 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Correction: Hypoxia inducible factor (HIF) as a model for studying inhibition of protein–protein interactions
George M.
Burslem
ab,
Hannah F.
Kyle
bc,
Adam
Nelson
ab,
Thomas A.
Edwards
bc and
Andrew J.
Wilson
*ab
aSchool of Chemistry, University of Leeds, Woodhouse Lane, Leeds LS2 9JT, UK. E-mail: a.j.wilson@leeds.ac.uk
bAstbury Centre for Structural Molecular Biology, University of Leeds, Woodhouse Lane, Leeds LS2 9JT, UK
cSchool of Molecular and Cellular Biology, Faculty of Biological Sciences, University of Leeds, Woodhouse Lane, Leeds LS2 9JT, UK
First published on 6th June 2017
Abstract
Correction for ‘Hypoxia inducible factor (HIF) as a model for studying inhibition of protein–protein interactions’ by George M. Burslem et al., Chem. Sci., 2017, DOI: 10.1039/c7sc00388a.
The authors regret that Fig. 3 is incorrect in the original manuscript. The chemical structure of VH298 was shown with an oxazole ring instead of the correct methyl-thiazole ring. The correct figure is displayed below.
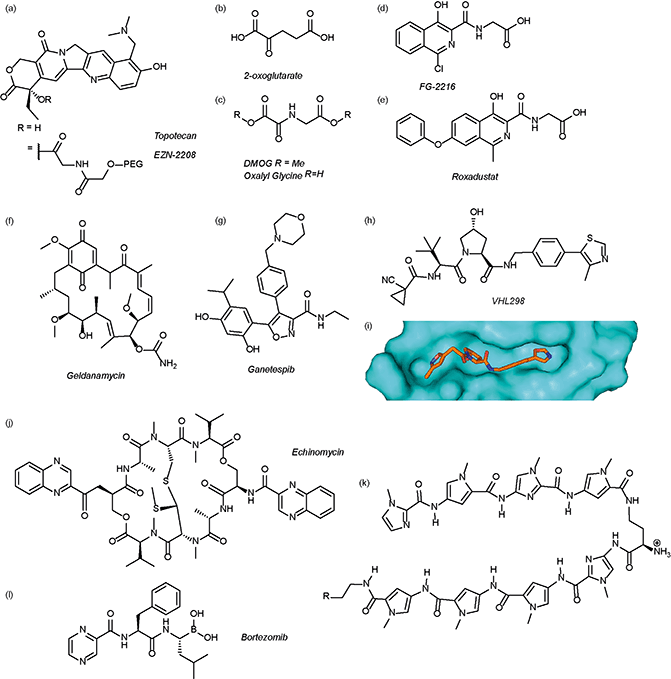 | ||
| Fig. 3 Modulators of the HIF pathway (a) topotecan and derivative EZN-2208 (b) 2-oxoglutarate (c) DMOG and oxalyl glycine (d) FG-2216 (e) roxadustat (f) geldanamycin (g) ganetespib (h) X-ray crystal structure of a hydroxyproline derived inhibitor (orange) bound to pVHL (cyan), PDB ID 3zrc (i) optimised hydroxyproline derived pVHL inhibitor VH298 (j) echinomycin (k) DNA sequence specific polyamide (l) bortezomib. | ||
The Royal Society of Chemistry apologises for these errors and any consequent inconvenience to authors and readers.
| This journal is © The Royal Society of Chemistry 2017 |
