Blood-based near-infrared spectroscopy for the rapid low-cost detection of Alzheimer's disease†
Maria
Paraskevaidi
 *a,
Camilo L. M.
Morais
*a,
Camilo L. M.
Morais
 a,
Daniel L. D.
Freitas
b,
Kássio M. G.
Lima
a,
Daniel L. D.
Freitas
b,
Kássio M. G.
Lima
 b,
David M. A.
Mann
c,
David
Allsop
d,
Pierre L.
Martin-Hirsch
e and
Francis L.
Martin
b,
David M. A.
Mann
c,
David
Allsop
d,
Pierre L.
Martin-Hirsch
e and
Francis L.
Martin
 *a
*a
aSchool of Pharmacy and Biomedical Sciences, University of Central Lancashire, Preston PR1 2HE, UK. E-mail: mparaskevaidi@uclan.ac.uk; flmartin@uclan.ac.uk
bInstitute of Chemistry, Biological Chemistry and Chemometrics, Federal University of Rio Grande do Norte, Natal 59072-970, Brazil
cClinical Neuroscience Research Group, Division of Medicine and Neuroscience, University of Manchester, Greater Manchester Neurosciences Centre, Hope Hospital, Salford M6 8HD, UK
dDivision of Biomedical and Life Sciences, Faculty of Health and Medicine, Lancaster University, Lancaster LA1 4YQ, UK
eDepartment of Obstetrics and Gynaecology, Central Lancashire Teaching Hospitals NHS Foundation Trust, Preston PR2 9HT, UK
First published on 15th August 2018
Abstract
Alzheimer's disease (AD) is currently under-diagnosed and is predicted to affect a great number of people in the future, due to the unrestrained aging of the population. An accurate diagnosis of AD at an early stage, prior to (severe) symptomatology, is of crucial importance as it would allow the subscription of effective palliative care and/or enrolment into specific clinical trials. Today, new analytical methods and research initiatives are being developed for the on-time diagnosis of this devastating disorder. During the last decade, spectroscopic techniques have shown great promise in the robust diagnosis of various pathologies, including neurodegenerative diseases and dementia. In the current study, blood plasma samples were analysed with near-infrared (NIR) spectroscopy as a minimally-invasive method to distinguish patients with AD (n = 111) from non-demented volunteers (n = 173). After applying multivariate classification models (principal component analysis with quadratic discriminant analysis – PCA-QDA), AD individuals were correctly identified with 92.8% accuracy, 87.5% sensitivity and 96.1% specificity. Our results show the potential of NIR spectroscopy as a simple and cost-effective diagnostic tool for AD. Robust and early diagnosis may be a first step towards tackling this disease by allowing timely intervention.
Introduction
Alzheimer's disease (AD), being responsible for 60–80% of the cases, constitutes the most common type of dementia. A risk factor for the development of AD is increasing age, which, in combination with the progressive increase in the number of elderly people, is expected to lead to ∼135 million affected individuals worldwide by 2050.1 Apart from the detrimental impact of this disorder on patients, their families and the society, the economic burden should also be considered; the worldwide cost for the treatment of this disease had been estimated to become a trillion dollar in 2018.2 Furthermore, AD is definitively diagnosed only after a post-mortem brain biopsy. It is therefore more than evident that effective means to diagnose AD accurately and at an early-stage is crucial in order to intervene with therapeutic strategies and recruit patients to clinical trials.Infrared (IR) spectroscopy has advanced significantly over the last few decades, specifically in the field of biomedical investigation.3,4 By exploiting the vibrational movements of the chemical bonds within a sample after excitation, IR spectroscopy can provide quantitative and qualitative information about a sample. Technological advancements have also simplified the previously expensive and complicated instrumentation, thus facilitating the wider use of these systems. In this study, near-IR (NIR) spectroscopy was employed to study the region of the electromagnetic spectrum ranging between ∼750–2500 nm. The most prominent bands in the NIR include overtones and combinations of fundamental vibrations of –CH, –NH, and –OH groups.5 It has been previously shown that NIR spectroscopy holds promise for biomedical applications,6 including the study of human skin (skin carcinomas, atopy and leprosy),7 diabetes,8 breast and colorectal cancers,9,10 Alzheimer's disease11 and chronic fatigue syndrome.12
Spectroscopic techniques have been previously employed by independent research groups for the investigation of neurodegenerative disorders, either by analysis of brain biopsies or biofluids, such as cerebrospinal fluid (CSF) and blood samples.13–17 The objective of the current study was to detect AD using a minimally-invasive, but at the same time rapid and inexpensive, blood test. Our aim was to use a large number of individuals and add further evidence to the current literature about the diagnostic capabilities of spectroscopy as a diagnostic tool.
The use of a suitable substrate in spectroscopy is also of major importance as it could distort the resultant spectral information and lead to falsified conclusions; for this reason, numerous studies have previously used costly and/or fragile substrates, such as calcium/barium fluoride or gold substrates, to avoid signal interference.18–22 At the same time, however, the substrate of choice should be relatively inexpensive in order to be welcomed to a clinical setting. Therefore, a secondary aim of this study was to investigate whether the signal from the commonly-used and inexpensive low-E glass slide23,24 could be removed from the samples’ spectra without affecting the diagnostic result.
Materials and methods
Patient cohort and sample collection
Our cohort included 111 patients with AD and 173 individuals with no symptoms of AD, who were designated as healthy controls (HC). The latter group mainly consisted of close relatives (e.g., spouses) escorting the patients at the time of examination. More information about the age and gender of the participants is provided in Table 1.| Alzheimer's disease | Healthy control | |
|---|---|---|
| Number of cases | ||
| 111 | 173 | |
| Age | ||
| <65 | 60/111 | 90/173 |
| ≥65 | 51/111 | 79/173 |
| Unknown | — | 4/173 |
| Gender | ||
| Female | 50/111 | 103/173 |
| Male | 61/111 | 68/173 |
| Unknown | — | 2/173 |
All participants were recruited at Salford Royal Hospital (Salford, UK) with informed consent obtained prior to enrolment in accordance with Local Ethical Approval (05/Q1405/24 conferred by North West 10 Research Ethics Committee Greater Manchester North). Blood samples were collected in EDTA tubes following standard operating procedures. To acquire the plasma, whole blood was centrifuged for 10 min at 2000 rpm and 4 °C and the supernatant was collected in new microtubes. Plasma samples were aliquoted and kept at −80 °C until needed for the spectroscopic analysis. Samples were thoroughly thawed before depositing 50 μL onto IR-reflective glass slides (MirrIR Low-E slides, Kevley Technologies, USA) and left to dry overnight at room temperature.
NIR spectroscopy
Spectra were acquired using an ARCoptix FT-NIR Rocket spectrometer (Arcoptix S.A., Switzerland) in the range of 900 to 2600 nm. Samples were interrogated using the transmission mode with 10 point spectra collected per sample (resolution of 8 cm−1). Each sample spectrum was subtracted from a low-E slide background spectrum in order to eliminate the signal resulting from the slide.Pre-processing and computational analysis
Data pre-processing and multivariate classification models were built using MATLAB R2014b software (MathWorks Inc., USA) with PLS Toolbox version 7.9.3 (Eigenvector Research Inc., USA) and lab-made routines. The 10 spectra collected per sample were initially averaged, and the following pre-processing steps were applied to the dataset: truncation in the biofingerprint region (1850–2150 nm) (highlighted in Fig. 1a), Savitzky–Golay (SG) smoothing to remove unwanted noise from the spectra (window = 15 points, 2nd order polynomial function), extended multiplicative signal correction (EMSC) to correct for light scattering and automatic weighted least squares baseline correction to remove baseline absorptions. The spectra were divided into training (70%) and test (30%) sets using the Kennard-Stone (KS) sample selection algorithm.25 The training set was used for construction of classification models, whereas the test set was only used for final model evaluation.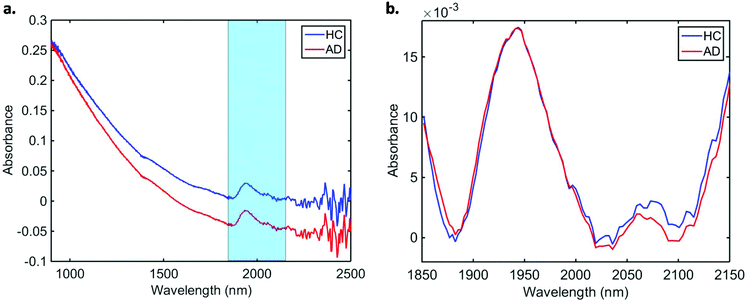 | ||
| Fig. 1 Average (a) raw and (b) pre-processed NIR spectra for healthy controls (HC) and Alzheimer's disease (AD) patients. | ||
Classification was performed using principal component analysis with quadratic discriminant analysis (PCA-QDA). A PCA-QDA model is based on a PCA decomposition followed by a Mahalanobis distance calculation. PCA reduces the original dataset into a few number of principal components (PCs) accounting for the majority of the variance across the spectra. As a result, a scores and a loading array are generated for each PC representing the variance on the sample and variable (e.g., wavelength) directions, respectively.26 PCA also solves problems with ill-conditioned data (data matrix with a large condition number) by reducing redundant information across the data and solving collinearity problems. For this reason, PCA is commonly employed prior to discriminant analysis, with the PCA scores used as input variables for the QDA algorithm.
As aforementioned, QDA is a classification algorithm based on a Mahalanobis distance calculation. QDA assumes classes having different variance structures, calculating an individual variance-covariance matrix for each class.27 This improves the classification capacity of QDA in comparison with linear methods (e.g., linear discriminant analysis – LDA) when classes with different variances are being analysed, which occurs often in complex datasets. The QDA classification scores were calculated in a non-Bayesian form in order to reduce the degree of overfitting, as follows:28
Qik = (xi − ![[x with combining macron]](https://www.rsc.org/images/entities/b_char_0078_0304.gif) k)TCk−1(xi − k)TCk−1(xi − ![[x with combining macron]](https://www.rsc.org/images/entities/b_char_0078_0304.gif) k) k) | (1) |
![[x with combining macron]](https://www.rsc.org/images/entities/b_char_0078_0304.gif) k is the mean vector for class k; Ck is the variance-covariance matrix of class k; and T represents the transpose matrix.
k is the mean vector for class k; Ck is the variance-covariance matrix of class k; and T represents the transpose matrix.
Outliers were identified using a Hotelling T2versus Q residual test.29 This test enables someone to create a chart containing the Hotelling T2 values in the x-axis and the Q residuals in the y-axis, where all samples far from the origin [0,0] are considered to be outliers. The Hotelling T2 values represent the sum of the normalised PCA scores, which is the distance from the multivariate mean to the projection of the sample onto the PCs; and the Q residuals are the sum of squares of each sample in the error matrix, which are the residuals between the sample and its projection via PCA.
Model validation
Validation was performed on a patient basis, meaning that each sample represents a different patient rather than an individual spectrum. The models were validated using quality parameters including accuracy (total number of samples correctly classified considering true and false negatives), sensitivity (proportion of positives correctly identified), specificity (proportion of negatives correctly identified), positive predictive value (proportion of test positives which are true positives) and negative predictive value (proportion of test negatives which are true negatives) (Table S1†).30In addition, a receiver operating characteristics (ROC) curve was generated using easyROC version 1.3 (http://www.biosoft.hacettepe.edu.tr/easyROC/),31 where the area under the curve (AUC) value was calculated as a general indicator of how well the model distinguished between the classes.
Results
In total, we acquired 1110 NIR spectra from AD patients (n = 111) and 1730 spectra from HC volunteers (n = 173). The absorption due to the low-E slide signal was subtracted from the samples’ signal in order to reduce glass interference (Fig. S1†). The average raw and pre-processed spectra (truncation at 1850–2150 nm, SG smoothing, EMSC and baseline correction) for each class are depicted in Fig. 1. Seven outliers (three due to AD and four due to HC samples) were detected using a Hotelling T2versus Q residual test (Fig. S2†). These samples were removed from the classification model. In total, 194 samples were used in the training set (118 HC, 76 AD) and 83 samples in the test set (51 HC, 32 AD), defined by the KS algorithm. After pre-processing, slight visual differences are evident between HC and AD patients (Fig. 1b). Significant differences were observed between HC and AD spectra (1850–2150 nm) using a two-tailed t-test with 95% confidence level (p < 0.001).For classification, the PCA-QDA algorithm was applied using 2 PCs (67.24% cumulative explained variance). PCA scores and loadings are depicted in Fig. 2a and b, respectively. The scores profile on the two PCs was superposed for HC and AD samples, with no clear separation observed between the classes. The loading profiles (Fig. 2b) indicated greater differences close to regions corresponding to a combination of O–H stretch/C–O stretch second overtone (∼1860 nm), second overtone C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching (H-bonded) in peptides (1908 nm), and a combination of bands consisting of N–H bend second overtone, C–H stretch/C
O stretching (H-bonded) in peptides (1908 nm), and a combination of bands consisting of N–H bend second overtone, C–H stretch/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretch, and C
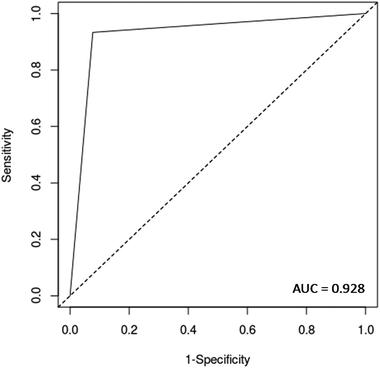
O stretch, and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretch/N–H in-plane bend/C–N stretching in proteins (2100 nm, 2111 nm, 2150 nm).32–34 These bands correspond to the most important spectral features used by the QDA classifier in PCA-QDA. The PCA-QDA model distinguished between AD and HC individuals with 92.8% accuracy, 87.5% sensitivity and 96.1% specificity (Table 2). The ROC curve and AUC value for PCA-QDA are shown in Fig. 3. The AUC value (0.928) is close to the maximum of 1, indicating its excellent predictive response.
O stretch/N–H in-plane bend/C–N stretching in proteins (2100 nm, 2111 nm, 2150 nm).32–34 These bands correspond to the most important spectral features used by the QDA classifier in PCA-QDA. The PCA-QDA model distinguished between AD and HC individuals with 92.8% accuracy, 87.5% sensitivity and 96.1% specificity (Table 2). The ROC curve and AUC value for PCA-QDA are shown in Fig. 3. The AUC value (0.928) is close to the maximum of 1, indicating its excellent predictive response.
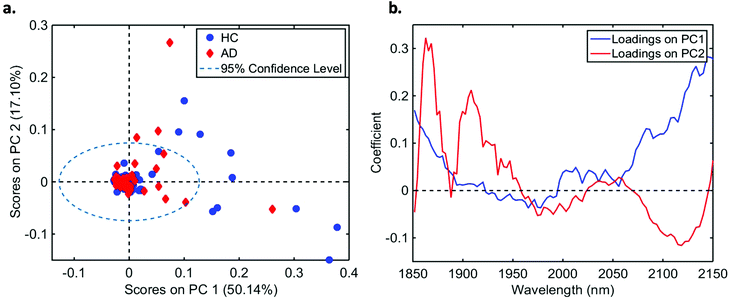 | ||
| Fig. 2 (a) PCA scores on PC1 and PC2 for healthy controls (HC) and Alzheimer's disease (AD) samples (explained variance for each PC inside parenthesis); (b) PCA loadings based on PC1 and PC2. | ||
| Parameter | Value (%) |
|---|---|
| Accuracy | 92.8 |
| Sensitivity | 87.5 |
| Specificity | 96.1 |
| PPV | 93.3 |
| NPV | 92.5 |
Discussion
With improved life conditions and health care, increased longevity has resulted in a greater number of elderly people and, thus, many cases of demented individuals worldwide. Numerous research groups have devoted substantial resources and co-ordinated their efforts to study dementias and provide an accurate diagnosis. For instance, the most studied biomarkers for AD are amyloid-β (Aβ) and tau protein (phosphorylated-tau and total-tau) in CSF. Collection of CSF, however, is an invasive procedure, making routine testing difficult. The understanding that the blood–brain barrier (BBB) is a semipermeable membrane, allowing the secretion of biological molecules between the brain and peripheral blood, as well as the fact that 500 mL CSF is daily absorbed into the bloodstream, has led to the characterization of blood as an “information-rich” sample.35Blood biomarkers indicative of disease constitute a developing field with great promise in the area of neurodegenerative disorders. A great number of new, blood-based molecular tests have emerged over the years, suggesting different biological markers for the detection of AD and other dementias.35–41 Even though the diagnostic capability of the above-mentioned tests is satisfactory, the high cost and laborious experimental work of these methods are great disadvantages for the development of a clinical test.
In contrast to conventional molecular techniques, spectroscopic tests allow cost-effective and rapid results. Previous studies using the mid-IR region for the diagnosis of AD have achieved comparable diagnostic results to the current NIR study. For instance, using ATR-FTIR in the mid-IR region, Paraskevaidi et al. achieved 86% sensitivity and specificity for individuals who carried one or two alleles of apolipoprotein e4 (APOE ε4);42 Carmona et al. used the mid-IR region (as well as Raman spectroscopy) to differentiate between healthy elderly and demented patients with a sensitivity of 89% and a specificity of 92%;14 Peuchant et al. also employed mid-IR spectroscopy and achieved 98.4% diagnostic accuracy.43 Other preliminary studies have also successfully applied spectroscopic approaches (IR or Raman spectroscopy) for the diagnosis of AD or other types of dementia;15,44,45 however, the small number of samples in these studies was a limitation, preventing more general conclusions. According to the NIR results of the present study, most of the differences between healthy and demented individuals seem to be related mainly to protein bands. Some of the well-known characteristics of AD include the building-up of Aβ plaques and neurofibrillary tangles (primarily consisting of tau protein) in the brain, and therefore we can speculate that the observed changes in the NIR region may be attributed to such protein changes.
The current study has achieved exceptionally high diagnostic accuracies using NIR spectroscopy in the transmission mode. The PCA-QDA classification model presented 92.8% accuracy, 87.5% sensitivity and 96.7% specificity which are comparable, and even superior, to current conventional diagnostic biomarkers. Using NIR spectroscopy also decreases the instrumental cost substantially, as instrumentation is much cheaper than other IR or Raman systems and it can be easily translated to portable systems. Our results, coming from a large cohort, add to the current literature by validating previous spectroscopic work, thus taking spectroscopy one step forward towards clinical implementation. Indeed, repetition and validation in independent research groups is of crucial importance for every new biomarker or diagnostic test prior to clinical trials. Herein, we have also shown that after appropriate spectral pre-processing, the low-E signal can be subtracted from the spectra, therefore allowing direct comparison of the sample information without interference from the slide. It should be noted here that the patient cohort in this study was already diagnosed with the disease, therefore the potential of blood-based NIR spectroscopy for pre-symptomatic detection remains to be further explored. Nevertheless, we are optimistic as previous spectroscopy studies have demonstrated segregation between early-stage/mild AD and healthy controls,15,44,46 but a larger number of early-stage patients are still required.
To conclude, this study detected a blood signature for AD, showing great promise in the accurate, simple and minimally-invasive diagnosis of the disease. Our main objective in the current study was to show whether NIR spectroscopy, in the transmission mode, could provide satisfactory diagnostic performance after removal of the substrate's signal. Future studies should focus on the recruitment of more participants, including asymptomatic individuals or patients with mild cognitive impairment (MCI). This would be the next big step in this field, as accurate identification of MCI individuals would allow immediate management and recruitment into clinical trials; the latter may also prove crucial for the development of new therapeutic strategies.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
MP acknowledges the Rosemere Cancer Foundation for funding. CLMM would like to thank CAPES-Brazil (Doutorado Pleno no Exterior, grant 88881.128982/2016-01) for financial support. KMGL would like to thank CNPq-Brazil (Bolsa de Produtividade, grant 303733/2017-9) for financial support.References
- Policy Brief for Head of Government, Alzheimer's Disease International (https://www.alz.co.uk/research/GlobalImpactDementia2013.pdf).
- P. Martin, W. Anders, G. Maëlenn, A. Gemma-Claire, W. Yu-Tzu and P. Matthew, World Alzheimer Report 2015: the global impact of dementia: an analysis of prevalence, incidence, cost and trends, Alzheimer's Disease International, 2015. (https://www.alz.co.uk/research/WorldAlzheimerReport2015.pdf) Search PubMed.
- L. Wang and B. Mizaikoff, Anal. Bioanal. Chem., 2008, 391, 1641–1654 CrossRef PubMed.
- D. I. Ellis and R. Goodacre, Analyst, 2006, 131, 875–885 RSC.
- G. Reich, Adv. Drug Delivery Rev., 2005, 57, 1109–1143 CrossRef PubMed.
- A. Sakudo, Clin. Chim. Acta, 2016, 455, 181–188 CrossRef PubMed.
- R. K. Lauridsen, H. Everland, L. F. Nielsen, S. B. Engelsen and L. Nørgaard, Skin Res. Technol., 2003, 9, 137–146 CrossRef PubMed.
- G. Pichler, B. Urlesberger, P. Jirak, H. Zotter, E. Reiterer, W. Müller and M. Borkenstein, Diabetes Care, 2004, 27, 1942–1946 CrossRef PubMed.
- M. K. Simick, R. A. Jong, B. C. Wilson and L. D. Lilge, J. Biomed. Opt., 2004, 9, 794–804 CrossRef PubMed.
- H. Chen, Z. Lin, L. Mo, T. Wu and C. Tan, BioMed Res. Int., 2015, 2015, 472197 Search PubMed.
- D. H. Burns, S. Rosendahl, D. Bandilla, O. C. Maes, H. M. Chertkow and H. M. Schipper, J. Alzheimer's Dis., 2009, 17, 391–397 Search PubMed.
- A. Sakudo, H. Kuratsune, Y. H. Kato and K. Ikuta, Clin. Chim. Acta, 2012, 413, 1629–1632 CrossRef PubMed.
- M. Paraskevaidi, P. L. Martin-Hirsch and F. L. Martin, Mol. Neurodegener., 2018, 13, 20 CrossRef PubMed.
- P. Carmona, M. Molina, M. Calero, F. Bermejo-Pareja, P. Martinez-Martin and A. Toledano, J. Alzheimer's Dis., 2013, 34, 911–920 Search PubMed.
- E. Ryzhikova, O. Kazakov, L. Halamkova, D. Celmins, P. Malone, E. Molho, E. A. Zimmerman and I. K. Lednev, J. Biophotonics, 2015, 8, 584–596 CrossRef PubMed.
- R. Michael, A. Lenferink, G. F. Vrensen, E. Gelpi, R. I. Barraquer and C. Otto, Sci. Rep., 2017, 7, 15603 CrossRef PubMed.
- M. Griebe, M. Daffertshofer, M. Stroick, M. Syren, P. Ahmad-Nejad, M. Neumaier, J. Backhaus, M. G. Hennerici and M. Fatar, Neurosci. Lett., 2007, 420, 29–33 CrossRef PubMed.
- M. Grimbergen, C. van Swol, R. van Moorselaar, J. Uff, A. Mahadevan-Jansen and N. Stone, J. Photochem. Photobiol., B, 2009, 95, 170–176 CrossRef PubMed.
- B. W. De Jong, T. C. Bakker Schut, K. Maquelin, T. van der Kwast, C. H. Bangma, D.-J. Kok and G. J. Puppels, Anal. Chem., 2006, 78, 7761–7769 CrossRef PubMed.
- L. Mikoliunaite, R. D. Rodriguez, E. Sheremet, V. Kolchuzhin, J. Mehner, A. Ramanavicius and D. R. Zahn, Sci. Rep., 2015, 5, 13150 CrossRef PubMed.
- J. De Meutter, K.-M. Derfoufi and E. Goormaghtigh, Biomed. Spectrosc. Imaging, 2016, 5, 145–154 Search PubMed.
- M. J. Pilling, P. Bassan and P. Gardner, Analyst, 2015, 140, 2383–2392 RSC.
- L. Cui, H. J. Butler, P. L. Martin-Hirsch and F. L. Martin, Anal. Methods, 2016, 8, 481–487 RSC.
- M. J. Baker, J. Trevisan, P. Bassan, R. Bhargava, H. J. Butler, K. M. Dorling, P. R. Fielden, S. W. Fogarty, N. J. Fullwood, K. A. Heys, C. Hughes, P. Lasch, P. L. Martin-Hirsch, B. Obinaju, G. D. Sockalingum, J. Sulé-Suso, R. J. Strong, M. J. Walsh, B. R. Wood, P. Gardner and F. L. Martin, Nat. Protoc., 2014, 9, 1771–1791 CrossRef PubMed.
- R. W. Kennard and L. A. Stone, Technometrics, 1969, 11, 137–148 CrossRef.
- R. Bro and A. K. Smilde, Anal. Methods, 2014, 6, 2812–2831 RSC.
- C. L. Morais and K. M. Lima, J. Braz. Chem. Soc., 2017, 31 Search PubMed.
- S. J. Dixon and R. G. Brereton, Chemom. Intell. Lab. Syst., 2009, 95, 1–17 CrossRef.
- J. Kuligowski, G. Quintás, C. Herwig and B. Lendl, Talanta, 2012, 99, 566–573 CrossRef PubMed.
- C. L. Morais and K. M. Lima, Chemom. Intell. Lab. Syst., 2017, 170, 1–12 CrossRef.
- D. Goksuluk, S. Korkmaz, G. Zararsiz and A. E. Karaagaoglu, R Journal, 2016, 8, e30 Search PubMed.
- J. J. Workman Jr., Appl. Spectrosc. Rev., 1996, 31, 251–320 CrossRef.
- S. Türker-Kaya and C. W. Huck, Molecules, 2017, 22, 168 CrossRef PubMed.
- M. Manley, Chem. Soc. Rev., 2014, 43, 8200–8214 RSC.
- A. Hye, S. Lynham, M. Thambisetty, M. Causevic, J. Campbell, H. L. Byers, C. Hooper, F. Rijsdijk, S. J. Tabrizi, S. Banner, C. E. Shaw, C. Foy, M. Poppe, N. Archer, G. Hamilton, J. Powell, R. G. Brown, P. Sham, M. Ward and S. Lovestone, Brain, 2006, 129, 3042–3050 CrossRef PubMed.
- S. Ray, M. Britschgi, C. Herbert, Y. Takeda-Uchimura, A. Boxer, K. Blennow, L. F. Friedman, D. R. Galasko, M. Jutel and A. Karydas, Nat. Med., 2007, 13, 1359 CrossRef PubMed.
- B. Olsson, R. Lautner, U. Andreasson, A. Öhrfelt, E. Portelius, M. Bjerke, M. Hölttä, C. Rosén, C. Olsson and G. Strobel, Lancet Neurol., 2016, 15, 673–684 CrossRef PubMed.
- N. Mattsson, U. Andreasson, H. Zetterberg, K. Blennow and I. for the Alzheimer's Disease Neuroimaging, JAMA Neurol., 2017, 74, 557–566 CrossRef PubMed.
- N. Mattsson, H. Zetterberg, S. Janelidze, P. S. Insel, U. Andreasson and E. Stomrud, Neurology, 2016, 87, 1827–1835 CrossRef PubMed.
- M. Mapstone, A. K. Cheema, M. S. Fiandaca, X. Zhong, T. R. Mhyre, L. H. MacArthur, W. J. Hall, S. G. Fisher, D. R. Peterson, J. M. Haley, M. D. Nazar, S. A. Rich, D. J. Berlau, C. B. Peltz, M. T. Tan, C. H. Kawas and H. J. Federoff, Nat. Med., 2014, 20, 415–418 CrossRef PubMed.
- A. Nakamura, N. Kaneko, V. L. Villemagne, T. Kato, J. Doecke, V. Doré, C. Fowler, Q.-X. Li, R. Martins and C. Rowe, Nature, 2018, 554, 249 CrossRef PubMed.
- M. Paraskevaidi, C. L. Morais, K. M. Lima, J. S. Snowden, J. A. Saxon, A. M. Richardson, M. Jones, D. M. Mann, D. Allsop and P. L. Martin-Hirsch, Proc. Natl. Acad. Sci. U. S. A., 2017, 201701517 Search PubMed.
- E. Peuchant, S. Richard-Harston, I. Bourdel-Marchasson, J. F. Dartigues, L. Letenneur, P. Barberger-Gateau, S. Arnaud-Dabernat and J. Y. Daniel, Transl. Res., 2008, 152, 103–112 CrossRef PubMed.
- S. Mordechai, E. Shufan, B. P. Katz and A. Salman, Analyst, 2017, 142, 1276–1284 RSC.
- P. Carmona, M. Molina, E. López-Tobar and A. Toledano, Anal. Bioanal. Chem., 2015, 407, 7747–7756 CrossRef PubMed.
- M. Paraskevaidi, C. L. Morais, D. E. Halliwell, D. M. Mann, D. Allsop, P. L. Martin-Hirsch and F. L. Martin, ACS Chem. Neurosci., 2018 DOI:10.1021/acschemneuro.8b00198.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8an01205a |
| This journal is © The Royal Society of Chemistry 2018 |