Acrylate-based materials for heart valve scaffold engineering†
Rosaria
Santoro‡
a,
Seshasailam
Venkateswaran‡
b,
Francesco
Amadeo
a,
Rong
Zhang
 bc,
Maura
Brioschi
d,
Anthony
Callanan
e,
Marco
Agrifoglio
f,
Cristina
Banfi
d,
Mark
Bradley
bc,
Maura
Brioschi
d,
Anthony
Callanan
e,
Marco
Agrifoglio
f,
Cristina
Banfi
d,
Mark
Bradley
 *b and
Maurizio
Pesce
*b and
Maurizio
Pesce
 *a
*a
aUnità di Ingegneria Tissutale Cardiovascolare, Centro Cardiologico Monzino, IRCCS, Milan, Italy. E-mail: maurizio.pesce@ccfm.it
bSchool of Chemistry, EaStCHEM, University of Edinburgh, Edinburgh, UK
cSchool of Materials Science & Engineering, Changzhou University, Changzhou, China
dUnità di Proteomica, Centro Cardiologico Monzino, IRCCS, Milan, Italy
eInstitute for Bioengineering, School of Engineering, University of Edinburgh, Edinburgh, UK
fDipartimento Di Scienze Cliniche e di Comunità (Sez. Cardiovascolare) Università di Milano, Milan, Italy
First published on 13th November 2017
Abstract
Calcific aortic valve disease (CAVD) is the most frequent cardiac valve pathology. Its standard treatment consists of surgical replacement either with mechanical (metal made) or biological (animal tissue made) valve prostheses, both of which have glaring deficiencies. In the search for novel materials to manufacture artificial valve tissue, we have conducted a high-throughput screening with subsequent up-scaling to identify non-degradable polymer substrates that promote valve interstitial cells (VICs) adherence/growth and, at the same time, prevent their evolution toward a pro-calcific phenotype. Here, we provide evidence that one of the two identified ‘hit’ polymers, poly(methoxyethylmethacrylate-co-diethylaminoethylmethacrylate), provided robust VICs adhesion and maintained the healthy VICs phenotype without inducing pro-osteogenic differentiation. This ability was also maintained when the polymer was used to coat a non-woven poly-caprolactone (PCL) scaffold using a novel solvent coating procedure, followed by bioreactor-assisted VICs seeding. Since we observed that VICs had an increased secretion of the elastin-maturing component MFAP4 in addition to other valve-specific extracellular matrix components, we conclude that valve implants constructed with this polyacrylate will drive the biological response of human valve-specific cells.
1. Introduction
Calcific aortic valve disease (CAVD) is a pathology of the heart with a continuously increasing incidence worldwide. Calcific valves display severe impairment of valve opening, with consequent variations in the trans-valve flow velocity and an insufficient recoil with consequence for regurgitation and heart failure.1 When damage becomes too severe, failing valves are replaced with mechanical or biological prostheses. About 300![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 heart valve procedures are performed annually worldwide, a number that is expected to triple by 2050 with the majority of the patients over the age of 65.2,3 Despite their acceptable mechanical performance, mechanical prostheses are prone to thromboembolic complications causing patients to require lifelong anti-coagulation therapy with a consequent increase of bleeding. On the other hand, ‘biological’ valves manufactured with animal-derived pericardium, undergo structural leaflet deterioration,4 especially in the young.5,6 Deterioration of biological implants is caused by chronic inflammation due to an incomplete detoxification of the fixative remnants in the tissue,7,8 or by failure of fixatives at removing xeno-antigens such as 1,3 α-Galactose (α-Gal).9–13 Unfortunately, this problem is likely to persist in mini-invasively implantable valve devices (the so called TAVI), which are also produced with animal-derived pericardium.14
000 heart valve procedures are performed annually worldwide, a number that is expected to triple by 2050 with the majority of the patients over the age of 65.2,3 Despite their acceptable mechanical performance, mechanical prostheses are prone to thromboembolic complications causing patients to require lifelong anti-coagulation therapy with a consequent increase of bleeding. On the other hand, ‘biological’ valves manufactured with animal-derived pericardium, undergo structural leaflet deterioration,4 especially in the young.5,6 Deterioration of biological implants is caused by chronic inflammation due to an incomplete detoxification of the fixative remnants in the tissue,7,8 or by failure of fixatives at removing xeno-antigens such as 1,3 α-Galactose (α-Gal).9–13 Unfortunately, this problem is likely to persist in mini-invasively implantable valve devices (the so called TAVI), which are also produced with animal-derived pericardium.14
Tissue-engineered heart valves (TEHVs)15 can be obtained by combining bio-absorbable scaffolds and cells of various origins.16,17 Multiple materials of natural or synthetic origin have been tested to this aim. For example, poly-caprolactone, poly-glycerol-sebacate, poly-lactic-acid – or combinations of them,18–21 are very common materials employed in TEHVs manufacturing. While mechanical performance of tissue engineered valve constructs can be improved via force-oriented extracellular matrix (ECM) deposition using bioreactors,22–26 TEHVs produced with bio-absorbable polymers have still major shortcomings such as an insufficient structural stability of the leaflets in the long term, due to retraction and thickening effects, which result in ultimate failure of the tissue constructs and recurrence of valve insufficiency.22,24,27 Possible solutions to circumvent these limitations have been sought by approaching the problem from a more systematic perspective28,29 and in particular by (i) adapting the chemistry and the biophysical properties of the materials to the required features of the valve-resident cells, which recently emerged as tissue-specific mechano-sensors29–34 or, (ii) adopting advanced micro- or nano-fabrication criteria35 to finely tune the scaffold mechanics to the natural three layered structure of the valve tissue and the anisotropic distribution of compression and bending forces acting in valve motion.20,21 A further simple alternative to resolve the bioactive remodeling of TEHVs scaffolds could be to identify non-bio absorbable materials that could be employed, as main structural components or coating materials, to manufacture valve scaffolds enabling adhesion, maintaining a quiescent phenotype and triggering synthesis of appropriate ECM molecules by valve cells.32,36,37
To test this hypothesis, in the present investigation we conducted a high-throughput polymer screen,38,39 and identified 7 acrylate materials that promoted adherence and growth of human aortic VICs. Upon scale-up processing, two of these polymers supported adhesion of human VICs and, at least in part, suppressed their pro-osteogenic activation.40 Finally, one of these two polymers, the poly(methoxyethylmethacrylate-co-diethylaminoethylmethacrylate), was incorporated into a PCL-based non-woven scaffold by a solvent coating procedure. The results of VICs seeding into polymer-coated scaffolds41 showed a reduction of αSMA expression, one of the principal VICs activation marker, and an enhanced expression of the MFAP4, a key extracellular matrix maturing component.42 Since this polymer is a poly-acrylate, it is easily tunable in terms of its mechanical properties and can also be readily functionalized e.g. with peptide ligands to promote selective cell adhesion and VICs phenotype regulation.43–47 The present investigation, thus, opens a new way to manufacture valve inspired scaffolds with non-biodegradable materials that will allow cell binding, will maintain an appropriate VICs phenotype and will provide and optimal synthesis of valve ECM.
2. Materials and methods
2.1 Preparation of polymer microarrays
Glass slides were soaked (4 h) in 1 M NaOH (1 M), rinsed thoroughly with water and acetone and finally dried (ambient) before immersing (2 h) in acetonitrile (15 mL) containing 1% (v/v) (3-aminopropyl)triethoxysilane, followed by cleaning with acetone (3 × 15 mL) and drying (100 °C, 1 h). The aminosilane-treated slides were dip-coated with agarose aqueous solution (1% v/v, 60 °C) before drying at room temperature (24 h). Polymer microarrays, containing 380 polyacrylates, each printed in quadruplicate, were fabricated as previously reported.39 The monomers that were used to obtain the polymers and the amines used for their functionalization are indicated, respectively in Tables S1 and S2.† Solutions (1% w/v) of the polymers in N-methylpyrrolidone (NMP) were placed into microwell plates and then printed on the agarose-coated slides using a contact printer (QArraymini, Genetix, UK) with 32 aQu solid pins (K2785, Genetix). The printing conditions were 5 stamps per spot, with a 100 s−3 inking timing and a 200 s−3 stamping time. The printed slides were dried in a vacuum oven (45 °C, overnight), sterilized (30 min, UV) light prior to cell culture.2.2 Cell isolation and expansion
Collection of valve leaflets was performed in the event of surgical replacement of stenotic aortic valves, according to GCP and National laws. The procedure was approved by the Ethical Committee of Centro Cardiologico Monzino. An informed consent was obtained by patients. Primary human VICs were isolated by enzymatic dissociation of pathologic AoV leaflets (65.2 ± 12.5 years). The isolation protocol has been previously described.40 Briefly, the non-calcific part of the leaflets were incubated (5 min, shaking, 37 °C) in Collagenase type II solution (1000 U ml−1, Worthington), to remove the endothelial cell layer. VICs were isolated by further incubation (2 h) under the same conditions and amplified on standard culture dishes (1% gelatin coated) using a “complete medium” (DMEM, Lonza) supplemented with penicillin/streptomycin (150 U ml−1, Sigma Aldrich), L-glutamine (2 mM, Sigma Aldrich) and bovine serum (10%, HyClone, Thermo Scientific) and expanded for up to four passages before use.2.2 Polymers microarray screening
Following expansion, aortic VICs isolated from 3 independent donors were seeded (3 × 105 cells per array) and cultured for 72 h onto the PAs microarrays in duplicate, housed in a purpose-made manufactured polycarbonate chamber, designed to circumscribe an area around the array. At the end of the culture period, arrays were fixed in 4% paraformaldehyde (4% PFA) for 20 minutes, washed in phosphate buffered saline (PBS) and stained for 4′,6-diamidin-2-fenilindole (DAPI), phalloidin, vimentin and alpha smooth muscle actin (αSMA). Immunofluorescence images were acquired using a Nikon Eclipse TE200 or a Zeiss Apotome fluorescence microscope (Carl Zeiss, Jena, Germany), through z-stack reconstruction. The adhesion of VICs on the different PAs after 72 hours of culture was evaluated using automated counting of the number of nuclei per spot: cell nuclei stained with DAPI were quantified by implementation of the Analyze Particles tool of ImageJ software (National Institute of Health, Bethesda, MD). In order to derive a priority list of the materials to be implemented in a secondary screening, criteria to obtain a ranking of the PA success to induce cell adhesion were established. PAs were then classified by assigning a score = 3 to PAs that promoted adhesion of the cells from all donors on at least 3 out of 4 materials replica spots averaged on all tested arrays; a score = 2 to PAs promoting adhesion on at least 2 out of 4 materials replica spots (23 polymers; Table S4†); a score = 1 to PAs promoting adhesion on at least 1 out of 4 materials replica spots (83 polymers), and a score = 0 to all the others. The polymers that were considered ‘hit’ materials in this screen had all a score = 3 (Table 1).| PA number | Monomer (1) | Monomer (2) | Monomer (3) | Ratio (mol) | Functionalization | ||
|---|---|---|---|---|---|---|---|
| M (1) | M (2) | M (3) | |||||
| PA98 | MEMA | DEAEMA | 50 | 50 | None | ||
| PA309 | MMA | GMA | 90 | 10 | DnHA | ||
| PA316 | MMA | GMA | 70 | 30 | DBnA | ||
| PA317 | MMA | GMA | 50 | 50 | DBnA | ||
| PA321 | MMA | GMA | 90 | 10 | cHMA | ||
| PA338 | MMA | GMA | 50 | 50 | MAn | ||
| PA426 | MEMA | DEAEA | BMA | 40 | 30 | 30 | None |
2.3 Scale-up and validation of “hit” polymer
The seven “hit” polymers identified from microarray primary screening were synthesized by free-radical polymerization and characterized by gel permeation chromatography (GPC) and infrared spectroscopy (IR). GPC was conducted on an Agilent 1100 instrument, fitted with a PLGel 5 μm MIXED-C column (300 × 7.5 mm), with NMP as the eluent (flow rate 1 mL min−1), pre-calibrated using polystyrene standards. IR analysis was conducted using a Brucker Tensor 27 spectrometer. Solutions of the ‘hit’ polymers prepared in THF (2% w/v) were spin-coated (2000 rpm, 10 s) onto glass coverslips cover slips (∅ 19 mm or ∅ 32 mm) using a spin-coater (6708D, Speedline technologies). The coated coverslips were dried (overnight, convection oven, 40 °C) and sterilised (UV, 30 min) prior to cell culture in either 6- or 12-well plates (coated with agarose (1% w/v) aqueous solution and dried overnight).2.4 VICs culture onto 2D scale-up coated coverslips; RNA expression analysis
In order to monitor the human VICs phenotype evolution at longer time points, cells from 3 independent donors (average age 64.7 ± 10.5) were cultured on coverslips (∅ 19 mm, cell density = 2000 cell per mm2) coated with the hit polymers for 7 and 14 days. For gene expression analysis, RNA was extracted from these cells at both time points using Tripure reagent (Roche Diagnostics). Immunofluorescence (Collagen type I and αSMA) and cell staining (Phalloidin, and DAPI) were instead performed only on VICs cultured for 14 days, followed by automated cell counting (ImageJ™) on confocal microscopy (LSM710; Carl Zeiss, Jena, Germany) images (area = 0.7 mm2). Quantitative real-time PCR (qRT-PCR) amplifications were performed for GAPDH, bone morphogenetic protein 2 (BMP2), alkaline phosphatase (ALP), osteopontin (OPN) and the transcription factor Runx2 (RUNX2) (primers details in following table), using Power SYBR Green PCR Master Mix (Applied Biosystems) on a 7900 Fast Real-Time PCR System (Applied Biosystems). Primers details are provided in Table S3.†2.5 Scaffolds preparation
PCL scaffolds (Mimetix® discs, Electrospinning Company, thickness 2 mm, diameter 2 cm and pore size ∼100 μm) were dip-coated (∼1 s) using a solution of PA98 in acetone (1% w/v, 900 μL per scaffold, ∼1 s) and placed on polypropylene 48-well plates with deep rectangular wells that allowed easier removal of the scaffold post-drying (under ambient conditions in a fume hood) followed by determination of polymer loading (weight increase of scaffold), scanning electron microscopy (SEM) and Infrared spectroscopy (FT-IR). For preparing acetone-treated scaffolds, acetone was used instead of PA98 solution. Coated scaffolds and uncoated controls were sterilized (48 h incubation in BASE.128 (AL.CHI.MIA s.r.l.), an EC certified decontamination solution containing an antibiotic/antifungal mixture containing Gentamicin, Vancomycin, Cefotaxime and Amphotericin B) prior to cell culture.2.6 Scaffold mechanical testing
For the compression analysis, electrospun PCL scaffolds samples (untreated or PA98-coated or acetone-treated) were tested using an Instron tester (Model 3367 running the software Bluehill 3 (Norwood, MA, USA), equipped with a 50 N load cell). The samples were also tested on a CellScale Microsquisher (Waterloo, Ontario, Canada) for flexure analysis. All sample measurements were taken using digital vernier callipers (0–25 mm range; 0.001 mm accuracy). Compression tests were performed on cylindrical samples (width: 3.34 ± 0.44 mm, 3.66 ± 0.75 mm and 3.31 ± 0.25 mm; thickness: 2.73 ± 0.30 mm, 3.39 ± 0.39 mm and 2.86 ± 0.25 mm for uncoated, acetone-treated and PA98-coated, respectively; mean ± Std, n = 4; P > 0.05 by one-way ANOVA with Bonferroni post-hoc test) using a stain rate of 5% min−1 until they reached a 20% strain with stiffness calculated (between 0–5%, 5–10% and 10–15% strain) as the incremental tangent modulus determined using an extrapolation method as previously described.48 The flexure tests were performed on rectangular samples (height: 2.37 ± 0.05 mm, 2.39 ± 0.03 mm and 2.43 ± 0.06 mm for uncoated, acetone-treated and PA98-coated, respectively; mean ± Std, n = 6; P > 0.05 by one-way ANOVA with Bonferroni post-hoc test) and the flexural elastic modulus was evaluated using a test method based on ASTM D790-02. The samples were simply supported across a 10 mm span, with an equivalent knife-edge load applied to the middle of the sample with a flexure strain rate of 33.33 μ s−1 and a deflection depth of 2 mm.2.7 Bioreactor cell seeding and culture; RNA expression analyses
Uncoated and coated scaffolds (8 mm diameter) were seeded and cultured statically or dynamically with VICs. In static seeding (n = 4, average age of donors = 72.4 ± 1.7), scaffolds were placed in agarose-coated multiwells, gently seeded with cell suspension (50 μl containing 1.5 × 105 cells) over the top surface and incubated (2 hours, under standard cell culture conditions) and cultured (1 day or 7 days, 2 ml of medium with the media changed twice per week). Dynamic culture (n = 9, average age of donors = 67.2 ± 10.5) was performed using the U-CUP (Cellec Biotek AG, Basel, CH) and the VICs (4.5 × 105 cells per scaffold), suspended in 10 ml of complete medium, were perfusion seeded into the scaffold at a flow rate of 3 ml min−1 for 24 hours. The scaffolds were then either harvested or, following complete medium renewal, further dynamically cultured (7 or 14 days, flow rate = 0.03 ml min−1 for with the media changed twice per week).49After static or dynamic culture, the samples were rinsed (PBS) followed by MTT staining, histology, gene expression or proteomic analysis. To qualitatively highlight cell distribution within the scaffolds, both statically and dynamically seeded scaffolds were incubated in MTT solution (0.12 mM, in PBS containing 10% FBS, 3 h) and imaged (seeded and reverse sides) with a Zeiss Stemi 2000-C microscope fitted with a KL1500 LCD light source and Axio CAM colour camera, using Axiovision software.
To perform histology and immunofluorescence analysis, the scaffolds seeded by both methods were fixed (4% PFA, 4 °C, overnight) and incubated (overnight, 4 °C, 15% sucrose solution in PBS), followed by embedding in a solution of sucrose (15%) and bovine skin gelatin (7.5%) in PBS. Cryo-sections (10 μm thickness) were stained with DAPI, phalloidin and αSMA/collagen I antibodies. Cell number was quantified by counting the number of nuclei (3 sections per donor and per seeding method).
For gene expression analysis in the 3D culture system, RNA was extracted and PCR was performed as described in section 2.4.
2.8 Sample preparation for mass spectrometry based analysis
Coated and uncoated scaffolds used for the culture of VICs from 3 different donors were cut in small pieces 1 mm2 and subsequently washed 3 times with PBS. In order to decellularize the scaffolds, the samples were incubated with 500 μl of 0.25% v/v Triton X-100 (Sigma Aldrich) at 37 °C for 15 minutes with gentle agitation. After removal of the supernatant, containing cells, scaffold samples were vigorously washed 7 times with ice cold water to completely eliminate Triton X-100. 100 μl of 25 mmol L−1 NH4HCO3 containing 0.1% w/v RapiGest SF were then added for tryptic digestion. Samples were reduced with 5 mmol L−1 TCEP (tris(2-carboxyethyl)phosphine), dissolved in 100 mmol L−1 NH4HCO3, at room temperature for 30 min, and then thio-modified with 10 mmol L−1 iodacetamide for 30 min at room temperature. Digestion was performed overnight at 37 °C using 0.5 μg of sequencing grade trypsin (Promega, Milan, Italy). After digestion, 2% v/v TFA was added to hydrolyse RapiGest SF and inactivate trypsin, and the solution was incubated at 37 °C for 40 min before being vortexed and centrifuged at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 minutes to eliminate RapiGest SF. Quantitative label-free LC-MSE was performed as indicated in the ESI† according to previously described methods.50,51
000g for 10 minutes to eliminate RapiGest SF. Quantitative label-free LC-MSE was performed as indicated in the ESI† according to previously described methods.50,51
2.9 Data representation and statistical analysis
Data are represented and mean ± SE. Statistical comparisons were made by ANOVA or one-tailed Student's t-test, where appropriate, using the GraphPad software. Clusterization analysis of differentially expressed proteins was performed using Multi Experimental Viewer software (MeV), following normalization and centering around the mean expression values using Pearson's correlation to calculate distance among clusters.3. Results
Fig. 1 summarizes the main experimental steps that were followed in the study, from the isolation of human aortic VICs (phase A), to their culturing onto polymer microarrays for primary polymer screening (phase B),39 followed by 2D scale-up of the VICs adhering ‘hit’ polymers (phase C) and the final validation of one of the two best identified polymers (PA98) using a 3D culture approach (phase D).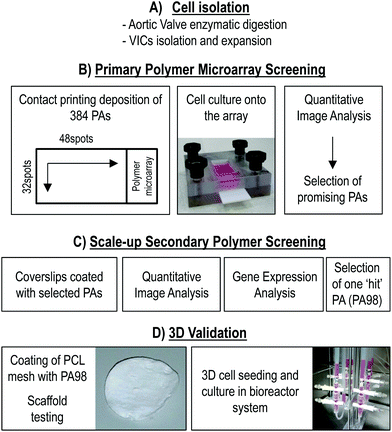 | ||
| Fig. 1 Experimental flowchart. In a first phase (A), cells from pathologic heart valves were isolated and amplified in culture. They were characterized as previously shown.40 In a second phase (B), 380 PAs-containing microarrays were screened with the amplified cells by cell adhesion screening and quantification. The selected ‘hit’ polymers were then scaled-up onto glass coverslips (C) and quantitative imaging analysis and gene expression analyses were performed. PA98 was selected to be transferred to 3D condition. The final phase (D) of the work consisted in the coating of a PCL 3D scaffold with PA98, followed by perfusion cell seeding, cell counting, and mRNA/proteins extraction. | ||
3.1 High throughput array screening reveals specific polymer compositions promoting hVIC-adhering
Human-derived VICs were cultured for 72 h on polymer microarrays, following which their adhesion was evaluated by counting the average number of cells adhered on each spot (Fig. 2A and C) by counting the DAPI stained nuclei. As shown in Fig. 2A seven ‘hit’ polymers reproducibly promoted VICs adhesion with a number of cells per spot (Fig. 2C) comprised between 18 ± 3 to 60 ± 6 (mean ± SE). Five of the ‘hits’ polymers contained methyl methacrylate (MMA) mixed at various molar ratios with glycidyl methacrylate (GMA), while two others contained methoxyethyl methacrylate (MEMA) (Table 1). Interestingly, a number of other polymeric blends with similar monomer composition were also scored as second choice materials inducing VICs adhesion on primary arrays (Table S4†).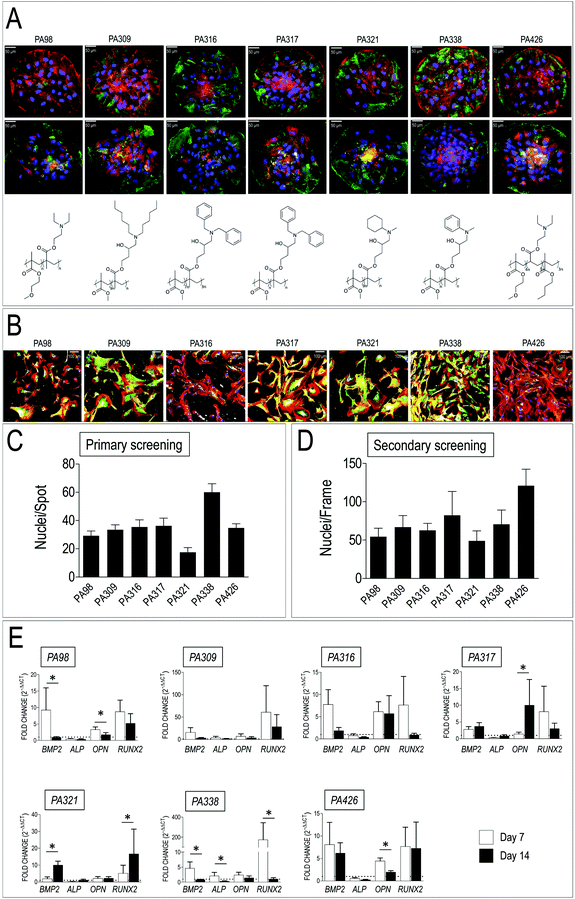 | ||
| Fig. 2 Results of primary and secondary material screening. (A) Representative images of VICs adhered onto the seven ‘hit’ PAs on polymer arrays, whose chemical structure is also shown below each micrograph (see also Table 1 for composition). Cells were stained with anti-vimentin (top row panels, green fluorescence) and anti-αSMA antibody (bottom row panels, green fluorescence). DAPI staining was used to visualize nuclei and count the cells. (B) Representative confocal images of VICs cultured on glass coverlips replicas of the seven hits polymers. Cells were stained with Phalloidin-TRITC, anti-αSMA (green) and anti-Collagen I (white fluorescence). (C) Results of primary screening of polymer spots in the arrays with human-derived VICs. Cell number was determined by nuclei counting on four polymer spot replicas in each array. Data are given as an average number of cells per spot. The observations were repeated with three independent donor-derived VICs primary cell lines. (D) Results of secondary polymer screening on coated glass slides. Cell counting was performed as for panel B, with the exception that the number of cells is given nuclei per frame. (E) Real-Time PCR for detection of osteogenic genes (BMP2, ALP, OPN, RUNX2) expression in VICs cultured for 7 and 14 days onto the indicated hit polymers. Data are plotted as a fold difference (2−ΔΔCt) in the expression of each of the genes in cells cultured onto each of the polymers vs. the expression level observed in VICs cultured on plastics. A dotted line corresponding to the plastics-cultured VICs (fold change = 1) without polymers is indicated in each of the graphs. * indicate statistical significance (P < 0.05) in day 7 vs. day 14 gene expression comparisons by paired t-test. | ||
In order to readily detect the primary response of the cells to the materials, cells adhering to polymer spots were also stained with antibodies directed against vimentin and αSMA, two VICs/activated VICs markers.52 The results showed a variable expression of the two markers in cells plated onto different polymers (Fig. 2A), thus anticipating a variable cell response to the materials chemistry. Taken together, these results indicated a specificity of VICs adhesion to polymers having a peculiar chemical composition and structure, although with a variable activation level.
3.2 Polymers scaling up and biological validation of the selected hit PAs
The seven selected polymers were employed to coat larger glass slides to perform long-term cell culture (up to 2 weeks) to confirm and validate adhesion data and assess VICs phenotype by immunofluorescence and RT-PCR, respectively (Fig. 2B, D and E). The number of nuclei per frame were again determined by counting DAPI-stained nuclei, confirming the ability of the selected PA blends to support human VICs adhesion. Immunofluorescence analysis allowed evaluation of αSMA and Collagen type I expression as indicators of VICs activation53 and their ability to secrete crucial components of the extracellular matrix components.54 Interestingly, as already noted for the primary screening, the expression level of these two proteins was remarkably different, again indicating a polymer-dependent variation of cells activation and secretion of extracellular matrix. Finally, since VICs activation normally precedes their differentiation into calcified matrix secreting cells,53,55–57 the expression of genes involved in calcific cells transformation (bone morphogenetic protein 2, BMP2; alkaline phosphatase, ALP; osteopontin, OPN, Runx2 transcription factor; RUNX2),58 was assessed by Q-RT-PCR at 7 and 14 days of culture onto each of the selected PAs. As shown in Fig. 2E, the results clearly indicated polymers onto which expression of osteogenic genes was highly variable (PA309, PA316, PA426), polymers onto which pro-pathologic genes were upregulated (PA317 and PA321), and polymers that maintained, especially at 14 days culture time, the expression level of the genes at comparable levels to those found in plastic cultured cells (PA98 and PA338).3.3 Coating of a nonwoven poly-caprolactone scaffold with PA98
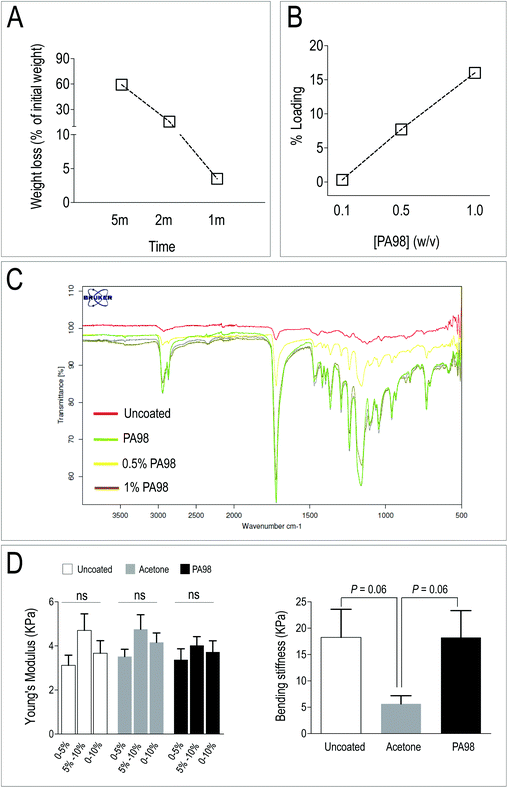
PCL scaffolds have been extensively employed in TEHVs due to its relatively slow in vivo degradation, and higher mechanical performance compared to other bio-absorbable polymers such as, for example, Poly-Glycolic Acid (PGA).59,60 In addition, PCL is very versatile and can be mixed or functionalized with biological-derived molecules – e.g. gelatin, fibrin, ECM-derived adhesion molecules and even mineral microparticles – to finely adjust scaffolds stiffness and promote cell growth and phenotypic adaptation.61–64 Despite, according to Q-RT-PCR results, PA338 maintained a lower expression level of three out of five of pro-osteogenic genes (Fig. 2E), immunofluorescence data of the primary and secondary screening indicated that VICs cultured onto PA98 expressed lower levels of the αSMA (Fig. 2A and B). PA98 poly(methoxyethylmethacrylate-co-diethylaminoethylmethacrylate) was therefore chosen to perform validation of polyacrylate materials for 3D VICs culturing, adopting a solvent coating procedure of a commercially available non-woven PCL scaffold.Acetone and THF were first tested for their ability to dissolve PA98 (visual inspection of 1% solutions) and also for the stability of the electrospun PCL scaffold when in contact with the solvents. Although both solvents dissolved PA98, THF disintegrated the PCL scaffold immediately. By contrast, acetone determined a time dependent weight loss between 59.2% (5 min) and 1.5% (5 s) (Fig. 3A), thus confirming that a short dipping time could be appropriate for scaffold solvent coating with PA98. Following the solvent screening, the influence of polymer concentration was investigated to avoid clogging the PCL mesh. Scaffolds were dip-coated for 1 second in either 0.1%, 0.5% or 1% solutions (w/v %) of PA98 in acetone and dried (Fig. 3B). Based on mass increase, PA98 loadings of 0.3%, 7.7% and 16% were achieved, respectively for the 0.1%, 0.5%, 1% coating solutions. IR spectroscopy (Fig. 3C) of the coated scaffolds confirmed a concentration-dependent increasing incorporation of PA98 with a 1 second dipping time.
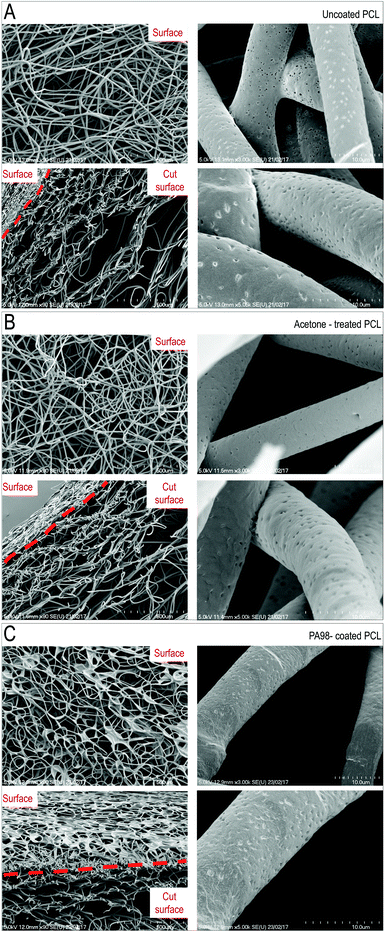
To unravel possible damages caused to scaffolds by the coating procedures, mechanical testing of the untreated, acetone-treated and PA98-treated scaffolds was performed. As shown in Fig. 3D, results of compression and flexural tests indicated no major differences in mechanical properties between coated and uncoated scaffolds, although an almost significant modification of the bending stiffness of the scaffold was noticed with the acetone-only treatment. Finally, the ultrastructure of the scaffold, before and after coating, or solvent only treatment, was investigated by scanning electron microscopy (SEM) (Fig. 4). The scaffolds treated with acetone (1 s) were virtually identical to the untreated samples confirming acetone as a suitable solvent for coating. By contrast an evident polymer gradient was found between the upper and the lower scaffold surfaces, likely caused by percolation of the polymer solutions under gravity during the overnight drying. Despite this, even when with the highest concentration (1%) of polymer solution, the scaffolds appeared to retain their pores. These results led us to define a final procedure for PCL scaffold functionalization consisting in a dipping time of ∼1 s using a 1% w/v solution of PA98 in acetone for further validation with valve cells.
3.4 Controlled seeding of human VICs in PA98-coated PCL scaffold
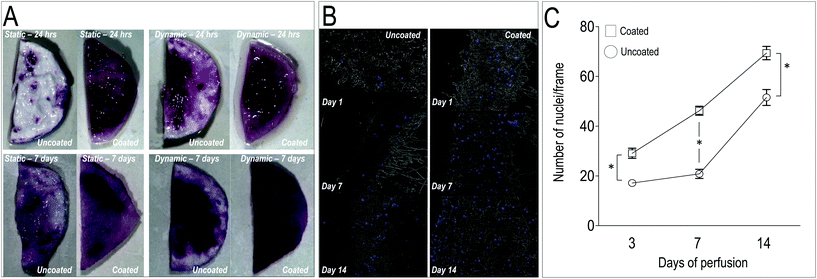
In order to test the ability of the scaffolds to support homing and growth of valve cells, we established a VICs seeding protocol into untreated and PA98-coated PCL scaffolds (PCL-PA). Instead of adopting a conventional cell deposition protocol into tissue culture plates, round shaped patches of uncoated, acetone-treated and PA98-coated scaffolds were also placed into a direct perfusion bioreactor system.65 In such a system, cell seeding homogeneity and efficiency depends on the scaffolds 3D structure, with particular regard to pore size and interconnections,66 and it is therefore more standardized. A static VICs culture conditions was anyway used as a control of the seeding efficiency in bioreactor-supported dynamic culture.As a first test to evaluate the cellularization of the scaffolds, a MTT staining of the uncoated and PCL-PA scaffolds was performed under static and dynamic conditions at 1 and 7 days. Results (Fig. 5A) showed that dynamic culturing of the cells into the PCL-PA scaffolds provided the most uniform distribution of the cells compared to the other conditions. Experiments conducted in parallel with acetone-treated scaffolds confirmed no differences in cell survival, thus showing that no solvent remnants persisted in the scaffold after the coating procedure (data not shown). To quantify cell dynamics in the scaffolds, longer time course experiments (up to 14 days) were performed, followed by cell counting in transversal sections (Fig. 5B). This again consolidated the superiority of the PCL-PA scaffold to trap VICs under the dynamic condition compared to the untreated scaffold (Fig. 5C). Finally, since the VICs increase in the coated scaffolds was not accompanied by a significant increase of cells expressing the Ki67 proliferation marker (data not shown), the higher number of cells was likely due to an increased VICs adhesion in the presence of the polymer. Altogether we conclude that coating the PCL mesh with PA98 enhanced seeding efficiency of cells into the PCL-PA scaffold during dynamic perfusion.
3.5 Regulation of the VICs phenotype in the PCL-PA scaffold
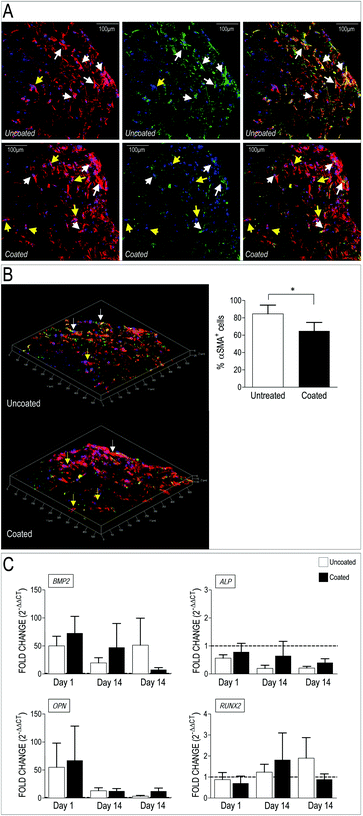
Valve interstitial cells respond to mechanical cues with enhanced activation toward a myofibroblast phenotype and calcification.30–32,34,36,53,55,67–69 Transition of these cells from quiescent to activated phenotype is however reversible, thus suggesting the polymer loading in the scaffold may revert their evolution toward pathologic cells. We therefore investigated whether the dynamic perfusion seeding system and the employment of the 3D scaffolds had any effect on VICs phenotype when the polymer was present. To do this, we first performed immunofluorescence on transversal sections of the PCL and PCL-PA scaffolds seeded with human VICs. As shown in Fig. 6A, staining for the αSMA and the actin cytoskeleton indicated that cells present in the PCL scaffold had higher levels of stress fibers-associated αSMA, compared to those present in the scaffold carrying PA98. This difference was particularly evident in 3D reconstructed optical sections and was supported by a significant reduction of αSMA+ cells (Fig. 6B) and, although only as a trend, by Q-RT-PCR showing that expression of pro-pathologic genes BMP2, ALP, OPN and RUNX2 returned at basal level at late time points (Fig. 6C).3.6 Culture on PA98-coated scaffolds influences ECM secreted by VICs
Appropriate formation of valve tissue depends not only by control of VICs quiescent phenotype, but also by deposition of appropriate extracellular matrix components. To assess deposition of extracellular matrix components by VICs in the 3D setting, we employed a label-free Mass Spectrometry-based proteomic approach (LC-MSE), which allowed us to obtain quantitative analysis of proteins released by the cells in the 3D environment. Data processing of a total of 1503 peptides led to identification of 100 human proteins present in uncoated and coated scaffolds (Table S5†). As indicated in the table, the majority of these were linked to metabolism and cytoskeleton; on the other hand, a number of ECM components released by the cells were also found, confirming the ability of the human VICs to secrete valve-specific matrix such as Collagen, Emilin, Fibronectin, Fibulin and Vitronectin.Quantitative analysis of the proteomic profiles led to a clear separation between the PA98 coated vs. the uncoated scaffolds, revealing a strong effect of the polymer loading on general protein expression by the cells in the 3D environment (Fig. 7A). In addition, pairwise ANOVA indicated 24 proteins that were differentially expressed, with 12 upregulated in PCL, and 12 significantly more abundant in PCL-PA scaffolds (Fig. 7B, Tables S6 and S7†). It was particularly interesting to observe that the presence of PA98 determined a sharp increase in the presence of the microfibril-associated glycoprotein 4 (MFAP4), a protein that has been recently reported to be involved in extracellular fibril organization and elastic fibers assembly.42,70 Together, these results suggest that presence of the PA98 coating promotes maturation of the ECM secreted by VICs.
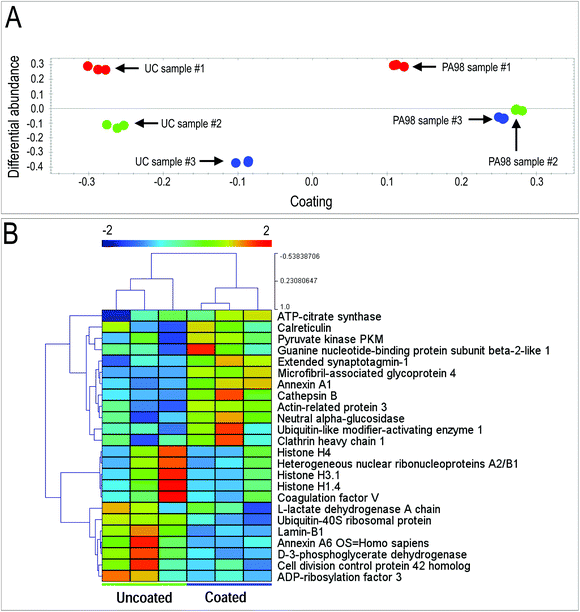 | ||
| Fig. 7 Results of mass spectrometry analysis on VICs-seeded scaffolds. Identification of proteins differentially released by human VICs in the PCL-PA scaffolds by Mass Spectrometry-based proteomic analysis. (A) Results of principal component analysis of differentially expressed proteins in the three VICs seeded uncoated and PA98-coated scaffolds (Table S5†). As shown, samples were clearly discriminated on the basis of the coating and the relative abundance of expression, highlighting a coherently different release of extracellular proteins in the PCL scaffolds coated with the polymer vs. the uncoated condition. (B) Clusterization analysis of the mass spectrometry data from uncoated vs. coated scaffolds identified a clear separation between the protein expression pattern in uncoated vs. PA98 coated scaffold (n = 3; proteins listed in Tables S6 and S7†). | ||
4. Discussion
In the present contribution we adopted a high-throughput screening system (polymer microarrays)39 to identify, for the first time, acrylate-based materials that could be implemented in a valve scaffold manufacturing process. Our choice to perform a PAs screening for valve engineering was driven by the translational potential of these materials in the clinical environment. Polyacrylates are indeed already approved for use in humans for various applications like, e.g. bone repair, tooth cements and contact lenses (discussed in ref. 71 and 72). In addition, compared to materials with variable bio-absorbability such as polyurethanes (PU), poly(hydroxyalkanoates), poly(ε-caprolactone), and poly(glycerol sebacate),73 PAs are non-absorbable materials that have been found to give rise to mild foreign body reactions and provide optimal blood compatibility especially adopting copolymerization74,75 or surface functionalization76,77 strategies. Although the employment of this class of polymers appears contra-intuitive due to drawbacks of the so called polymeric valves,78 the very flexible PA chemistry may be implemented to manufacture scaffolds able to recruit cells into a finely tuned mechanical environment, and keep permanently under control the stiffness- and strain-dependent cell myofibroblast differentiation.30,34 Another potential advantage may, finally, be derived by the flexibility of these polymers to be functionalized with various biologics, such as adhesion molecules, binding repeats, extracellular matrix components and cytokines, which may locally instruct the cell behavior44,45 or promote endothelialization at their surface.43The basic adhesion assays, confirmed by the secondary screening data (Fig. 2) showed a propensity of human-derived VICs to adhere to polyacrylates with a specific composition of MEMA or MMA as primary monomers and GMA or DEAE(M)A as secondary monomers (Table 1), thus highlighting a clear a selectivity of the VICs adhesion process on acrylates. This is also supported by the composition of the second choice polymers, whose formulation was similar to the materials identified as ‘hits’ (Table S4†). Remarkably, Q-RT-PCR data (Fig. 2E) showed that not all the materials that best supported VICs adhesion also maintained a good control of their differentiation into myofibroblasts. This result may depend upon the specific polymer chemical composition, but also on peculiar physical characteristics of the polymers such as the wettability, roughness and water contact angle,79 which might create an optimal adherence surface for the cells compared to naturally occurring polymers. Particularly relevant, in this respect, is also the possibility that coating with PA98 determined an overall decrease of the PCL fibers surface stiffness, leading to a partial repression of the VICs ‘activated’ phenotype,69 whose control is linked to elasticity of the surrounding matrix.30,80 This latter aspect is particularly interesting, considering that valve-specific adhesion repeats and GAGs known to control the VICs phenotype have been identified,81–85 and thereby might be included in advanced material scaffold formulations to suppress cellular pathologic evolution,35 promote ECM deposition and definitive valve tissue maturation.86,87
Our data showed that when one of two best identified materials (PA98) was employed to coat a PCL bio-reabsorbable scaffold, higher VICs numbers were found at all the time points compared to uncoated scaffolds (Fig. 5C), and that these cells exhibited lower polymerization of the αSMA VICs activation marker (Fig. 6A and B). Finally, the analysis of the expression of two key pro-osteogenic genes BMP2 and OPN involved in VICs pathologic evolution showed a return to basal levels in bioreactor based-maturation of cellularized polymer loaded scaffolds (Fig. 6C). This finding, along with the evidence that PA98 was associated to an increased MFAP4 protein expression (Fig. 7, Tables S5 and S6†), strongly suggests that the presence of the polyacrylate coating on the PCL scaffold may serve as a selective functionalization of the bioabsorbable polymer promoting the capture of valve-competent cells and inducing a higher maturation level of the extracellular matrix, in particular the elastic fibres.42,88,89 How and whether PA coating will affect the degradation of the PCL scaffolds when in contact with VICs or circulating monocytes, or at what extent PA98 functionalization lasts after the scaffold is implanted in vivo, and what it will be the final hemocompatability of the selected materials is matter of future investigations that will have to be conducted using appropriate models.
In summary, identification of acrylate materials able to control calcific differentiation of VICs and, potentially, other myofibroblast evolving cells (e.g. mesenchymal cells) could pave the way for the generation of scaffolds for in situ tissue engineering applications based on the ability to provide full mechanical support for the implant and instructing cues for mature valve leaflets tissue development. In this regard, the exciting possibility to proceed with a finely tuning of these materials to have defined stiffness and resistance to cyclic stretch,47 will be particularly relevant to avoid pathologic programming of scaffold populating cells and, thus, failure of the valve implants in the long term due to retraction and compaction effects typical of TEHVs.90 Our findings support, therefore, the adoption of these materials in cardiac valve scaffold manufacturing with hopes for overcoming the limitations of the current biological devices and classical tissue engineered implants.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The present work has been funded by an Institutional project grant at Centro Cardiologico Monzino (Ricerca Corrente), issued to MP and from a Ministry of Health Grant (Ricerca Finalizzata, 196/GR-2011-02348707) issued to RS. Cellec AG, Basel Switzerland, is acknowledged for assistance with the use of the U-CUP® bioreactor system. MBR thanks the ERC (ADREEM 340469).References
- A. Small, D. Kiss, J. Giri, S. Anwaruddin, H. Siddiqi, M. Guerraty, J. A. Chirinos, G. Ferrari and D. J. Rader, Arterioscler., Thromb., Vasc. Biol., 2017, 37, 623–632 CrossRef CAS PubMed.
- T. E. David, Nat. Rev. Cardiol., 2013, 10, 375–386 CrossRef PubMed.
- A. Kheradvar, E. Groves, C. Goergen, S. H. Alavi, R. Tranquillo, C. Simmons, L. Dasi, K. J. Grande-Allen, M. K. Mofrad, A. Falahatpisheh, B. Griffith, F. Baaijens, S. Little and S. Canic, Ann. Biomed. Eng., 2014, 1–14, DOI:10.1007/s10439-014-1191-5.
- D. Reineke, F. Gisler, L. Englberger and T. Carrel, Expert Rev. Cardiovasc. Ther., 2016, 14, 423–430 CrossRef CAS PubMed.
- J. Forcillo, M. Pellerin, L. P. Perrault, R. Cartier, D. Bouchard, P. Demers and M. Carrier, Ann. Thorac. Surg., 2013, 96, 486–493 CrossRef PubMed.
- S. F. Saleeb, J. W. Newburger, T. Geva, C. W. Baird, K. Gauvreau, R. F. Padera, P. J. del Nido, M. J. Borisuk, S. P. Sanders and J. E. Mayer, Circulation, 2014, 130, 51–60 CrossRef PubMed.
- M. Grabenwoger, J. Sider, F. Fitzal, C. Zelenka, U. Windberger, M. Grimm, A. Moritz, P. Bock and E. Wolner, Ann. Thorac. Surg., 1996, 62, 772–777 CAS.
- R. F. Siddiqui, J. R. Abraham and J. Butany, Histopathology, 2009, 55, 135–144 CrossRef PubMed.
- K. Z. Konakci, B. Bohle, R. Blumer, W. Hoetzenecker, G. Roth, B. Moser, G. Boltz-Nitulescu, M. Gorlitzer, W. Klepetko, E. Wolner and H. J. Ankersmit, Eur. J. Clin. Invest., 2005, 35, 17–23 CrossRef CAS PubMed.
- F. Naso, A. Gandaglia, T. Bottio, V. Tarzia, M. B. Nottle, A. J. F. d'Apice, P. J. Cowan, E. Cozzi, C. Galli, I. Lagutina, G. Lazzari, L. Iop, M. Spina and G. Gerosa, Xenotransplantation, 2013, 20, 252–261 CrossRef PubMed.
- F. Naso, A. Gandaglia, L. Iop, M. Spina and G. Gerosa, Xenotransplantation, 2012, 19, 215–220 CrossRef PubMed.
- U. Galili, Immunol. Cell Biol., 2005, 83, 674–686 CrossRef CAS PubMed.
- J. Hülsmann, K. Grün, S. El Amouri, M. Barth, K. Hornung, C. Holzfuß, A. Lichtenberg and P. Akhyari, Xenotransplantation, 2012, 19, 286–297 CrossRef PubMed.
- C. W. Hamm, M. Arsalan and M. J. Mack, Eur. Heart J., 2016, 37, 803–810 CrossRef PubMed.
- T. Shinoka, C. K. Breuer, R. E. Tanel, G. Zund, T. Miura, P. X. Ma, R. Langer, J. P. Vacanti and J. E. Mayer, Jr., Ann. Thorac. Surg., 1995, 60, S513–S516 CrossRef CAS PubMed.
- O. M. J. A. Stassen, D. E. P. Muylaert, C. V. C. Bouten and J. Hjortnaes, Curr. Treat. Options Cardiovasc. Med., 2017, 19, 71 CrossRef CAS PubMed.
- B. Weber, M. Y. Emmert and S. P. Hoerstrup, Swiss Med. Wkly, 2012, 142, w13622 Search PubMed.
- G. C. Engelmayr Jr., E. Rabkin, F. W. H. Sutherland, F. J. Schoen, J. E. Mayer Jr. and M. S. Sacks, Biomaterials, 2005, 26, 175–187 CrossRef PubMed.
- N. Masoumi, K. L. Johnson, M. C. Howell and G. C. Engelmayr Jr., Acta Biomater., 2013, 9, 5974–5988 CrossRef CAS PubMed.
- N. Masoumi, N. Annabi, A. Assmann, B. L. Larson, J. Hjortnaes, N. Alemdar, M. Kharaziha, K. B. Manning, J. E. Mayer Jr. and A. Khademhosseini, Biomaterials, 2014, 35, 7774–7785 CrossRef CAS PubMed.
- N. Masoumi, B. L. Larson, N. Annabi, M. Kharaziha, B. Zamanian, K. S. Shapero, A. T. Cubberley, G. Camci-Unal, K. B. Manning, J. E. Mayer, Jr. and A. Khademhosseini, Adv. Healthcare Mater., 2014, 3, 929–939 CrossRef CAS PubMed.
- I. A. van Loosdregt, G. Argento, A. Driessen-Mol, C. W. Oomens and F. P. Baaijens, J. Biomech., 2014, 47, 2064–2069 CrossRef PubMed.
- S. Loerakker, G. Argento, C. W. Oomens and F. P. Baaijens, J. Biomech., 2013, 46, 1792–1800 CrossRef CAS PubMed.
- M. A. van Vlimmeren, A. Driessen-Mol, C. J. Oomens and F. T. Baaijens, Biomech. Model. Mechanobiol., 2012, 11, 1015–1027 CrossRef PubMed.
- A. Balguid, N. J. B. Driessen, A. Mol, J. P. J. Schmitz, F. Verheyen, C. V. C. Bouten and F. P. T. Baaijens, J. Biomech., 2008, 41, 2612–2617 CrossRef PubMed.
- J. B. Schmidt and R. T. Tranquillo, Ann. Biomed. Eng., 2016, 44, 1785–1797 CrossRef PubMed.
- M. A. van Vlimmeren, A. Driessen-Mol, C. W. Oomens and F. P. Baaijens, Tissue Eng., Part C, 2011, 17, 983–991 CrossRef CAS PubMed.
- E. S. Fioretta, P. E. Dijkman, M. Y. Emmert and S. P. Hoerstrup, J. Tissue Eng. Regener. Med., 2016 DOI:10.1002/term.2326.
- J. Usprech, W. L. K. Chen and C. A. Simmons, Wiley Interdiscip. Rev.: Syst. Biol. Med., 2016, 8, 169–182 CrossRef PubMed.
- H. Ma, A. R. Killaars, F. W. DelRio, C. Yang and K. S. Anseth, Biomaterials, 2017, 131, 131–144 CrossRef CAS PubMed.
- H. Wang, L. A. Leinwand and K. S. Anseth, Nat. Rev. Cardiol., 2014, 11, 715–727 CrossRef PubMed.
- H. Wang, S. M. Haeger, A. M. Kloxin, L. A. Leinwand and K. S. Anseth, PLoS One, 2012, 7, e39969 CAS.
- M. Pesce and R. Santoro, Pharmacol. Ther., 2017, 171, 75–82 CrossRef CAS PubMed.
- C. Fisher, J. Chen and W. D. Merryman, Biomech. Model. Mechanobiol., 2013, 12, 5–17 CrossRef PubMed.
- A. Hasan, J. Saliba, H. Pezeshgi Modarres, A. Bakhaty, A. Nasajpour, M. R. K. Mofrad and A. Sanati-Nezhad, Biomaterials, 2016, 103, 278–292 CrossRef CAS PubMed.
- H. Wang, M. W. Tibbitt, S. J. Langer, L. A. Leinwand and K. S. Anseth, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 19336–19341 CrossRef CAS PubMed.
- J. Hjortnaes, G. Camci-Unal, J. D. Hutcheson, S. M. Jung, F. J. Schoen, J. Kluin, E. Aikawa and A. Khademhosseini, Adv. Healthcare Mater., 2015, 4, 121–130 CrossRef CAS PubMed.
- R. S. Tare, F. Khan, G. Tourniaire, S. M. Morgan, M. Bradley and R. O. Oreffo, Biomaterials, 2009, 30, 1045–1055 CrossRef CAS PubMed.
- G. Tourniaire, J. Collins, S. Campbell, H. Mizomoto, S. Ogawa, J. F. Thaburet and M. Bradley, Chem. Commun., 2006, 2118–2120 RSC.
- R. Santoro, F. Consolo, M. Spiccia, M. Piola, S. Kassem, F. Prandi, M. C. Vinci, E. Forti, G. Polvani, G. B. Fiore, M. Soncini and M. Pesce, J. Biomed. Mater. Res., Part B, 2016, 104, 345–356 CrossRef CAS PubMed.
- R. Santoro, A. L. Olivares, G. Brans, D. Wirz, C. Longinotti, D. Lacroix, I. Martin and D. Wendt, Biomaterials, 2010, 31, 8946–8952 CrossRef CAS PubMed.
- B. Pilecki, A. T. Holm, A. Schlosser, J. B. Moeller, A. P. Wohl, A. V. Zuk, S. E. Heumüller, R. Wallis, S. K. Moestrup, G. Sengle, U. Holmskov and G. L. Sorensen, J. Biol. Chem., 2016, 291, 1103–1114 CrossRef CAS PubMed.
- S. Pernagallo, O. Tura, M. Wu, K. Samuel, J. J. Diaz-Mochon, A. Hansen, R. Zhang, M. Jackson, G. J. Padfield, P. W. F. Hadoke, N. L. Mills, M. L. Turner, J. P. Iredale, D. C. Hay and M. Bradley, Adv. Healthcare Mater., 2012, 1, 646–656 CrossRef CAS PubMed.
- F. Khan, R. S. Tare, R. O. Oreffo and M. Bradley, Angew. Chem., Int. Ed., 2009, 48, 978–982 CrossRef CAS PubMed.
- R. Zhang, H. K. Mjoseng, M. A. Hoeve, N. G. Bauer, S. Pells, R. Besseling, S. Velugotla, G. Tourniaire, R. E. B. Kishen, Y. Tsenkina, C. Armit, C. R. E. Duffy, M. Helfen, F. Edenhofer, P. A. de Sousa and M. Bradley, Nat. Commun., 2013, 4, 1335 CrossRef PubMed.
- J. P. Gong, Y. Katsuyama, T. Kurokawa and Y. Osada, Adv. Mater., 2003, 15, 1155–1158 CrossRef CAS.
- J.-Y. Sun, X. Zhao, W. R. K. Illeperuma, O. Chaudhuri, K. H. Oh, D. J. Mooney, J. J. Vlassak and Z. Suo, Nature, 2012, 489, 133–136 CrossRef CAS PubMed.
- A. Callanan, N. F. Davis, T. M. McGloughlin and M. T. Walsh, J. Biomech., 2014, 47, 1885–1893 CrossRef CAS PubMed.
- D. Wendt, A. Marsano, M. Jakob, M. Heberer and I. Martin, Biotechnol. Bioeng., 2003, 84, 205–214 CrossRef CAS PubMed.
- M. Roverso, M. Brioschi, C. Banfi, S. Visentin, S. Burlina, R. Seraglia, P. Traldi and A. Lapolla, Eur. J. Mass Spectrom., 2016, 22, 71–82 CrossRef CAS PubMed.
- M. Brioschi, S. Eligini, M. Crisci, S. Fiorelli, E. Tremoli, S. Colli and C. Banfi, Anal. Bioanal. Chem., 2014, 406, 2817–2825 CrossRef CAS PubMed.
- A. M. Porras, N. C. A. van Engeland, E. Marchbanks, A. McCormack, C. V. C. Bouten, M. H. Yacoub, N. Latif and K. S. Masters, J. Am. Heart Assoc., 2017, 6(3) Search PubMed , e005041.
- J. H. Chen, C. Y. Yip, E. D. Sone and C. A. Simmons, Am. J. Pathol., 2009, 174, 1109–1119 CrossRef CAS PubMed.
- A. Balguid, M. P. Rubbens, A. Mol, R. A. Bank, A. J. Bogers, J. P. van Kats, B. A. de Mol, F. P. Baaijens and C. V. Bouten, Tissue Eng., 2007, 13, 1501–1511 CrossRef CAS PubMed.
- C. Y. Yip, J. H. Chen, R. Zhao and C. A. Simmons, Arterioscler., Thromb., Vasc. Biol., 2009, 29, 936–942 CrossRef CAS PubMed.
- A. C. Liu, V. R. Joag and A. I. Gotlieb, Am. J. Pathol., 2007, 171, 1407–1418 CrossRef CAS PubMed.
- P. M. Taylor, P. Batten, N. J. Brand, P. S. Thomas and M. H. Yacoub, Int. J. Biochem. Cell Biol., 2003, 35, 113–118 CrossRef CAS PubMed.
- A. C. Liu, V. R. Joag and A. I. Gotlieb, Am. J. Pathol., 2007, 171, 1407–1418 CrossRef CAS PubMed.
- M. M. Brugmans, R. S. Soekhradj-Soechit, D. van Geemen, M. Cox, C. V. Bouten, F. P. Baaijens and A. Driessen-Mol, Tissue Eng., Part A, 2016, 22, 123–132 CrossRef CAS PubMed.
- M. M. Brugmans, A. Driessen-Mol, M. P. Rubbens, M. A. Cox and F. P. Baaijens, J. Tissue Eng. Regener. Med., 2015, 9, E289–E301 CrossRef CAS PubMed.
- C. Klopsch, R. Gäbel, A. Kaminski, P. Mark, W. Wang, A. Toelk, E. Delyagina, G. Kleiner, L. Koch, B. Chichkov, P. Mela, S. Jockenhoevel, N. Ma and G. Steinhoff, J. Tissue Eng. Regener. Med., 2015, 9, E177–E190 CrossRef CAS PubMed.
- C. Rentsch, B. Rentsch, S. Heinemann, R. Bernhardt, B. Bischoff, Y. Förster, D. Scharnweber and S. Rammelt, BioMed Res. Int., 2014, 2014, 217078 CAS.
- M. S. Savelyeva, A. A. Abalymov, G. P. Lyubun, I. V. Vidyasheva, A. M. Yashchenok, T. E. L. Douglas, D. A. Gorin and B. V. Parakhonskiy, J. Biomed. Mater. Res., Part A, 2017, 105, 94–103 CrossRef CAS PubMed.
- H. A. Strobel, E. L. Calamari, A. Beliveau, A. Jain and M. W. Rolle, J. Biomed. Mater. Res., Part B, 2017 DOI:10.1002/jbm.b.33871.
- D. Wendt, A. Marsano, M. Jakob, M. Heberer and I. Martin, Biotechnol. Bioeng., 2003, 84, 205–214 CrossRef CAS PubMed.
- A. L. Olivares and D. Lacroix, Tissue Eng., Part C, 2012, 18, 624–631 CrossRef CAS PubMed.
- K. Wyss, C. Y. Y. Yip, Z. Mirzaei, X. Jin, J.-H. Chen and C. A. Simmons, J. Biomech., 2012, 45, 882–887 CrossRef PubMed.
- X. Yang, D. A. Fullerton, X. Su, L. Ao, J. C. Cleveland, Jr. and X. Meng, J. Am. Coll. Cardiol., 2009, 53, 491–500 CrossRef CAS PubMed.
- A. K. Schroer and W. D. Merryman, J. Cell Sci., 2015, 128, 1865–1875 CrossRef CAS PubMed.
- B. Pilecki, A. T. Holm, A. Schlosser, J. B. Moeller, A. P. Wohl, A. V. Zuk, S. E. Heumuller, R. Wallis, S. K. Moestrup, G. Sengle, U. Holmskov and G. L. Sorensen, J. Biol. Chem., 2016, 291, 1103–1114 CrossRef CAS PubMed.
- K. Modjarrad and S. Ebnesajjad, Handbook of Polymer Applications in Medicine and Medical Devices, Elsevier Science, 2013 Search PubMed.
- M. Ramalingam, S. Ramakrishna and S. Best, Biomaterials and Stem Cells in Regenerative Medicine, Taylor & Francis, 2012 Search PubMed.
- Y. Xue, V. Sant, J. Phillippi and S. Sant, Acta Biomater., 2017, 48, 2–19 CrossRef CAS PubMed.
- J. H. Lee, J. Y. Oh and D. M. Kim, J. Mater. Sci. Mater. Med., 1999, 10, 629–634 CrossRef CAS PubMed.
- J. H. Lee, S. H. Oh and W. G. Kim, J. Mater. Sci. Mater. Med., 2004, 15, 155–159 CrossRef CAS PubMed.
- L. Liu, G. Chen, T. Chao, B. D. Ratner, E. H. Sage and S. Jiang, J. Biomater. Sci., Polym. Ed., 2008, 19, 821–835 CrossRef CAS PubMed.
- M. Tanaka and A. Mochizuki, J. Biomater. Sci., Polym. Ed., 2010, 21, 1849–1863 CrossRef CAS PubMed.
- H. Ghanbari, H. Viatge, A. G. Kidane, G. Burriesci, M. Tavakoli and A. M. Seifalian, Trends Biotechnol., 2009, 27, 359–367 CrossRef CAS PubMed.
- Y. Mei, K. Saha, S. R. Bogatyrev, J. Yang, A. L. Hook, Z. I. Kalcioglu, S. W. Cho, M. Mitalipova, N. Pyzocha, F. Rojas, K. J. Van Vliet, M. C. Davies, M. R. Alexander, R. Langer, R. Jaenisch and D. G. Anderson, Nat. Mater., 2010, 9, 768–778 CrossRef CAS PubMed.
- K. M. Mabry, R. L. Lawrence and K. S. Anseth, Biomaterials, 2015, 49, 47–56 CrossRef CAS PubMed.
- K. J. Rodriguez, L. M. Piechura, A. M. Porras and K. S. Masters, BMC cardiovasc. Disord., 2014, 14, 29 CrossRef PubMed.
- K. J. Rodriguez, L. M. Piechura and K. S. Masters, Matrix Biol., 2011, 30, 70–82 CrossRef CAS PubMed.
- K. J. Rodriguez and K. S. Masters, J. Biomed. Mater. Res., Part A, 2009, 90A, 1043–1053 CrossRef CAS PubMed.
- X. Gu and K. S. Masters, J. Biomed. Mater. Res., Part A, 2010, 93A, 1620–1630 CAS.
- X. Gu and K. S. Masters, Am. J. Physiol., 2011, 300, H448–H458 CrossRef CAS PubMed.
- A. J. Engler, H. L. Sweeney, D. E. Discher and J. E. Schwarzbauer, J. Musculoskeletal Neuronal Interact., 2007, 7, 335 CAS.
- G. C. Reilly and A. J. Engler, J. Biomech., 2010, 43, 55–62 CrossRef PubMed.
- T. Martín-Rojas, F. Gil-Dones, L. F. Lopez-Almodovar, L. R. Padial, F. Vivanco and M. G. Barderas, J. Proteome Res., 2012, 11, 1537–1550 CrossRef PubMed.
- C. K. Breuer, B. A. Mettler, T. Anthony, V. L. Sales, F. J. Schoen and J. E. Mayer, Tissue Eng., 2004, 10, 1725–1736 CrossRef CAS PubMed.
- M. Pesce and R. Santoro, in Regenerative Medicine - from Protocol to Patient: 3. Tissue Engineering, Biomaterials and Nanotechnology, ed. G. Steinhoff, Springer International Publishing, Cham, 2016, pp. 1–12, DOI:10.1007/978-3-319-28274-9_1.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7bm00854f |
| ‡ Equal contribution. |
| This journal is © The Royal Society of Chemistry 2018 |