Branched peptides for enzymatic supramolecular hydrogelation†
Hongjian
He
,
Huaimin
Wang
 ,
Ning
Zhou
,
Dongsik
Yang
and
Bing
Xu
,
Ning
Zhou
,
Dongsik
Yang
and
Bing
Xu
 *
*
Department of Chemistry, Brandeis University, 415 South St., Waltham, MA 02454, USA. E-mail: bxu@brandeis.edu
First published on 6th December 2017
Abstract
Here we show the use of branched peptides (i.e., isopeptides) as the substrates of proteases for generating supramolecular hydrogels via enzymatically clipping the branch off the peptides. As the first report of using a protease to cut branched peptides to enable a molecular process for converting nanostructures, this work illustrates a fundamentally new molecular motif for exploring the applications of protease-instructed self-assembly for soft materials.
Integrating catalytic reactions and molecular self-assembly for functions, especially in generating higher-order protein assemblies for functions,1 is a prevailing process in biology. Such a fundamental fact in biology, as a source of bioinspiration,2 has coincided with the employment of catalysis and self-assembly for developing molecular processes to generate materials and medicine.3,4 Because enzymes, as an important category of proteins, are the most efficient natural catalysts involved in biological signalling and metabolic processes, the use of enzymatic reactions to regulate molecular assemblies has received considerable attentions and made significant progresses. For example, inspired by the use of transglutaminase to crosslink polymers to form polymeric hydrogels,5 we used enzymatic dephosphorylation to generate supramolecular hydrogels,6 which has led to a versatile approach that integrates enzymatic reaction with molecular self-assembly. As a multiple step molecular process, enzyme-instructed self-assembly (EISA) has shown exciting promises in several areas, such as responsive gels for sensing,7,8 imaging enzyme activities,9–11 selective inhibition of cancer cells,12–15 cell purification after differentiating iPS cell,16 self-delivery drugs,17–19 and adjuvants for vaccines.20
Among the substrates used for EISA, the most extensively investigated ones are linear peptides.3,21 Branched peptides received less attention, though they have served as building blocks for polymeric hydrogels,5 capture agent of proteins, and peptide amphiphiles.22,23 Thus, we decide to explore enzymatic self-assembly involving the proteolysis of branched peptides, which are the substrates of enterokinase (ENTK). Being discovered by Pavlov,24 ENTK, as an enteropeptidase, specifically cleaves the peptide sequence DDDDK from proteins and has led to the development of FLAG-tag (DYKDDDK).25 Though being extensively used for proteins purification, the applications of FLAG-tag for generating soft biomaterials such as supramolecular hydrogels has yet to be explored. As shown by our results, connecting the FLAG-tag as a peptidic branch to a self-assembling motif26 affords the precursors that form micelles. Enzymatic cleavage of the hydrophilic FLAG branch by ENTK turns the micelles to nanofibers, which results in supramolecular hydrogels. As the first case of using a protease to cut branched peptides to enable a molecular process for generating supramolecular hydrogels, this work illustrates a fundamentally new molecular motif for exploring the applications of protease instructed self-assembly15,27–30 for soft materials, such as supramolecular hydrogels.3,30–33
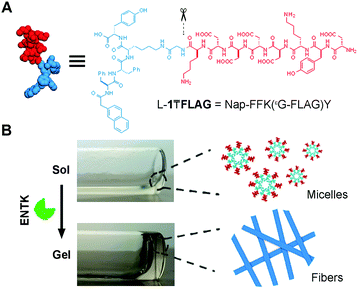
Fig. 1 shows the structure of the designed substrate of ENTK. In this work, we use the symbol “₸” to represent the branching so that the substrate is denoted as L-1₸FLAG. The branched hydrogelator precursors consist of (i) the FLAG-tag as the substrate of ENTK for enzymatic recognition and cleavage, (ii) a self-assembling peptide sequence L-1 (or D-1) composed of a 2-acetylnaphthyl group and a tetrapeptide (Phe–Phe–Lys–Tyr), and (iii) a glycine as the spacer for (i) and (ii). Using Fmoc-based solid-phase peptide synthesis,34 we first produce the peptide segments (i.e., Gly-FLAG and L-1 (or D-1), Scheme S1, ESI†). After synthesis and activating the C-terminus of Gly-FLAG, we connect it to the side chain of lysine residue in L-1 (or D-1). Finally, the removal of all protecting groups and the purification by high pressure liquid chromatography (HPLC) produce the designed branch peptides (i.e., L-1₸FLAG and D-1₸FLAG).
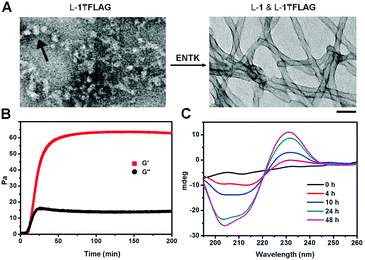
After obtaining the precursors, we examine enzymatic gelation catalysed by ENTK. L-1₸FLAG dissolves in PBS buffer at pH 7.4 to form a clear solution, which becomes a hydrogel upon the addition of ENTK (Fig. 1B). Transmission electron microscopy (TEM) reveals the solution of L-1₸FLAG (200 μM) containing nanoparticles of 21 ± 2 nm in diameter. Upon the addition of ENTK, the nanoparticles transform to the nanofibers of 24 ± 2 nm in diameter (Fig. 2A). This result indicates that L-1₸FLAG forms micelles in PBS buffer, ENTK generates L-1 from L-1₸FLAG, and the self-assembly of L-1 results in the nanofibers. Using dynamic light scattering (DLS), we measure the hydrodynamic diameter of the micelles formed by L-1₸FLAG (200 μM). According to DLS, the hydrodynamic diameter of the micelles mainly is around 47 nm (Fig. S1, ESI†), at the same magnitude of the diameters measured by TEM. Thus, both TEM and DLS results indicate that L-1₸FLAG (200 μM) self-assembles to form micelles in PBS buffer.
After the addition of ENTK to the solution of L-1₸FLAG (500 μM), the time-dependent storage moduli (G′) and loss moduli (G′′) exhibit a crossover of G′ and G′′ around 12 min, suggesting that cutting the branch (i.e., the FLAG-tag) off L-1₸FLAG quickly leads to the gelation point (Fig. 2B), even though significant amount of L-1₸FLAG remains. Circular dichroism (CD) spectra reveal that the enzymatic cleavage of L-1₸FLAG increases the intensity of the CD signal over time, indicating that more assemblies form with more enzymatic proteolysis (Fig. 2C and Fig. S4, ESI†). We measured the CD spectra of L-1, FLAG tag, and the mixture of L-1 and FLAG-tag (molar ratio 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), respectively. Interestingly, the CD of L-1 exhibits a β-sheet like curve with a negative peak around 220 nm, while the CD of FLAG tag shows a random-coil like cure with a negative peak at 195 nm (Fig. S8, ESI†). Thus, though the superposition of the signals of L-1 and FLAG-tag illusively implies α-helix conformation, hardly any α-helix structure exists in this mixture. This result agrees with that THT binds to the fibers formed by L-1 (β-sheet, Fig. 3C and D). Moreover, the absorbance of the naphthalene ring is around 235 nm, which unlikely would overlap with the signals from the peptide bonds.
1), respectively. Interestingly, the CD of L-1 exhibits a β-sheet like curve with a negative peak around 220 nm, while the CD of FLAG tag shows a random-coil like cure with a negative peak at 195 nm (Fig. S8, ESI†). Thus, though the superposition of the signals of L-1 and FLAG-tag illusively implies α-helix conformation, hardly any α-helix structure exists in this mixture. This result agrees with that THT binds to the fibers formed by L-1 (β-sheet, Fig. 3C and D). Moreover, the absorbance of the naphthalene ring is around 235 nm, which unlikely would overlap with the signals from the peptide bonds.
Reversing the chirality of the main chain (i.e., from L-1 to D-1) generates D-1₸FLAG, which, as a diastereomer of L-1₸FLAG, is able to act as the substrate of ENTK (Fig. S2, ESI†). Similar to the case of L-1₸FLAG, ENTK, within 24 h, turns the solution of D-1₸FLAG to a hydrogel (Fig. S3A, ESI†), which exhibits CD spectra that are almost mirror images to the CD of the hydrogel formed by L-1 (Fig. S5A, ESI†). This result confirms that D-1 (or L-1) as the major components to drive the formation of nanofibers and the hydrogelation. Moreover, upon the addition of ENTK to the solution of D-1₸FLAG (200 μM), TEM shows that the nanoparticles (15 ± 3 nm in diameter) in the solution become the nanofibers (17 ± 2 nm in diameter) (Fig. S3B, ESI†), and rheological measurement determines that G′ dominates G′′ at about 8 min (Fig. S5B, ESI†). We conducted frequency and strain sweeps on the gel formed by D-1₸FLAG (500 μM) with ENTK (10 U ml−1) after time-dependent G′ and G′′ measurement (2.5 h, Fig. S6, ESI†). Results show that the stress and frequency applied in the time-sweep experiments are located in the linear viscoelastic region of the gel, ensuring that the time-dependent G′ and G′′ are collected under appropriate conditions. Because the strength of supramolecular hydrogel depends on concentration, the hydrogels from 500 μM hydrogel precursors appear weak during rheology test. When the concentration increases to 2.5 wt% (about 13.7 mM) of the precursors, ENTK (10 U ml−1) catalysis generates a gel that is strong enough to maintain its shape (Fig. 1B and Fig. S3, ESI†). These results confirm that the chirality of the main chain hardly hampers the proteolytic cleavage of the FLAG-tag on the branches of the precursors.
To understand the rheological behaviour of the branched peptide during enzymatic reaction, we use TEM to monitor the morphology change of L-1₸FLAG (500 μM) solution after adding ENTK (10 U ml−1). At 500 μM (over 2.5 fold of the CMC, Fig. 3B) in PBS buffer, L-1₸FLAG forms amorphous aggregates (Fig. S7, ESI†), resulting in a network that may offer certain stiffness to the system and make the G′ to be almost same as G′′ even at time zero. The hydrolysis of L-1₸FLAG by ENTK generates L-1 that self-assembles to form nanofibers, providing more rigidity. Because G′ ≈ G′′ at time zero, a small fraction of L-1 generated from the hydrolysis of L-1₸FLAG would quickly result in G′ dominating G′′. Moreover, because of the co-assembly (discussed later) between the hydrogel precursors and hydrogelators, the amorphous aggregates mostly transformed to nanofibers (the main factor of gel rigidity) within 1 hour with little subsequent morphology change by enzymatic reaction. Thus, further hydrolysis hardly affect the mechanical property of the gels, explaining the quick plateaus of G′ and G′′.
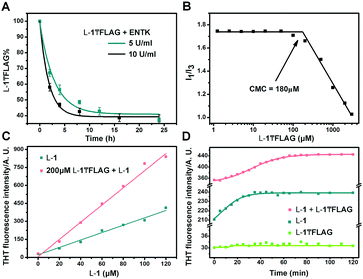
Finally, we use HPLC to investigate the proteolysis of L-1₸FLAG by ENTK at ambient condition. The amount of L-1₸FLAG decreases rapidly within 4 h and becomes constant around 40% after 8 h (Fig. 3A). The incomplete hydrolysis of L-1₸FLAG likely originates from the formation of nanofibers, which result in the hydrogel to slow down the diffusion of ENTK and its substrates. We determine the critical micelle concentration (CMC) of L-1₸FLAG using pyrene as the fluorescence probe.35 The I1/I3 ratio of pyrene on L-1₸FLAG sharply decreases at the L-1₸FLAG concentration of 180 μM, indicating the CMC (Fig. 3B). To better understand the intermolecular interaction, we utilize thioflavin T (THT), a known probe that fluoresces upon binding to amyloid fibers,36 to study the self-assembly of L-1 and L-1₸FLAG. Although the solution of L-1₸FLAG produces little fluorescence, the mixture of L-1₸FLAG and L-1 exhibits higher fluorescence than L-1 itself at the same concentration of L-1 (Fig. 3C), indicating that the mixture are able to generate more nanofibers. Meanwhile, the THT fluorescence change of the mixture (200 μM L-1₸FLAG + 60 μM L-1) takes a longer time (60 min) to reach plateau (Fig. 3D) than the time (30 min) needed by L-1 (60 μM) after dissolution. These results confirm the co-assembly of L-1₸FLAG and L-1, which likely results in the incomplete proteolysis of L-1₸FLAG catalysed by ENTK. In fact, the co-assembly and incomplete hydrolysis allow the assembled nanostructures to be dependent on the amount of enzymes.
In conclusion, we demonstrate the branched peptides (i.e., isopeptides) as a novel type of substrates for enzymatic self-assembly and hydrogelation. The branched peptides are able to form micelles in solution and transform to nanofibers in response to ENTK. Moreover, the branch peptides exhibit better gelation property by EISA than their linear analogue does (e.g., L-1-FLAG, Fig. S9, ESI†). This work lays the molecular foundation for exploring EISA of branched peptides in cellular milieu.37
Conflicts of interest
There are no conflicts to declare.Notes and references
- H. Wu and M. Fuxreiter, Cell, 2016, 165, 1055 CrossRef CAS PubMed.
- G. M. Whitesides, Interface Focus, 2015, 5, 20150031 CrossRef PubMed.
- X. Du, J. Zhou, J. Shi and B. Xu, Chem. Rev., 2015, 115, 13165 CrossRef CAS PubMed.
- Z. M. Yang, G. L. Liang and B. Xu, Soft Matter, 2007, 2, 515 RSC.
- B. Hu and P. B. Messersmith, J. Am. Chem. Soc., 2003, 125, 14298 CrossRef CAS PubMed.
- Z. Yang, H. Gu, D. Fu, P. Gao, J. K. Lam and B. Xu, Adv. Mater., 2004, 16, 1440 CrossRef CAS.
- M. Ikeda, R. Ochi, A. Wada and I. Hamachi, Chem. Sci., 2010, 1, 491 RSC.
- A. Wada, S.-i. Tamaru, M. Ikeda and I. Hamachi, J. Am. Chem. Soc., 2009, 131, 5321 CrossRef CAS PubMed.
- J. Zhou, X. Du, C. Berciu, H. He, J. Shi, D. Nicastro and B. Xu, Chem, 2016, 1, 246 CAS.
- D. Ye, A. J. Shuhendler, L. Cui, L. Tong, S. S. Tee, G. Tikhomirov, D. W. Felsher and J. Rao, Nat. Chem., 2014, 6, 519 CrossRef CAS PubMed.
- Z. Zheng, A. Tang, Y. Guan, L. Chen, F. Wang, P. Chen, W. Wang, Y. Luo, Y. Tian and G. Liang, Anal. Biochem., 2016, 88, 11982 CAS.
- Y. Kuang, J. Shi, J. Li, D. Yuan, K. A. Alberti, Q. Xu and B. Xu, Angew. Chem., Int. Ed., 2014, 53, 8104 CrossRef CAS PubMed.
- R. A. Pires, Y. M. Abul-Haija, D. S. Costa, R. Novoa-Carballal, R. L. Reis, R. V. Ulijn and I. Pashkuleva, J. Am. Chem. Soc., 2015, 137, 576 CrossRef CAS PubMed.
- X. Zhang, C. Li, Y. Wang, C. Ou, S. Ji, M. Chen and Z. Yang, RSC Adv., 2016, 6, 56903 RSC.
- A. Tanaka, Y. Fukuoka, Y. Morimoto, T. Honjo, D. Koda, M. Goto and T. Maruyama, J. Am. Chem. Soc., 2015, 137, 770 CrossRef CAS PubMed.
- Y. Kuang, K. Miki, C. J. Parr, K. Hayashi, I. Takei, J. Li, M. Iwasaki, M. Nakagawa, Y. Yoshida and H. Saito, Cell Chem. Biol., 2017, 24, 685 CrossRef CAS PubMed.
- Y. Gao, Y. Kuang, Z.-F. Guo, Z. Guo, I. J. Krauss and B. Xu, J. Am. Chem. Soc., 2009, 131, 13576 CrossRef CAS PubMed.
- J. Li, Y. Gao, Y. Kuang, J. Shi, X. Du, J. Zhou, H. Wang, Z. Yang and B. Xu, J. Am. Chem. Soc., 2013, 135, 9907 CrossRef CAS PubMed.
- Y. Wang, A. G. Cheetham, G. Angacian, H. Su, L. Xie and H. Cui, Adv. Drug Delivery Rev., 2017, 110, 112 CrossRef PubMed.
- Y. Tian, H. Wang, Y. Liu, L. Mao, W. Chen, Z. Zhu, W. Liu, W. Zheng, Y. Zhao and D. Kong, Nano Lett., 2014, 14, 1439 CrossRef CAS PubMed.
- Z. Yang, G. Liang and B. Xu, Acc. Chem. Res., 2008, 41, 315 CrossRef CAS PubMed.
- M. O. Guler, S. Soukasene, J. F. Hulvat and S. I. Stupp, Nano Lett., 2005, 5, 249 CrossRef CAS PubMed.
- L. L. Lock, C. D. Reyes, P. Zhang and H. Cui, J. Am. Chem. Soc., 2016, 138, 3533 CrossRef CAS PubMed.
- I. P. Pavlov and W. H. Thompson, The work of the digestive glands, Charles Griffin, 1902 Search PubMed.
- T. P. Hopp, K. S. Prickett, V. L. Price, R. T. Libby, C. J. March, D. P. Cerretti, D. L. Urdal and P. J. Conlon, Nat. Biotechnol., 1988, 6, 1204 CrossRef CAS.
- J. Zhou, X. Du, Y. Gao, J. Shi and B. Xu, J. Am. Chem. Soc., 2014, 136, 2970 CrossRef CAS PubMed.
- Z. Yang, M. Ma and B. Xu, Soft Matter, 2009, 5, 2546 CAS.
- S. Toledano, R. J. Williams, V. Jayawarna and R. V. Ulijn, J. Am. Chem. Soc., 2006, 128, 1070 CrossRef CAS PubMed.
- Y. Shi, Y. Hu, G. Ochbaum, R. Lin, R. Bitton, H. Cui and H. S. Azevedo, Chem. Commun., 2017, 53, 7037 RSC.
- S. C. Bremmer, A. J. McNeil and M. B. Soellner, Chem. Commun., 2014, 50, 1691 RSC.
- J. Aw, F. Widjaja, Y. Ding, J. Mu, Y. Liang and B. Xing, Chem. Commun., 2017, 53, 3330 RSC.
- X. Qin, W. Xie, S. Tian, J. Cai, H. Yuan, Z. Yu, G. L. Butterfoss, A. C. Khuong and R. A. Gross, Chem. Commun., 2013, 49, 4839 RSC.
- H. Zhang, C. Xu, J. Liu, X. Li, L. Guo and X. Li, Chem. Commun., 2015, 51, 7031 RSC.
- W. C. Chan and P. D. White, Fmoc solid phase peptide synthesis, Oxford University Press, 2000 Search PubMed.
- A. Domínguez, A. Fernández, N. González, E. Iglesias and L. Montenegro, J. Chem. Educ., 1997, 74, 1227 CrossRef.
- H. Naiki, K. Higuchi, M. Hosokawa and T. Takeda, Anal. Biochem., 1989, 177, 244 CrossRef CAS PubMed.
- H. Wang, Z. Feng and B. Xu, Chem. Soc. Rev., 2017, 46, 2421 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7cc08421h |
| This journal is © The Royal Society of Chemistry 2018 |