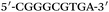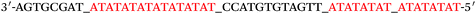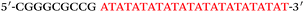Novel ultrasensitive homogeneous electrochemical aptasensor based on dsDNA-templated copper nanoparticles for the detection of ractopamine
Feifan
Sheng
a,
Xiaojun
Zhang
a and
Guangfeng
Wang
*ab
aKey Laboratory of Chem-Biosensing, Anhui Province, Key Laboratory of Functional Molecular Solids, Anhui Province, College of Chemistry and Materials Science, Center for Nano Science and Technology, Anhui Normal University, Wuhu 241000, P. R. China. E-mail: wangyuz@mail.ahnu.edu.cn
bState Key Laboratory of Chemo/Biosensing and Chemometrics, Hunan University, Changsha 410082, P. R. China
First published on 15th September 2016
Abstract
Herein, we describe a novel homogenous electrochemical aptasensor for the ultrasensitive detection of ractopamine (RAC) based on the signal amplification of a hairpin DNA cascade amplifier (HDCA) and electrocatalysis of dsDNA-templated copper nanoparticles. The present electrochemical aptasensor employs a label-free “turn-on” strategy with enzyme-free amplification. Briefly, the target RAC triggers the HDCA autocatalytic process of two ingeniously designed complementary hairpin DNA, which results in the formation of nucleic-acid-stabilized copper nanoparticles (dsDNA/CuNPs). Thus, with the mimic oxidase catalytic character of the dsDNA/CuNPs and enzyme-free hairpin DNA cascade amplifier reactions, a remarkable electrochemical response can be achieved, which is dependent on the concentration of the target RAC. Taking advantage of the highly amplified efficiency of trigger recycling and the excellent electrochemical catalytic response of dsDNA/CuNPs, the proposed strategy is capable of detecting RAC ultrasensitively. Under optimal conditions, the electrochemical signal increases with an increase in the target RAC concentration in the wide dynamic range from 1 × 10−12 M to 3 × 10−7 M with a detection limit (LOD) of 3 × 10−13 M. The proposed system does not need any sophisticated operation procedures such as electrode immobilization and expensive labeling. Therefore, the devised label-free and enzyme-free amplification homogeneous electrochemical aptasensor strategy may become an alternative method for the simple, sensitive, and selective detection of biological small molecules, proteins, nucleic acids and nuclease activity.
1. Introduction
Ractopamine (RAC) is one of the typical β-agonists, which originally are drugs that can promote vasodilatation, bronchodilation, increase heart rate, and are usually used for the treatment of asthma and pulmonary disease.1 They are also widely used to feed farm animals in high doses due to their ability to improve growth rate and increase the percentage of lean meat.2 However, the residues of these excessive drugs readily accumulate in animal tissue, and may cause acute poisoning when consumed by humans, with symptoms of muscle tremors, cardiac palpitation, tachycardia, confusion and nervousness.3,4 Since 1998, it has been estimated that more than 3000 people have been poisoned by eating swine and pork organs contaminated by the residues of β-agonists in China. To ensure food safety, the use of RAC has been banned in animal breeding in many countries such as China, Russia and the European Union.5 The Codex Alimentarius Commission recommends the adoption of the maximum residue limit (MRL) of 10 μg kg−1 of RAC in pork or beef, 40 μg kg−1 of RAC in animal liver and 90 μg kg−1 of RAC in animal kindey.6 Therefore, it is necessary to develop a facile, sensitive and selective method for the detection of RAC residues for routine monitoring.To date, many methods have been developed for the detection of RAC. One of the most frequently used methods is the traditional chromatographic method, which includes high performance liquid chromatography (HPLC), gas chromatography-mass spectrometry (GC-MS), and high performance liquid chromatography-mass spectrometry (HPLC-MS).6–9 However, these chromatography methods are susceptible to the same intrinsic limitations, such as complicated specimen preparation procedures, which are time consuming and often require large and sophisticated apparatus, thus making them unsuitable for field routine operation. Although some rapid detection methods, including enzyme immunoassays,10 immuno chromatographic assays,11,12 surface-enhanced Raman scattering immunoassays,13 and visual probes,14 have been developed to overcome these shortcomings, these methods either demand antibodies, which are difficult to acquire, or are unable to possess the sensitivity to reach the maximum residue limit (MRL) grade. Due to the high sensitivity, low cost and fast response of electrochemical aptasensors, they have been designed to monitor RAC in real sample matrices.15–18 However, electrochemical RAC aptasensors involve the immobilization of DNA probes on the electrode surface, and further detection in the solution phase, and the immobilization processes are time-consuming and intricate. Moreover, the existence of local steric hindrance and limited reaction area may lead to a relatively low binding efficiency. The molecular activity also may be affected by the processes used for electrode modification.19,20 To solve these problems, it is urgent to fabricate novel electrochemical RAC aptasensors which could avoid the process of electrode immobilization or modification.
Homogeneous electrochemistry is a simple assay process which usually involves no electrode immobilization step. In homogenous electrochemical sensing, the recognition reaction, such as hybridization between the probe DNA and target DNA, occurs in the solution phase instead of on the surface of the electrode.19 It has the advantages of rapid response, simplicity, and improved recognition efficiency compared with heterogeneous methods.19–26 Therefore, it is highly desirable to develop homogeneous electrochemical RAC aptasensors.
However, although recently, homogeneous electrochemical strategies have been developed for the detection of various targets, including small biological molecules, metal ions, DNA and enzyme activities,19–30 most of these homogeneous electrochemical signals originate from intercalated methylene blue (MB)23,31–33 or the ferrocene (Fc) label,19,34,35 which may be confronted with some limitations such as the toxicity of MB dye, or expensive and complex labeling processes. Moreover, in order to enhance sensitivity in homogenous electrochemistry, enzyme amplification is usually used to amplify signals,23,33,35 which mostly involves unfavourable conditions such as controlled reaction temperatures and long reaction times. Thus, it is significant to develop a biocompatible and label-free probe for enzyme-free amplified homogenous electrochemical sensors.
Very recently, Wang et al. reported that poly-adenine–thymine (poly AT) can be employed as a promising template for the synthesis of copper nanoparticles (CuNPs) through the accumulation of Cu(0) at a low concentration in the major groove of dsDNA scaffolds.36–38 Attractively, compared with other nucleic acid-templated metal nanoparticles, the dsDNA-templated CuNPs (dsDNA/CuNPs) could be synthesized via a simple and fast process in ∼3 h. Also, the synthetic conditions are mild without any rigorous operations, including vigorous agitation, heating/cooling and dark treatment, which result in good repeatability. In addition, due to the fact that copper is a significant and essential micronutrient for all living animals and plants,39–41 CuNPs is safer than other heavy-metal nanoparticles and quantum dots.42,43 Therefore, dsDNA/CuNPs with synthetic rapidity, simplicity, high efficiency and hypotoxicity, may become a competent biosensing probe for chemical/biological analysis. However, to date, the numerous effective applications of dsDNA/CuNPs are still based on their fluorescent properties, which may limit their further applications due to the disadvantages of fluorimetry, including the requirement of expensive instruments and sensitivity to interferences.44 Considering that copper has good conductivity, the novel electrochemical character of dsDNA/CuNPs and their further electrochemical application are worth exploring.
Inspired by the urgent requirement of label-free and enzyme-free amplified homogeneous electrochemical detection of RAC and the desirable exploration of the electrochemical character of dsDNA/CuNPs, herein, we report, for the first time, the electrocatalysis response of dsDNA/CuNPs and their further use as a label-free and biocompatible probe for the homogeneous electrochemical biosensing of RAC with enzyme-free amplification. The signal is based on the electrochemical catalysis of the dsDNA/CuNPs and signal amplification is realized by an enzyme-free hairpin DNA cascade amplifier (HDCA).45 Taking advantage of the highly amplified efficiency of trigger recycling and excellent electrochemical catalytic signals of dsDNA/CuNPs, the proposed strategy is capable of detecting RAC ultrasensitively. The features of this “turn-on” system for RAC detection is simple, immobilization-free, label-fee and sensitive, which indicate a high performance for the detection of RAC. The developed strategy could be easily expanded for the exploration of a wide range of analytes including biological small molecules, proteins, nucleic acids and nuclease activity.
2. Experimental
2.1. Materials and chemicals
HPLC-purified oligonucleotide sequences were purchased from Sangon Biotech. Co., Ltd (Shanghai, China), which are listed in Table 1. Ractopamine (RAC), clenbuterol (CLB), salbuterol (SAL) and chloramphenicol (CAP) were purchased from Aldrich Company. 3-(N-Morpholino) propanesulfonic acid (MOPS) and sodium ascorbate (SA) were purchased from Sangon Biotech. Co., Ltd (Shanghai, China). 3,3′,5,5′-Tetramethylbenzidine (TMB), methanol, copper sulfate (CuSO4·5H2O) and hydrogen peroxide (H2O2) were purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). All other chemicals were of analytical grade and used as received.2.2. Apparatus
A CHI 660B electrochemical workstation (Shanghai Chenhua Instruments Co., Shanghai, China) was employed to conduct the electrochemical experiments. The conventional three-electrode configuration was used, with a bare glassy carbon working electrode (GCE) (2 mm in diameter), a saturated calomel electrode (SCE) as the reference electrode, and a platinum wire as the auxiliary electrode. All fluorescence spectra and real-time scan were measured on an LS-55 fluorescence spectrophotometer (PerkinElmer Company, USA) at room temperature. The detection solution of 200 μL was placed in a 0.1 × 1 cm2 micro quartz cuvette with a total volume of 350 μL. Ultraviolet-visible (UV-vis) absorption characterization was performed on a UV-3010 spectrophotometer (Hitachi, Japan). High-resolution transmission electron micrograph (HRTEM) observations for the morphological measurements of CuNPs were obtained with a Tecnai G220S-TWIN transmission electron microscope (FEI). All pH measurements were done with a Model pHs-3c meter (Shanghai, China).2.3. Preparation of dsDNA/CuNPs and electrochemical catalysis investigation
Prior to use, all oligonucleotide stock solutions (100 μM) were prepared in 10 mM MOPS buffer (pH = 7.4) and stored at −4 °C overnight. The working solution was obtained by diluting the stock solution with 10 mM MOPS buffer. First, to obtain double-strand DNA, the corresponding complementary single-strand DNA (aptamer, trigger, HP1, and HP2) solution was heated to 90 °C for 5 min to remove aggregates, and then cooled slowly to room temperature. Subsequently, 20 μL aptamer (2 μM) mixed with 16 μL trigger (2 μM) was added to the above solution and incubated for 20 min at 37 °C. Then, 7 μL RAC (30 nM) was added to the above mixture and incubated for 20 min at 37 °C. To avoid steric hindrance, which prevents the HDCA process, we added HP1 and HP2 until the reaction of RAC finished. Then, 40 μL HP1 (4 μM) and 40 μL HP2 (4 μM) were added to the above mixture. To achieve high sensitivity, the above solution was incubated for 2 h at 37 °C in 10 mM MOPS (pH = 7.4). Typically, 12 μL sodium ascorbate (100 mM) was added to the above solution. After incubation for 10 min at room temperature, 12 μL CuSO4 (10 mM) was introduced to the solution and incubated for another 10 min. The final volume was 200 μL and the final concentrations were 200 nM of c, 160 nM of trigger, 1 nM of RAC, 800 nM of HP1, 800 nM of HP2, 6 mM of sodium ascorbate and 600 μM of CuSO4. Finally, the dsDNA/CuNPs solution (200 μL) was placed in 3 mL 10 mM MOPS buffer (pH = 7.4), which is denoted as dsDNA/CuNPs resulting solution for further electrochemical experiments.All electrochemical experiments were carried out at room temperature. The electrocatalysis toward H2O2 was carried out in 3 mL 10 mM MOPS buffer (pH = 7.4) containing the resulting dsDNA/CuNPs solution. Cyclic voltammetry (CV) was performed within the potential range of 0 to −0.4 V using the scan rate of 50 mV s−1. Different concentrations of the target RAC was investigated under the same conditions.
2.4. Colorimetric catalysis assay
In order to demonstrate the peroxidase-like activity of dsDNA/CuNPs, experiments were performed with TMB. With 1 nM of RAC, the obtained resulting dsDNA/CuNPs solution (200 μL) was tested in 1 mL 0.5 mM TMB and 10 mM H2O2 and the colorimetric changes were visually observed and recorded in the range of 370 nm to 650 nm using UV-Vis spectroscopy.2.5. Selectivity study and stability of resulting dsDNA/CuNPs solution
Different concentrations of 1.0 nM of the target RAC, 2.0 nM of the single-base-mismatched DNA, 2.0 nM of the double base-mismatched DNA or 2.0 nM noncomplementary DNA were added to the aptamer–trigger mixture and incubated for 20 min at 37 °C. HP1 and HP2 were added and incubated for 2 h at 37 °C, and then Cu2+ and sodium ascorbate were added. Then, the cyclic voltammetry signal from each mixture was measured and compared. With 1 nM of RAC, the obtained dsDNA/CuNPs solution (200 μL) was used for the investigation of storage stability. The solution was sealed and stored at 4 °C. Cyclic voltammetry of the solution was tested once per day and tested for 7 days.2.6. Sensing of RAC in blank swine urine samples
High quality swine from a local feeding factory was detected to be free of RAC by HPLC-MS and used to perform spiking and recovery studies. Real samples were prepared according to the ref. 46. Five blank swine urine samples (4.0 g) were packed into a 100 mL polypropylene centrifuge tube and exactly weighed and spiked with suitable amounts of RAC standard solutions. Then, 20 mL of a methanol/H2O = 9/1 (v/v) extraction solution was added into the centrifuge tube. After vigorous shaking for 10 min, the mixture solution was centrifuged for 20 min at 3000 rpm and the supernatants collected, and the extraction procedure was repeated twice. The collected supernatants were dried at 60 °C under reduced pressure by rotary evaporation. The dried extract was re-dissolved in 4.0 mL methanol and filtered using a 0.22 μm nylon filter and diluted to 20 mL with methanol. Finally, the obtained dilute RAC samples were added to the aptamer–trigger mixture and incubated for 20 min at 37 °C. HP1 and HP2 were added and incubated for 2 h at 37 °C, in the presence of Cu2+ and sodium ascorbate, and CuNPs were formed on the dsDNA structure. The final volume was 200 μL and the final concentrations of RAC were 10, 50, 100, 500 and 1000 pM. Then, the current intensity of each solution was measured.3. Results and discussion
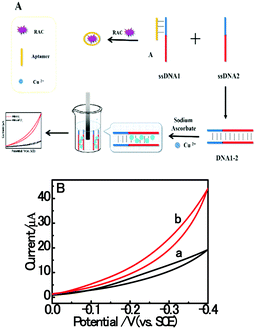
3.1. Principle of homogeneous electrochemical RAC assay
Based on the electrocatalysis of the dsDNA/CuNPs resulting solution (experimental study in the following section), a label-free homogeneous electrochemical strategy for the ultrasensitive detection of RAC with enzyme-free amplification is demonstrated in Scheme 1. In principle, the two DNA hairpin structures HP1 and HP2, initially do not hybridize with each other because of the effective block created by complementary domains.45 Nevertheless, in the presence of another ssDNA sequence, which is termed hairpin DNA cascade amplifier, HP1 and HP2 can form a stable duplex without consuming the trigger (T).47 As shown in Scheme 1, HP1, HP2 and T contain a few domains, which are labeled in common letters. Complementarity between letters b c d is denoted by an asterisk, b* c* d*. Upon the addition of the target RAC, specific binding between the aptamer (A) and the target RAC results in the dissociation of the A–T complex. Subsequently, T (f) hybridization of the exposed toehold domain a of HP1 gradually initiates a strand displacement, thus generating an HP1-T intermediate through domain hybridization (a, b and f, g), however HP1-T has an exposed ssDNA domain, d, which is able to hybridize with the exposed domain d* in HP2. Therefore, after hybridization of d and d*, the sequence b will undergo branch migration and displace the T sequence (g) to form the HP1-2 duplex (b c d and b* c* d*), followed by displacement of T (f, g) further hybridizations of HP1 and HP2 for the next catalytic round. The poly (AT) sequence was designed on the domain b c d and b* c* d*of HP1–HP2. In the presence of Cu2+ and sodium ascorbate, CuNPs are formed on the dsDNA structure. Based on the peroxidase-like character of the dsDNA/CuNPs resulting solution, as shown in Scheme 1C, RAC could be detected. Due to the fact that T can initiate hybridization between hairpins HP1 and HP2 for multiple cycles only in the presence of RAC, a label-free and enzyme-free HDCA autocatalytic signal amplification homogeneous electrochemical strategy for the monitoring of RAC is obtained.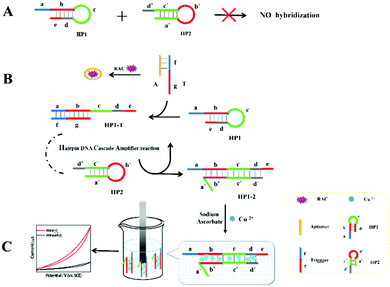 | ||
| Scheme 1 Principle of homogeneous electrochemical aptasensor based on dsDNA-templated copper nanoparticles for the detection of ractopamine. | ||
3.2. Characterization of dsDNA/CuNPs and their catalysis investigation
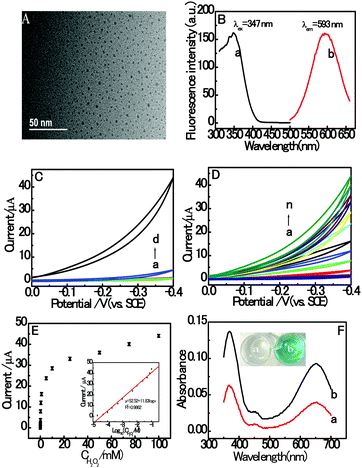
The morphology characterization with typical HRTEM images of the prepared dsDNA/CuNPs is shown in Fig. 1A. As can be seen from the HRTEM image, the dsDNA/CuNPs were individually spherical and uniform. Although, a small number of Cu nanoparticles in larger or aggregated clusters is detected as well. Moreover, the AT-rich dsDNA/CuNPs were characterized by fluorescence spectroscopy and the results are shown in Fig. 1B. The maximum excitation and emission wavelengths of the CuNPs are 347 nm and 593 nm, respectively (curves a and b of Fig. 1B), which are in accordance with the literature.37In addition, in order to explore the novel electrochemical properties of dsDNA/CuNPs, their CV response towards H2O2 was measured in blank buffer or buffer containing the dsDNA/CuNPs solution. As can be seen in Fig. 1C, a negligible CV signal on the bare GCE was obtained in MOPS (pH 7.4) with (curve c) and without (curve a) H2O2, which suggests that there was no catalytic activity. However, a large electrocatalytic response was obtained in the dsDNA/CuNPs resulting solution in the presence of 100 mM H2O2, shown as curve d in Fig. 1C, which may have resulted from the electrocatalysis of dsDNA/CuNPs. In the control experiments, the cyclic voltammogram response of the dsDNA/CuNPs resulting solution without H2O2 was also investigated. No obvious electrocatalytic response could be observed (curve b of Fig. 1C), which confirms that the electrochemical catalysis was from the dsDNA/CuNPs resulting solution toward H2O2 not dsDNA/CuNPs itself.
Based on the cyclic voltammogram results in Fig. 1C, the electrochemical catalysis of the dsDNA/CuNPs resulting solution towards different concentrations of H2O2 was further studied. As shown in Fig. 1D, the electrocatalytic current intensity increased with an increase in the concentration of H2O2 from 0 to 100 mM. Fig. 1E depicts the relationship between peak current and H2O2 concentrations, which shows a sigmoid shape. As shown in the inset of Fig. 1E, the peak current has a linear correlation with the H2O2 concentration in the range of 10 μM to 100 mM. The obtained regression equation is expressed as y μA−1 = 52.52 + 11.83![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) x μM−1, R2 = 0.9862, where y is the electrocatalytic signal, x is the concentration of H2O2 and R is the regression coefficient. The detection limit was calculated to be 3 μM (3 n/s, where n is the standard deviation of the intercept and s is the slope of the calibration curve), which displays the excellent catalytic activity of dsDNA/CuNPs.
x μM−1, R2 = 0.9862, where y is the electrocatalytic signal, x is the concentration of H2O2 and R is the regression coefficient. The detection limit was calculated to be 3 μM (3 n/s, where n is the standard deviation of the intercept and s is the slope of the calibration curve), which displays the excellent catalytic activity of dsDNA/CuNPs.
Furthermore, the catalysis of dsDNA/CuNPs was also investigated via a colorimetric experiment. The TMB oxidation reaction was performed in solutions containing hydrogen peroxide (H2O2) with and without dsDNA/CuNPs. As shown in Fig. 1F, in the absence of dsDNA/CuNPs, the solution does not display any color change (the inset, a). However, it was observed that the TMB–H2O2 solution became blue (the inset, b) in the presence of dsDNA/CuNPs and the maximal absorption wavelength at 370 nm and 650 nm (curve b of Fig. 1F) was higher than that in the absence of dsDNA/CuNPs (curve a of Fig. 1F), thus proving that dsDNA/CuNPs possess intrinsic enzyme mimetic activity.
3.3. Feasibility investigation of the RAC assay
In this paper, the proposed homogeneous electrochemical biosensor for amplified RAC detection is based on the electrocatalysis and HDCA of dsDNA/CuNPs. Firstly, we investigated the electrocatalytic response of different resulting solutions to prove our design. As shown in curve h of Fig. 2A, in the presence of RAC, a large electrocatalytic response was obtained in the dsDNA/CuNPs resulting solution which may have resulted from the intrinsic enzyme mimetic activity of dsDNA/CuNPs, and implies the principle that the target triggered the hairpin to form dsDNA/CuNPs. However, in the absence of RAC, as we can see in curve g of Fig. 2A, the electrocatalytic response decreased remarkably compared with that in the presence of RAC, which further confirms that the conjugation between HP1 and HP2 was blocked without the addition of the released trigger and it could not form dsDNA/CuNPs. Despite the large decrease in electrocatalysis, it was found that there was still a small response, which may be due to the stem of the hairpin structure with some poly (AT) units to form a few dsDNA/CuNPs. In the presence of RAC and without the addition of HP2, HP1-T cannot be liberated. We also studied the electrocatalysis of the HP1-T template CuNPs. As shown in curve f of Fig. 2A, it was found that the electrocatalysis can be neglected compared with that of the HP1–HP2 dsDNA-templated CuNPs (curve h). In the control experiments, the cyclic voltammogram response of SA or Cu2+ or SA and Cu2+ without DNA was also investigated. No obvious electrocatalytic response could be observed (curve c, d and e of Fig. 2A), which implies that the system has no ability to catalyze H2O2 without the formation of dsDNA/CuNPs. The corresponding fluorescent emission images with UV-light excitation are also consistent with the above results (inset in Fig. 2B). From the histogram of Fig. 2B we can more clearly comprehend the control experiments of Fig. 2A. Furthermore, trigger was also used to confirm the feasibility and whether the electrocatalytic response of dsDNA/CuNPs relied on the concentrations of trigger. As shown in Fig. 2C, the CV response of dsDNA/CuNPs increased with the concentration of trigger. As shown in the inset of Fig. 2D, the calibration plot was constructed by plotting the current responses against the trigger concentration. Thus, the aforementioned results clearly demonstrate the feasibility of our strategy for an RAC assay.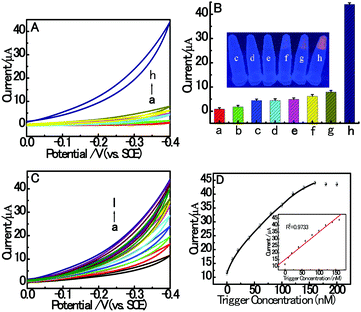 | ||
| Fig. 2 (A) CV responses in 3 mL MOPS (pH 7.4) on bare GCE without (a) or with 100 mM H2O2 (b–g) in the presence of 10 mM SA (c), 800 μM Cu2+ (d), 10 mM SA and 800 μM Cu2+ (e), 6 mM SA, 600 μM Cu2+, 1 nM RAC and DNA (200 nM aptamer, 160 nM trigger and 800 nM HP1) (f), 6 mM SA, 600 μM Cu2+ and DNA (200 nM aptamer, 160 nM trigger, 800 nM HP1 and 800 nM HP2) (g), 6 mM SA, 600 μM Cu2+, 1 nM RAC and DNA (200 nM aptamer, 160 nM trigger, 800 nM HP1 and 800 nM HP2) (h). (B) Histograms of CV responses corresponding to Fig. 2A. The inset shows the corresponding fluorescent emission images with UV-light excitation. (C) CV response towards different concentrations of trigger: (a) 0, (b) 10, (c) 20, (d) 40, (e) 60, (f) 80, (g) 100, (h) 120, (i) 140, (j) 160, (k) 180 and (l) 200 nM. (D) Relationship between the CV response and trigger concentration. Inset: Linear range between the response and concentrations of trigger from 0 to 160 nM. The error bars are the standard deviation of three repeated measurements. | ||
3.4. HDCA role in homogeneous electrochemical RAC assay
In order to study the signal amplification effect, dsDNA/CuNPs without the HDCA autocatalytic signal amplification process were designed and discussed. The schematic biosensing process is shown in Fig. 3A. Upon the addition of the target RAC, the specific binding between the aptamer (A) and target RAC results in the dissociation of the A–ssDNA1 complex. Subsequently, ssDNA1 hybridization to ssDNA2 generates a double-stranded structure (DNA1-2). The poly (AT) sequence, which is the same with that of HP1–HP2 duplex, is ingeniously designed on the red domain of ssDNA1 and ssDNA2. In the presence of Cu2+ and sodium ascorbate, CuNPs are formed on the DNA1-2 structure. Based on the peroxidase-like character of dsDNA/CuNPs, RAC could also be detected. As shown in curve a of Fig. 3B, under the same target concentration, the signal based on dsDNA/CuNPs without the HDCA autocatalytic signal amplification process is remarkably less than that with HDCA amplification (curve b), which may be due to the fact that the amplification process forms larger units of dsDNA/CuNPs.3.5. Optimization of experimental conditions
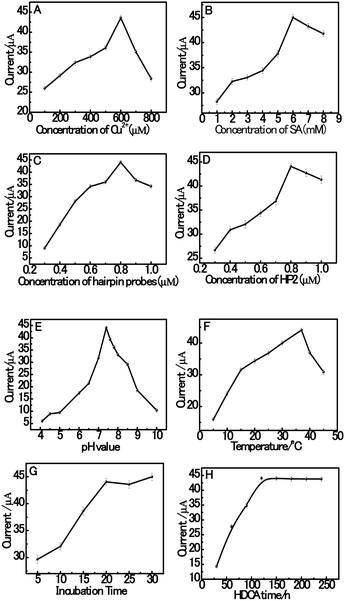
Herein, in order to achieve high assay sensitivity, a number of experimental conditions were optimized. The optimization of the variables of the system is shown in Fig. 4, including the concentration of Cu2+ and SA, the hybridization concentration of HP1 and HP2 in the hybridization buffer solution, pH value, the incubation temperature and time in solution. The effect of the concentration of Cu2+ was investigated since the concentration of Cu2+ is a crucial parameter that influences the current intensity of dsDNA/CuNPs. As shown in Fig. 4A, with an increase in the concentration of Cu2+, the current increases rapidly at first and then decreases when the concentration of Cu2+ is higher than 600 μM. This result may be attributed to the fact that Cu2+ complexes can cleave DNA sequences via oxygen-based radicals, which have also been reported previously.48 Thus, when the concentration of Cu2+ is higher than 600 μM, a mixture of Cu2+ might produce hydroxyl radicals (˙OH), which cause the destruction of the dsDNA templates. Furthermore, the concentration of sodium ascorbate also has a great effect on the electrochemical signal of the dsCuNPs. As can be seen from Fig. 4B, with an increase in the concentration of sodium ascorbate, it is observed that the current increases obviously and then reaches the highest when the concentration of SA is 6 mM.Hairpin probes (HP1 and HP2) were prepared with the same concentrations in the range of 0.3 to 1.0 μM, followed by measurement of their current signal. As shown in Fig. 4C, the concentration of hairpin probes at 0.8 μM achieves a better electrochemical signal than that at other concentrations. Then, we fixed the HP1 concentration at 0.8 μM and varied the concentration of HP2 from 0.3 μM to 1.0 μM as well. As can be seen from Fig. 4D, upon increasing the concentration of HP2 from 0.3 μM to 0.8 μM, it is observed that the current increases obviously followed by a slight decrease over the HP2 concentration of 0.8 μM. The high concentration of HP2 could enhance the hybridization efficiency of HP1 with HP2 during HDCA.
In this system, sensitivity can also be influenced by the pH value of the buffer solution. Thus, the pH value has a vital effect on the efficiency of the assay. Fig. 4E shows the effect of pH value on the catalytic response of dsDNA/CuNPs in MOPS solution over the pH range of 4.1 to 10.0. Obviously, the current increased with an increase in the pH value from 4.1 to 7.4. However, when the pH was higher than 7.4, the current decreased rapidly. The suggested reason for this is that copper may be complexed by OH− when the pH value is relatively higher, which also because of its reduced binding with poly (AT).49 The effect of incubation temperature in the HDCA system of dsDNA/CuNPs was studied at varying temperatures ranging from 5 to 45 °C, as shown in Fig. 4F. It can be observed that the current responses increase with an increase in the incubation temperature from 5 to 37 °C but then decreases gradually with an incubation temperature increase from 37 to 45 °C.
The effect of HDCA incubation time was investigated for different time periods ranging from 30 to 240 min. As seen in Fig. 4G, the electrocatalytic response intensifies as the incubation time increases and remains constant to a saturation value after about 120 min, which indicates that 120 min incubation time is efficient for the assay. The effect of incubation time for aptamer conjugation with the trigger was investigated for different time periods ranging from 5 min to 30 min. As can be seen in Fig. 4H, the current intensity increases as the incubation time increases and reaches a saturation value after about 20 min, which means that 20 min incubation time is efficient.
Thus, 600 μM Cu2+, 6 mM SA, 800 nM HP1, 800 nM HP2, 0.01 M MOPS buffer (pH 7.4), 37 °C incubation temperature, 20 min aptamer–trigger incubation time and 120 min. HDCA incubation time were chosen as the optimized experimental conditions in subsequent experiments.
3.6. Homogenous electrochemical detection of RAC
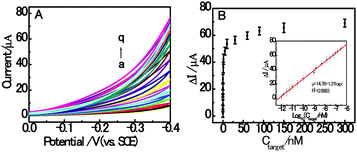
Under the optimized experimental conditions, the sensitivity detection performance of the fabricated autocatalytic homogenous electrochemical sensing system was investigated towards different concentrations of RAC. As shown in Fig. 5A, the CV response current increased with the concentration of target RAC ranging from 0 to 300 nM, which indicates that the formation of dsDNA/CuNPs is highly dependent on the concentration of target RAC. This also confirms the working principle that the target RAC is hybridized with the aptamer to trigger the formation of double-stranded DNA. Fig. 5B depicts the relationship between the peak current and RAC concentration, which shows a sigmoid shape. Moreover, as shown in the inset of Fig. 5B, a linear dependence between the currents and the logarithm of RAC concentration is obtained. The regression equation is ΔI μA−1 = 14.78 + 1.21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ctarget nM−1, R2 = 0.9983, where ΔI (the difference in the currents detected in the presence and absence of the target) is an enhancement of the cyclic voltammogram current, Ctarget is the RAC concentration, and R is the regression coefficient. The detection limit is estimated to be 0.3 pM (S/N = 3). To the best of our knowledge, this is the best detection limit achieved among all the reported RAC detectors and it is even lower than most of the existing electrochemical RAC sensors (as shown in Table 3). Furthermore, the homogeneous incubation and catalytic reaction greatly reduce the operational difficulty and time consumption since DNA does not need to be immobilized onto the sensing interface. Although the DNA is not reusable, the disposable DNA consumption is affordable because of the small volume (200 μL) and low concentration (800 nM) of DNA needed, without modification.
Ctarget nM−1, R2 = 0.9983, where ΔI (the difference in the currents detected in the presence and absence of the target) is an enhancement of the cyclic voltammogram current, Ctarget is the RAC concentration, and R is the regression coefficient. The detection limit is estimated to be 0.3 pM (S/N = 3). To the best of our knowledge, this is the best detection limit achieved among all the reported RAC detectors and it is even lower than most of the existing electrochemical RAC sensors (as shown in Table 3). Furthermore, the homogeneous incubation and catalytic reaction greatly reduce the operational difficulty and time consumption since DNA does not need to be immobilized onto the sensing interface. Although the DNA is not reusable, the disposable DNA consumption is affordable because of the small volume (200 μL) and low concentration (800 nM) of DNA needed, without modification.
3.7. Interference and stability study
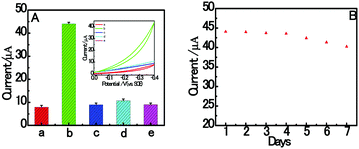
The specificity of the proposed aptasensor was investigated by replacing RAC with different common interferences (CLB, SAL and CAP). As shown in Fig. 6A, the electrochemical signal is negligible, even with the addition of interfering reagents at a concentration of 2.0 nM. However, it was found that the proposed biosensor has an obvious response with respect to 1.0 nM target RAC. These results clearly indicate that the homogenous electrochemical aptasensor has a high distinction ability to RAC. The reason for this lies in the high specificity between the RAC aptamer and RAC. As shown in Fig. 6B, it is obvious that only a 8.69% decrease was found after 7 days storage in a refrigerator at 4 °C. This result suggests that the homogeneous dsDNA/CuNPs aptasensor shows good stability.3.8. Application in real samples
To demonstrate the practical application potential of this homogenous electrochemical aptasensor, some spiked swine urine samples with RAC at five levels were tested. All contaminated samples with RAC were determined by three replicate measurements and the results are shown in Table 2. The recoveries for the spiked RAC were 87.4 to 117.0% with RSD from 5.8 to 9.4% (n = 3), which confirm that this homogenous electrochemical aptasensor based on dsDNA/CuNPs is reliable and sensitive to determine RAC residue in real samples.| Samples | Added (pM) | Found (pM) | Recovery (%) | RSD (%) n = 3 |
|---|---|---|---|---|
| 1 | 10 | 11.7 | 117.0 | 5.8 |
| 2 | 50 | 43.8 | 87.4 | 8.3 |
| 3 | 100 | 106.9 | 106.9 | 6.9 |
| 4 | 500 | 520.7 | 104.1 | 7.5 |
| 5 | 1000 | 905.6 | 90.6 | 9.4 |
Many methods for RAC have been reported and the representative methods are shown in Table 3. Compared with the published methods in Table 3, the advantages of the developed homogenous electrochemical aptasensor based on RAC aptamer are seen. Firstly, the detection limit of our homogenous electrochemical aptasensor is obviously better than the other methods. Importantly, the five orders of magnitude dynamic range in lower concentration level obtained in this study is wider than most of the reported RAC methods. Thus, the superior performance of the homogenous electrochemical aptasensor over the reported methods highlights the benefits that can be attributed to the novel fabrication strategy of the aptasensor.
| Methods | Detectable range (nM) | LODs (pM) | Ref. |
|---|---|---|---|
| Electroluminescence sensor | 0.07–8.9 | 25 | 17 |
| Electrochemical biosensor | 0.001–1 | 0.5 | 18 |
| Quartz crystal microbalance | 2.5 × 103–1.5 × 106 | 1.16 × 106 | 46 |
| Molecularly imprinted polymer | 0.15–14.8 | 30 | 50 |
| Electrochemical | 17–962 | 1.16 × 104 | 51 |
| Molecularly imprinted polymer | 1.21–3030 | 757 | 52 |
| Chemiluminescence | 2.96–1479 | 1.5 × 103 | 53 |
| Surface plasmon resonance | 59.2–2959 | 1.5 × 104 | 54 |
| Electrochemical biosensor | 0.03–295.9 | 4.5 | 55 |
| Electrochemical sensor | 0.1–1000 | 100 | 56 |
| Homogeneous electrochemical | 0.001–300 | 0.3 | This study |
4. Conclusions
In summary, a novel simple, rapid, ultrasensitive, label-free and enzyme-free amplified homogeneous electrochemical aptasensor platform has been developed for target RAC on the basis of an ingeniously designed autocatalytic target recycling strategy. The proposed strategy presents multifaceted advantages over the currently available assays. Primarily, by taking advantage of its simplicity in synthesis, favorable biocompatibility and good conductivity properties, dsDNA/CuNPs were first exploited as an effective signal indicator for the construction of a new homogeneous electrochemical strategy for RAC sensing. Then, more significantly, the RAC target could catalyze the regeneration of multiple HP1–HP2 duplexes in repeated cycles, thus achieving enzyme-free signal amplification in situ, which avoids the shortcomings of enzyme instability. Thirdly, the detection limit towards the target RAC was as low as 0.3 pM, which could rival or even exceed that of the reported excellent approaches. Additionally, it presents the obvious advantages of simplicity in biosensor fabrication and rapidness in the recognition and detection process. Moreover, the designed autocatalytic and homogeneous aptasensor has the potential to detect a wide spectrum of analytes, including biological small molecules, proteins, nucleic acids and nuclease activity. Hence, there is tremendous potential for the development of dsDNA–CuNPs based autocatalytic and immobilization-free electrochemical aptasensors for applications in bioanalysis and clinical biomedicine.Acknowledgements
This work was financially supported by a project (no. 21371007) from the National Natural Science Foundation of China, Anhui Provincial Natural Science Foundation (1208085QB28), Anhui Provincial Natural Science Foundation for Distinguished Youth (1408085J03), The Key Program in the Youth Elite Support Plan in Universities of Anhui Province (gxyqZD2016023), Natural Science Foundation of Anhui (KJ2012A139 and 1608085MB46) and the Program for Innovative Research Team at Anhui Normal University.Notes and references
- H. A. Kuiper, M. Y. Noordam, M. M. H. Dooren-Filpsen, R. Schilt and A. H. Roos, J. Anim. Sci., 1998, 76(1), 195–207 CrossRef CAS PubMed.
- Suherman, K. Morita and T. Kawaguchi, Biosens. Bioelectron., 2015, 67, 356–363 CrossRef CAS PubMed.
- Y. P. Wu, Y. F. Bi, G. L. Bingga, X. W. Li, S. X. Zhang, J. C. Li, H. Li and S. Y. Ding, J. Chromatogr. A, 2015, 1400, 74–81 CrossRef CAS PubMed.
- T. Anh Huong Nguyen, T. Ngoc Mai Pham, T. Tuoi Doan, J. Sáiz, T. Quynh Hoa Nguyen, P. C. Hauser and T. Duc Mai, J. Chromatogr. A, 2014, 1360, 305–311 CrossRef PubMed.
- S. Fan, H. Miao, Y. F. Zhao, H. J. Chen and Y. Wu, J. Agric. Food Chem., 2012, 60, 1898–1905 CrossRef PubMed.
- A. C. Valese, G. A. P. Oliveira, C. R. Kleemann, L. Molognoni and E. Daguer, J. Food Compos. Anal., 2016, 47, 38–44 CrossRef CAS.
- G. L. Ding, D. G. Li, J. Qin, J. L. Zhu, B. T. Wang, Q. Q. Geng, M. C. Guo, D. Punyapitak and Y. S. Cao, Meat Sci., 2015, 106, 55–60 CrossRef CAS PubMed.
- L. M. He, Y. J. Su, Z. L. Zeng, Y. H. Liu and X. H. Huang, Anim. Feed Sci. Technol., 2007, 132, 316–323 CrossRef CAS.
- C. Li, Y. L. Wu, T. Yang, Y. Zhang and W. G. HuangFu, J. Chromatogr. A, 2010, 1217, 7873–7877 CrossRef CAS PubMed.
- C. T. Elliott, C. S. Thompson and C. J. M. Arts, Analyst, 1998, 123, 1103–1107 RSC.
- M. Z. Zhang, M. Z. Wang, Z. L. Chen, J. H. Fang, M. M. Fang, J. Liu and X. P. Yu, Anal. Bioanal. Chem., 2009, 395(8), 2591–2599 CrossRef CAS PubMed.
- P. L. Wang, Z. Wang and X. Su, Biosens. Bioelectron., 2015, 64, 511–516 CrossRef CAS PubMed.
- G. Zhu, Y. Hu, J. Gao and L. Zhong, Anal. Chim. Acta, 2011, 697(1–2), 61–66 CrossRef CAS PubMed.
- Y. Zhou, P. L. Wang, X. Su, H. Zhao and Y. J. He, Talanta, 2013, 112, 20–25 CrossRef CAS PubMed.
- X. Yang, B. Feng, P. Yang, Y. Ding, Y. Chen and J. Fei, J. Food Chem. Nutr., 2014, 145, 619–624 CrossRef CAS PubMed.
- Q. H. Wei, Q. Wang, H. Y. Wang, H. W. Gu, Q. Q. Zhang, X. Gao and B. Qi, Mater. Lett., 2015, 147, 58–60 CrossRef CAS.
- Z. Li, Y. Wang, W. Kong, C. Li, Z. Wang and Z. Fu, Biosens. Bioelectron., 2013, 39, 311–314 CrossRef CAS PubMed.
- F. Yang, P. L. Wang, R. G. Wang, Y. Zhou, X. O. Su, Y. J. He, L. Shi and D. S. Yao, Biosens. Bioelectron., 2016, 77, 347–352 CrossRef CAS PubMed.
- S. F. Liu, Y. Lin, L. Wang, T. Liu, C. B. Cheng, W. J. Wei and B. Tang, Anal. Chem., 2014, 86, 4008–4015 CrossRef CAS PubMed.
- W. Li, X. J. Liu, T. Hou, H. Y. Li and F. Li, Biosens. Bioelectron., 2015, 70, 304–309 CrossRef CAS PubMed.
- F. Xuan, X. T. Luo and I. M. Hsing, Anal. Chem., 2012, 84, 5216–5220 CrossRef CAS PubMed.
- F. Xuan, X. T. Luo and I. M. Hsing, Anal. Chem., 2013, 85, 4586–4593 CrossRef CAS PubMed.
- F. T. Zhang, J. Nie, D. W. Zhang, J. T. Chen, Y. L. Zhou and X. X. Zhang, Anal. Chem., 2014, 86, 9489–9495 CrossRef CAS PubMed.
- X. F. Wei, X. M. Ma, J. J. Sun, Z. Y. Lin, L. H. Guo, B. Qiu and G. N. Chen, Anal. Chem., 2014, 86, 3563–3567 CrossRef CAS PubMed.
- R. Miranda-Castro, D. Marchal, B. Limoges and F. Mavre, Chem. Commun., 2012, 48, 8772–8774 RSC.
- S. F. Liu, Y. Wang, C. X. Zhang, Y. Lin and F. Li, Chem. Commun., 2013, 49, 2335–2337 RSC.
- L. F. Zhang, T. Hou, H. Y. Li and F. Li, Analyst, 2015, 140, 4030–4036 RSC.
- X. J. Liu, W. Li, T. Hou, S. S. Dong, G. H. Yu and F. Li, Anal. Chem., 2015, 87, 4030–4036 CrossRef CAS PubMed.
- X. Z. Wang, X. L. Liu, T. Hou, W. Li and F. Li, Sens. Actuators, B, 2015, 208, 575–580 CrossRef CAS.
- X. T. Luo, T. M. H. Lee and I. M. Hsing, Anal. Chem., 2008, 80, 7341–7346 CrossRef CAS PubMed.
- T. Hou, W. Li, X. J. Liu and F. Li, Anal. Chem., 2015, 87, 11368–11374 CrossRef CAS PubMed.
- Y. Tan, X. F. Wei, Y. Zhang, P. L. Wang, B. Qiu, L. G. Guo, Z. Y. Lin and H. H. Yang, Anal. Chem., 2015, 87, 11826–11831 CrossRef CAS PubMed.
- Y. Tan, X. F. Wei, M. M. Zhao, B. Qiu, L. H. Guo, Z. Y. Lin and H. H. Yang, Anal. Chem., 2015, 87, 9204–9208 CrossRef CAS PubMed.
- F. Xuan, T. W. Fan and I. M. Hsing, ACS Nano, 2015, 9(5), 5027–5033 CrossRef CAS PubMed.
- J. Y. Zhuang, D. P. Tang, W. Q. Lai, G. N. Chen and H. H. Yang, Anal. Chem., 2014, 86, 8400–8407 CrossRef CAS PubMed.
- T. P. Qing, Z. H. Qing, Z. G. Mao, X. X. He, F. Z. Xu, L. Wen, D. G. He, H. Shi and K. M. Wang, RSC Adv., 2014, 4, 61092–61095 RSC.
- A. Rotaru, S. Dutta, E. Jentzsch, K. Gothelf and A. Mokhir, Angew. Chem., Int. Ed., 2010, 49, 5665–5667 CrossRef CAS PubMed.
- Q. W. Song, Y. Shi, D. C. He, S. H. Xu and J. Ouyang, Chem. – Eur. J., 2015, 21,, 2417–2422 CrossRef PubMed.
- J. Plastino, E. L. Green, J. Sanders-Loehr and J. P. Klinman, Biochemistry, 1999, 38, 8204–8216 CrossRef CAS PubMed.
- I. A. Koval, P. Gamez, C. Belle, K. Selmeczi and J. Reedijk, Chem. Soc. Rev., 2006, 35, 814–840 RSC.
- B. C. Yin, B. C. Ye, W. Tan, H. Wang and C. Xie, J. Am. Chem. Soc., 2009, 131, 14624–14625 CrossRef CAS PubMed.
- X. Zhu, H. Shi, Y. Shen, B. Zhang, J. Zhao and G. Li, Nano Res., 2015, 8(8), 2714–2720 CrossRef CAS.
- Z. Qing, T. Qing, Z. Mao, X. He, K. Wang, Z. Zou, H. Shi and D. He, Chem. Commun., 2014, 50, 12746–12748 RSC.
- L. B. Zhang, J. B. Zhu, S. J. Guo, T. Li, J. Li and E. K. Wang, J. Am. Chem. Soc., 2013, 135, 2403–2406 CrossRef CAS PubMed.
- P. Yin, H. M. T. Choi, C. R. Calvert and N. A. Pierce, Nature, 2008, 451, 318–323 CrossRef CAS PubMed.
- L. J. Kong, M. F. Pan, G. Z. Fang, X. L. He, Y. K. Yang, J. Dai and S. Wang, Biosens. Bioelectron., 2014, 51, 286–292 CrossRef CAS PubMed.
- C. C. Wu, S. Cansiz, L. Q. Zhang, I. T. Teng, L. P. Qiu, J. Li, Y. Liu, C. S. Zhou, R. Hu, T. Zhang, C. Cui, L. Cui and W. H. Tan, J. Am. Chem. Soc., 2015, 137, 4900–4903 CrossRef CAS PubMed.
- D. S. Sigman, D. R. Graham, V. D. D. Aurora and A. M. J. Stern, Biol. Chem., 1979, 254, 12269 CAS.
- H. Wang, Y. X. Wang, J. Y. Jin and R. H. Yang, Anal. Chem., 2008, 80, 9021–9028 CrossRef CAS PubMed.
- Y. W. Tang, G. Z. Fang, S. Wang and J. L. Li, Anal. Bioanal. Chem., 2011, 401, 2275–2282 CrossRef CAS PubMed.
- M. Y. Wang, W. Zhu, L. Ma, J. J. Ma, D. E. Zhang, Z. W. Tong and J. Chen, Biosens. Bioelectron., 2016, 78, 259–266 CrossRef CAS PubMed.
- H. L. Liu, G. Z. Fang and S. Wang, Biosens. Bioelectron., 2014, 55, 127–132 CrossRef CAS PubMed.
- J. Han, H. F. Gao, W. W. Wang, Z. X. Wang and Z. F. Fu, Biosens. Bioelectron., 2013, 48, 39–42 CrossRef CAS PubMed.
- T. Yao, X. Gu, T. F. Li, J. G. Li, J. Li, Z. Zhao, J. Wang, Y. C. Qin and Y. G. She, Biosens. Bioelectron., 2016, 75, 96–100 CrossRef CAS PubMed.
- H. Wang, Y. Zhang, H. Li, B. Du, H. M. Ma, D. Wu and Q. Wei, Biosens. Bioelectron., 2013, 49, 14–19 CrossRef CAS PubMed.
- Y. Zhou, P. L. Wang, X. O. Su, H. Zhao and Y. J. He, Microchim. Acta, 2014, 181, 1973–1979 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2017 |