Am. Chem. Soc., 2014, 136, 9838–9841 CrossRef CAS PubMed.
- F. Biedermann, E. Elmalem, I. Ghosh, W. M. Nau and O. A. Scherman, Angew. Chem., Int. Ed., 2012, 51, 7739–7743 CrossRef CAS PubMed.
- H. J. Kim, D. R. Whang, J. Gierschner and S. Y. Park, Angew. Chem., Int. Ed., 2016, 55, 15915–15919 CrossRef CAS PubMed.
- R. N. Dsouza, U. Pischel and W. M. Nau, Chem. Rev., 2011, 111, 7941–7980 CrossRef CAS PubMed.
- L. Li, P. Rose and P. K. Moore, Annu. Rev. Pharmacol. Toxicol., 2011, 51, 169–187 CrossRef CAS PubMed.
- J. M. Fukuto, S. J. Carrington, D. J. Tantillo, J. G. Harrison, L. J. Ignarro, B. A. Freeman, A. Chen and D. A. Wink, Chem. Res. Toxicol., 2012, 25, 769–793 CrossRef CAS PubMed.
- R. Wang, Physiol. Rev., 2012, 92, 791–896 CrossRef CAS PubMed.
- P. K. Yadav, M. Martinov, V. Vitvitsky, J. Seravalli, R. Wedmann, M. R. Filipovic and R. Banerjee, J. Am. Chem. Soc., 2016, 138, 289–299 CrossRef CAS PubMed.
- K. Ono, T. Akaike, T. Sawa, Y. Kumagai, D. A. Wink, D. J. Tantillo, A. J. Hobbs, P. Nagy, M. Xian, J. Lin and J. M. Fukuto, Free Radical Biol. Med., 2014, 77, 82–94 CrossRef CAS PubMed.
- K. Eto, T. Asada, K. Arima, T. Makifuchi and H. Kimura, Biochem. Biophys. Res. Commun., 2002, 293, 1485–1488 CrossRef CAS PubMed.
- M. Lee, C. Schwab, S. Yu, E. McGeer and P. L. McGeer, Neurobiol. Aging, 2009, 30, 1523–1534 CrossRef CAS PubMed.
- G. J. Mao, T. T. Wei, X. X. Wang, S. Y. Huan, D. Q. Lu, J. Zhang, X. B. Zhang, W. Tan, G. L. Shen and R. Q. Yu, Anal. Chem., 2013, 85, 7875–7881 CrossRef CAS PubMed.
- Y. Qian, L. Zhang, S. Ding, X. Deng, C. He, X. E. Zheng, H.-L. Zhu and J. Zhao, Chem. Sci., 2012, 3, 2920–2923 RSC.
- C. A. Wagner, J. Nephrol., 2009, 22, 173–176 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7sc03977h |
| This journal is © The Royal Society of Chemistry 2017 |