Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Evaluation of Cu(I) binding to the E2 domain of the amyloid precursor protein – a lesson in quantification of metal binding to proteins via ligand competition†
Tessa R.
Young
 a,
Anthony G.
Wedd
a and
Zhiguang
Xiao
a,
Anthony G.
Wedd
a and
Zhiguang
Xiao
 *ab
*ab
aSchool of Chemistry and Bio21 Molecular Science and Biotechnology Institute, The University of Melbourne, Parkville, Victoria 3010, Australia
bMelbourne Dementia Research Centre, Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Parkville, Victoria 3052, Australia. E-mail: zhiguang.xiao@florey.edu.au; Tel: +61 3 9035 6072
First published on 28th November 2017
Abstract
The extracellular domain E2 of the amyloid precursor protein (APP) features a His-rich metal-binding site (denoted as the M1 site). In conjunction with surrounding basic residues, the site participates in interactions with components of the extracellular matrix including heparins, a class of negatively charged polysaccharide molecules of varying length. This work studied the chemistry of Cu(I) binding to APP E2 with the probe ligands Bcs, Bca, Fz and Fs. APP E2 forms a stable Cu(I)-mediated ternary complex with each of these anionic ligands. The complex with Bca was selected for isolation and characterization and was demonstrated, by native ESI-MS analysis, to have the stoichiometry E2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Cu(I)
Cu(I)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Bca = 1
Bca = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Formation of these ternary complexes is specific for the APP E2 domain and requires Cu(I) coordination to the M1 site. Mutation of the M1 site was consistent with the His ligands being part of the E2 ligand set. It is likely that interactions between the negatively charged probe ligands and a positively charged patch on the surface of APP E2 are one aspect of the generation of the stable ternary complexes. Their formation prevented meaningful quantification of the affinity of Cu(I) binding to the M1 site with these probe ligands. However, the ternary complexes are disrupted by heparin, allowing reliable determination of a picomolar Cu(I) affinity for the E2/heparin complex with the Fz or Bca probe ligands. This is the first documented example of the formation of stable ternary complexes between a Cu(I) binding protein and a probe ligand. The ready disruption of the complexes by heparin identified clear ‘tell-tale’ signs for diagnosis of ternary complex formation and allowed a systematic review of conditions and criteria for reliable determination of affinities for metal binding via ligand competition. This study also provides new insights into a potential correlation of APP functions regulated by copper binding and heparin interaction.
1. Formation of these ternary complexes is specific for the APP E2 domain and requires Cu(I) coordination to the M1 site. Mutation of the M1 site was consistent with the His ligands being part of the E2 ligand set. It is likely that interactions between the negatively charged probe ligands and a positively charged patch on the surface of APP E2 are one aspect of the generation of the stable ternary complexes. Their formation prevented meaningful quantification of the affinity of Cu(I) binding to the M1 site with these probe ligands. However, the ternary complexes are disrupted by heparin, allowing reliable determination of a picomolar Cu(I) affinity for the E2/heparin complex with the Fz or Bca probe ligands. This is the first documented example of the formation of stable ternary complexes between a Cu(I) binding protein and a probe ligand. The ready disruption of the complexes by heparin identified clear ‘tell-tale’ signs for diagnosis of ternary complex formation and allowed a systematic review of conditions and criteria for reliable determination of affinities for metal binding via ligand competition. This study also provides new insights into a potential correlation of APP functions regulated by copper binding and heparin interaction.
Significance to metallomicsThe APP E2 protein domain forms stable Cu(I)-mediated ternary complexes with several commonly used competing ligands which, however, are disrupted upon addition of heparin, a physiological interacting partner of the APP. This provides new insights into the interactions of the APP with metals and heparin, which may both be involved in mediating the physiological functions of APP in the human brain. In addition, this study also allows validation of several simple control experiments which can be applied generally to confirm or disprove the presence of stable ternary complexes in metal-binding competition experiments. |
Introduction
The affinity for a metal ion M binding to a protein ligand P is defined by the dissociation constant KD (eqn (1)).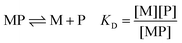 | (1) |
| MLn + P ⇌ MP + nL | (2) |
 | (3) |
A criticism of this approach is the possible formation of a ternary complex in which the metal ion M is coordinated simultaneously to both ligand L and protein P. Short-lived ternary complexes may facilitate transfer of the metal ion M between ligand L and protein P but, provided that they are transient, eqn (2) and (3) remain valid at equilibrium.
Copper is an essential but potentially toxic metal and must be bound tightly at all times.6–8 Mishandling in cells is associated with multiple disease states.9–11 Free Cu+ is unstable in aqueous solution and so quantification of Cu(I) binding to biomolecules is accomplished most reliably via the ligand competition of eqn (2) and (3). The four bidentate ligands ferrene S (Fs), ferrozine (Fz), bicinchoninic acid (Bca) and bathocuproine disulfonate (Bcs) (Fig. 1) have been employed widely for in vitro quantification of Cu(I) binding to biomolecules and been established as a set of complementary probes with versatile in vitro applications.1,4,12–15 They each bind Cu(I) to form a stable chromophoric 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 complex [CuIL2]3− with different log β2 values and distinct spectroscopic fingerprints (see Table 1) that allow quantification of Cu(I) affinities in the nanomolar to attomolar range.4,16 Stable ternary complexes with protein ligands have not been reported.
2 complex [CuIL2]3− with different log β2 values and distinct spectroscopic fingerprints (see Table 1) that allow quantification of Cu(I) affinities in the nanomolar to attomolar range.4,16 Stable ternary complexes with protein ligands have not been reported.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a
a
| Ligand (L) | λ max (nm) | ε max (cm−1 M−1) | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) β2 β2 |
|---|---|---|---|
| a Data quoted from ref. 4 and 16. b At low copper concentrations (<10 μM), the [CuI(Bca)2]3− complex was quantified by absorbance at 358 nm with reference to copper-free ligand solution. | |||
| Fs | 484 | 6700 | 13.7 |
| Fz | 470 | 4320 | 15.1 |
| Bca | 562 | 7900 | 17.2 |
| 358b | 42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900b 900b |
||
| Bcs | 483 | 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
19.8 |
We now report formation of stable ternary complexes between Cu(I), the E2 domain of the amyloid precursor protein (APP) and each of the above four ligands. These complexes are disrupted in the presence of heparin, known to interact with and bind to APP. This permitted reliable determination of a picomolar Cu(I) affinity for the E2/heparin complex, but not for the APP E2 domain itself in the absence of heparin. Consequently, this provided an opportunity to systematically review the general conditions and criteria necessary for reliable quantification of metal affinity via ligand competition.
APP is a type I transmembrane glycoprotein with a large extracellular domain (ectodomain) and a short amyloid intracellular domain (AICD) (Fig. 2a).17,18 The former includes two structured domains E1 (comprising subdomains D1 and D2) and E2 that are linked by a flexible domain (AcD) composed largely of aspartic and glutamic acid residues. A second flexible linker, the juxtamembrane region (JMR) connects the extracellular domain to the single transmembrane helix (TM). The ectodomain interacts with components of the extracellular matrix (ECM) while AICD interacts with many adaptor proteins in the cytosol.19,20 Protease processing of APP generates amyloid β peptides (Aβ) of 36–43 amino acids which are critically involved in Alzheimer's disease as the main components of the amyloid plaques found in the brains of Alzheimer patients.21
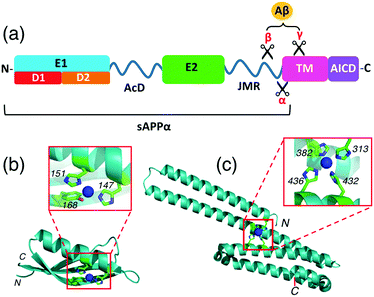 | ||
| Fig. 2 (a) Schematic representation of the structure and subdomains of APP-695; (b) crystal structure of APP D2 (pdb: 2FK2) with a close-up view of the metal-binding site with Cu(I) or Cu(II) coordinated; (c) crystal structure of APP E2 (pdb: 3UMK) with a close-up view of the metal-binding M1 site with Cu(II) coordinated. | ||
Interactions between copper ions and APP are a subject of current interest as they may play a role in normal synaptic function and in Alzheimer's disease pathogenesis, as reviewed recently.22,23 X-ray crystallographic studies have characterized a Cu(I)/Cu(II) binding site in the APP D2 domain and a Cu(II)-binding site in the APP E2 domain.24,25 The former site features the imidazole side-chains of two His ligands (H147, H151) with the nearby Tyr168 being claimed to be another possible metal ligand also (Fig. 2b).24 We have demonstrated that this site binds Cu(I) with ∼0.1 nM affinity and binds Cu(II) more weakly, by about 2 orders of magnitude.26 The Cu(II) site in APP E2 features four His ligands (H313, H382, H432, H436) and was denoted the M1 metal-binding site as it can also bind other divalent metal ions such as Zn(II) and Cd(II) (Fig. 2c).25 We hypothesized that the same site may be capable of coordinating Cu(I) as well. This could be of physiological significance, considering the dynamic redox environment in the synaptic cleft.27
APP is known to interact strongly with glycosaminoglycan (GAG) ligands including heparins and heparan sulfate proteoglycans (HSPGs).28–30 The highest-affinity heparin binding site is located within the E2 domain.31 Structural characterization of E2/heparin complexes for homologous amyloid precursor-like proteins 1 (APLP1) from human and from the worm Caenorhabditis elegans have mapped the heparin binding sites to a region involving the M1 site.32–35 A recent study showed that Cu(II) binding to the M1 site in APP E2 enhances the heparin binding affinity, as does Zn(II) to a lesser extent.36 Given the abundance of GAGs in the extracellular matrix, it is likely that a high proportion of cell-surface APP is heparin-bound.37 Consequently, the metal-binding properties of the E2/heparin complex are of interest.
Experimental section
Materials and general methods
Reagents including buffers (Mops, ammonium acetate), ligands (Fs, Fz, Bca, Bcs), metal salts (cupric sulfate and acetate), reductants (NH2OH, ascorbic acid (Asc)) and heparin (H3393, from porcine intestinal mucosa) were purchased from Sigma-Aldrich and were all used as received. Concentrations of heparin were estimated by mass, assuming an average molar mass of 19 kDa for the H3393 mixture.38 Cu(II) standards were prepared by dissolving cupric salts in Milli-Q water. Their concentrations were calibrated via reaction with excess ligand Bcs in Mops buffer containing reductant NH2OH. Under such conditions, all copper ions were converted quantitatively to the well-defined chromophoric complex anion [CuI(Bcs)2]3− (Table 1).39Construction of expression plasmids
Genes encoding the APP E2 domain (residues 295–498 of APP695) and a quadruple mutant of E2 (E2-qm; the four His residues of the M1 site were converted to Ala: H313, 382, 432, 436A) were synthesized by Bio Basic Inc. (Canada) with DNA genetic code optimized for expression in E. coli cells. These genes included an N-terminal sequence coding for residues ‘MN’ to facilitate recombinant protein expression and N-terminal acetylation in the E. coli host. The modified pET20b expression vector (see Table S1, ESI†) included a C-terminal (His)6-tag and a tobacco etch virus (TEV) protease cleavage site to facilitate protein purification and tag-cleavage, respectively. A pNatB expression vector, encoding subunits Naa20 and Naa25 of the fission yeast NatB acetylation complex40 was obtained from Addgene (plasmid ID: 53613).Protein expression and purification
Protein domains APP E2 and E2-qm were expressed with N-terminal acetylation to mimic the continuation of the polypeptide backbone in the complete APP. To this end, each E2 expression plasmid was transformed into E. coli BL21(DE3) cells that were co-transfected with an expression vector pNatB that encodes the two NatB acetylation complex subunits (Naa20 and Naa25). This allows endogenous N-terminal acetylation of a target protein during expression.40 Transformed cells were grown in 2YT medium containing ampicillin (100 mg L−1) and chloramphenicol (34 mg L−1) in shaking flasks at 37 °C until growth media reached an OD ∼ 1. Protein expression was induced by addition of IPTG (0.4 mM) with overnight agitation at room temperature and cells were harvested by low-speed centrifugation at 4 °C. The clarified cell lysate was prepared in buffer A (20 mM sodium phosphate, pH 7.4, 500 mM NaCl, 5 mM PMSF) and loaded directly onto a pre-equilibrated IMAC (Ni-NTA) resin. His-tagged proteins were eluted in buffer A with imidazole (250 mM), incubated with TEV protease (in approx 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 ratio) for 24–48 hours at 4 °C, and buffer-exchanged (to 20 mM imidazole in buffer A) by dialysis. The solution was reapplied to a Ni-NTA resin for removal of uncleaved protein and (His-tagged) TEV protease. The fully-cleaved protein was incubated with EDTA (≥2 × excess of protein concentration) to remove any contaminating metal ions, before final purification by size-exclusion chromatography on a Superdex-75 gel filtration column (HR 10/300, Pharmacia) in Mops buffer (20 mM, pH 7.4, 150 mM NaCl). Purified fractions were pooled, concentrated using centrifugal filters and stored at −80 °C.
100 ratio) for 24–48 hours at 4 °C, and buffer-exchanged (to 20 mM imidazole in buffer A) by dialysis. The solution was reapplied to a Ni-NTA resin for removal of uncleaved protein and (His-tagged) TEV protease. The fully-cleaved protein was incubated with EDTA (≥2 × excess of protein concentration) to remove any contaminating metal ions, before final purification by size-exclusion chromatography on a Superdex-75 gel filtration column (HR 10/300, Pharmacia) in Mops buffer (20 mM, pH 7.4, 150 mM NaCl). Purified fractions were pooled, concentrated using centrifugal filters and stored at −80 °C.
Two control proteins, APP D2 domain (encompassing APP695 residues 133–189; see Fig. 2a and b) and human copper chaperone Atox1 were also expressed and purified from Pichia pastoris and E. coli hosts, respectively, as previously reported.4,41 Protein concentrations were estimated based on solution absorbance at 280 nm using molar extinction coefficients calculated from their respective amino acid compositions: ε(280) = 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 930 cm−1 M−1 for APP E2 (after TEV-cleavage); 7365 cm−1 M−1 for APP D2; 2980 cm−1 M−1 for Atox1.
930 cm−1 M−1 for APP E2 (after TEV-cleavage); 7365 cm−1 M−1 for APP D2; 2980 cm−1 M−1 for Atox1.
Spectroscopic methods
UV-visible spectra were recorded on a Varian Cary 300 spectrophotometer in dual beam mode with quartz cuvettes of 1.0 cm path length. All titrations with metal ions were performed in appropriate buffers and corrected for baseline. Circular dichroism (CD) spectra were recorded on a Chirascan-plus spectrometer (Applied Photophysics) using a quartz cell (0.1 cm). Spectra were averaged from three scans.Quantification of Cu(I) binding
Quantification of Cu(I) binding was conducted by competition with probe ligand L based on eqn (4) and (5) with eqn (6) derived from them via the mass balance for each competing component (charges omitted in eqn (4)–(6) for clarity). Eqn (6) was employed for curve-fitting of the experimental data to derive the conditional dissociation constant KD of CuIP. The fitting was based on the known total concentrations of [Cu(I)]tot, [L]tot and [P]tot and the experimental equilibrium concentration of [CuIL2]3− that was determined under each condition from the characteristic solution absorbance of the complex (Table 1).16| CuIL2+ P ⇌ CuIP + 2L | (4) |
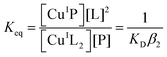 | (5) |
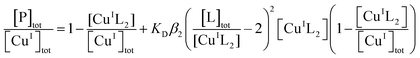 | (6) |
The spread of the β2 values for different probe complexes [CuIL2]3− (Table 1) mean that, by varying the identity and relative concentration of the selected probe ligand, the free Cu+ concentration (expressed as pCu+ = −log[Cuaq+]) can be buffered anywhere in the range −9 < pCu+ < −19. Consequently, reaction 4 may be tuned to be non-competitive for estimation of the stoichiometry of Cu(I) binding to the protein or to be competitive for determination of KD.1,16 In the former case, the target metal site in protein P removes Cu(I) quantitatively from [CuIL2]3−. In the latter case, the target metal site in protein P competes for Cu(I) effectively with metal fraction being controlled within a range of 20–80% of total Cu(I) content for sensitive and reliable estimation.
The experiments were conducted in deoxygenated buffers containing reductants NH2OH and/or Asc in an anaerobic glovebox ([O2] < 1 ppm) according to reported protocols.4,16 Briefly, a set of solutions were prepared that contained identical total concentrations of Cu(I) (produced in situ from reduction of CuSO4 by excess NH2OH/Asc) and a probe ligand L, but varying total concentrations of the target protein P. The equilibrium concentrations of the probe complex [CuIL2]3− were determined for each set of solutions from their stable spectra and were fitted to eqn (6) to derive an average KD for the CuIP complex.
Ternary complex isolation
Samples of reaction mixtures (∼1.0 mL) were applied to a desalting gel filtration column (Econo-Pac 10DG packed with P6-DG gel, Bio-Rad) equilibrated in Mops buffer (pH 7.4, 100 mM NaCl). Continued addition of buffer induced gravity flow through the resin. Elution fractions (1.0 mL) were collected and the UV-visible absorbance spectrum of each was recorded.Mass spectrometry
Intact protein masses (for denatured proteins) were acquired on an Agilent 6220 ESI-TOF LC/MS mass spectrometer coupled to an Agilent 1200 LC system (Agilent, Palo Alto, CA) and reference mass corrected via a dual-spray electrospray ionisation (ESI) source. Acquisition was performed using the Agilent Mass Hunter Acquisition software version B.02.01 (B2116.30) and analysed using Mass Hunter version B.05.00 (B5.0.519.0). Mass spectrometer conditions: ionisation mode: electrospray ionisation; drying gas flow: 7 L min−1; nebuliser: 35 psi; drying gas temperature: 325 °C; capillary voltage (Vcap): 4000 V; fragmentor: 250 V; skimmer: 65 V; OCT RFV: 250 V; scan range acquired: 100–3200 m/z; internal reference ions: Positive ion mode: m/z = 121.050873 and 922.009798. Protein samples were desalted prior to MS analysis using a reverse phase Aeris 3.6 μm WIDEPORE XB-C18 150 × 2.1 mm column (Phenomenex) and a linear acetonitrile elution gradient (5–95% v/v; 0.1% formic acid) over 10 min at 0.2 mL min−1.Native MS spectra were acquired on a Synapt HDMS system (Waters, Manchester, U.K.), using nanoESI in the positive ion mode. Instrument settings: capillary voltage, 1.7 kV; cone voltage, 60 V; trap collision energy, 40 V; source temperature, 50 °C. Prior to analysis, protein samples were buffer-exchanged into 10 mM ammonium acetate (pH 7.4) using centrifugal filters.
Results
Expression, purification and characterization of APP E2 domains
The APP E2 domain and a quadruple variant E2-qm (in which the four His residues of the M1 site were mutated to Ala; see Fig. 2a and c) were expressed and isolated from E. coli cells with N-terminal acetylation (see Experimental section). Protein purity was confirmed by SDS-PAGE (Fig. S1, ESI†) and identity by ESI-MS (Table S2, ESI†). The proteins emerged with the same elution volume from an analytical size exclusion column at a position consistent with the monomeric form (Fig. S2a, ESI†). Circular dichroism (CD) experiments were consistent with the α-helical structure persisting in solution (Fig. 2c and Fig. S2b, ESI†).Samples of APP D2 (Fig. 2b) and the copper metallo-chaperone Atox1 were expressed and isolated as reported.4,26 They were used as controls for Cu(I) binding experiments of APP E2.
APP E2 distorts the solution spectra of [CuIL2]3−
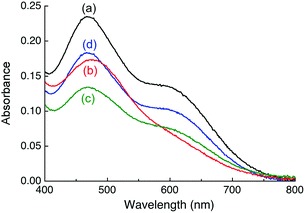
Titration of APP E2 into a solution of [CuI(Fz)2]3− in Mops buffer at pH 7.4 led to changes in the absorption envelope (Fig. 3bversusFig. 3a). Similar changes were detected for equivalent experiments with [CuI(Bca)2]3−, [CuI(Fs)2]3− and [CuI(Bcs)2]3− (Fig. 4a and Fig. S3a, b, S4a, b, ESI†), suggesting formation of a Cu(I)-mediated ternary complex E2–Cu(I)–L in each case (such as eqn (7); charges omitted for clarity):| CuIL2 + E2 → E2–Cu(I)–L + L | (7) |
Notably, upon titration with APP E2, the positions of absorbance maxima for [CuI(Bca)2]3− and [CuI(Bcs)2]3− in the visible region appeared to remain unchanged, but two tight isosbestic points at 405 and 455 nm for the former and one at 385 nm for the latter were detected (Fig. 4a and Fig. S4, ESI†), suggesting a clean reaction (such as eqn (7)) in these two cases.
Intriguingly, upon addition of one equiv. of heparin (relative to APP E2) into each of the above solutions, the solution spectra were restored to the fingerprints for the complex anions [CuIL2]3− (Fig. 3c, 4b and Fig. S3c, S4c, ESI†). For the Bcs system, even the original spectral intensity in the absence of APP-E2 was restored (Fig. S4c, ESI†). This suggests that heparin may have modified APP E2 to allow competition for Cu(I) binding with each of the four probe ligands according to eqn (4)–(6) and that the affinity of Bcs for Cu(I) is too strong to allow APP E2/heparin to compete.
Heparin is a physiological partner of APP and known to interact with and bind to the E2 domain specifically.32–35 Addition of heparin into each of the [CuIL2]3− probe solutions caused no change to the solution spectra (Fig. 3a, 4 and Fig. S3a, S4a, ESI†), indicating that heparin itself cannot extract Cu(I) from these probe complexes, nor interact with these probes via a Cu(I)-bridge.
Further controls were conducted with the APP D2 domain (that has been referred to as the ‘copper binding domain’ CuBD; Fig. 2b). Its Cu(I) binding affinity has been determined to be log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD = −10.1 at pH 7.0.26 As observed previously, addition of APP D2 into solutions containing complex [CuI(Fz)2]3− or [CuI(Fs)2]3− decreased the spectral intensities but did not distort the spectral envelope (see Fig. 3d and Fig. S3d, ESI†), consistent with the competition reaction according to eqn (4). Addition of heparin into both systems induced no further change to their spectra (Fig. 3d and Fig. S3d, ESI†). It is apparent that heparin does not alter the affinity of Cu(I) binding to the APP D2 domain.
KD = −10.1 at pH 7.0.26 As observed previously, addition of APP D2 into solutions containing complex [CuI(Fz)2]3− or [CuI(Fs)2]3− decreased the spectral intensities but did not distort the spectral envelope (see Fig. 3d and Fig. S3d, ESI†), consistent with the competition reaction according to eqn (4). Addition of heparin into both systems induced no further change to their spectra (Fig. 3d and Fig. S3d, ESI†). It is apparent that heparin does not alter the affinity of Cu(I) binding to the APP D2 domain.
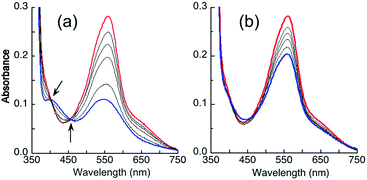
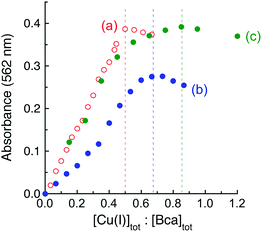
It was noted that the absorbance maxima of both [CuI(Bca)2]3− and [CuI(Bcs)2]3− in the visible region are indistinguishable from those of their putative ternary complexes E2–Cu(I)–L (L = Bca or Bcs) (see Fig. 4 and Fig. S4, ESI†). This may give a misleading impression of no ternary complex formation. An additional experiment is necessary to test for ternary complex formation. Fig. 5 for L = Bca shows a titration of Cu(I) into a solution containing both APP E2 and ligand L with a control titration of the ligand L only. Titration of Bca (100 μM) with Cu(I) generated a titration curve with a sharp turning point, as expected, at [Cu]tot![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [Bca]tot = 0.5
[Bca]tot = 0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Fig. 5a). An equivalent titration in the presence of APP E2 (30 μM) generated an apparent endpoint at [Cu]tot
1 (Fig. 5a). An equivalent titration in the presence of APP E2 (30 μM) generated an apparent endpoint at [Cu]tot![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [Bca]tot ∼ 0.67
[Bca]tot ∼ 0.67![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 but with an absorbance intensity at 562 nm of only ∼70% that of the control (compare Fig. 5bversusFig. 5a). In contrast, the same experiments with further addition of one equiv. of heparin relative to APP E2 led to a titration curve with an apparent endpoint at [Cu]tot
1 but with an absorbance intensity at 562 nm of only ∼70% that of the control (compare Fig. 5bversusFig. 5a). In contrast, the same experiments with further addition of one equiv. of heparin relative to APP E2 led to a titration curve with an apparent endpoint at [Cu]tot![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [Bca]tot ∼ 0.85
[Bca]tot ∼ 0.85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and a full absorbance intensity at 562 nm of that of the control (Fig. 5cversusFig. 5a). These experiments indicated that, at each Cu(I) saturation titration point, quantitative formation of the probe complex [CuI(Bca)2]3− was prevented in the absence of heparin but occurred in the presence of heparin. The poorly-defined titration endpoint in Fig. 5c indicated that the APP E2/heparin complex competes weakly with ligand Bca for Cu(I). This is also the case for the system of Bcs (Fig. S4, ESI†).
1 and a full absorbance intensity at 562 nm of that of the control (Fig. 5cversusFig. 5a). These experiments indicated that, at each Cu(I) saturation titration point, quantitative formation of the probe complex [CuI(Bca)2]3− was prevented in the absence of heparin but occurred in the presence of heparin. The poorly-defined titration endpoint in Fig. 5c indicated that the APP E2/heparin complex competes weakly with ligand Bca for Cu(I). This is also the case for the system of Bcs (Fig. S4, ESI†).
These experiments demonstrate that: (i) APP E2 forms a stable ternary complex with each of the four probe ligands, mediated by Cu(I) viaeqn (7); (ii) heparin disrupts the ternary complexes, enabling an effective competition for Cu(I) between APP E2 and each ligand L according to eqn (4); (iii) heparin itself has no detectable affinity for Cu(I). These conclusions were consolidated by further experiments.
Formation of stable ternary complexes invalidates quantification of metal binding affinity
Quantification of Cu(I) binding to APP E2 was hampered by the formation of a stable ternary complex with each of the four probe ligands, but the difficulty was lifted upon addition of heparin that disrupts these ternary complexes. The low affinity probe ligand Fs competes weakly for Cu(I) with the E2/heparin complex‡ and was employed to determine the Cu(I) binding stoichiometry of the complex under non-competitive conditions. Titration of the E2/heparin complex into a solution of [CuI(Fs)2]3− ([Cu]tot, 30 μM; [Fs]tot, 70 μM) led to an initial linear decrease of the [CuI(Fs)2]3− concentration with an extrapolated intercept at [E2]tot![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [Cu]tot = 0.33 (Fig. 6a(i)), consistent with a binding stoichiometry of Cu(I)
[Cu]tot = 0.33 (Fig. 6a(i)), consistent with a binding stoichiometry of Cu(I)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) E2 = 3
E2 = 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 under these conditions. Control experiments with variant APP E2-qm provided [E2]tot
1 under these conditions. Control experiments with variant APP E2-qm provided [E2]tot![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [Cu]tot = 0.50 under the same conditions (Fig. 6a(ii)). Consequently, it is concluded that a single equiv. of Cu(I) binds at the M1 site that features four His ligands (Fig. 2c).
[Cu]tot = 0.50 under the same conditions (Fig. 6a(ii)). Consequently, it is concluded that a single equiv. of Cu(I) binds at the M1 site that features four His ligands (Fig. 2c).
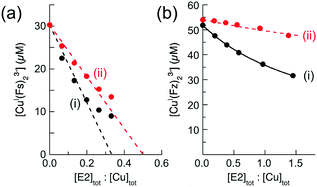 | ||
Fig. 6 Characterization of Cu(I) binding to APP E2 in the presence of heparin. (a) Quantification of Cu(I) binding stoichiometries of APP E2 (i) and its mutant APP E2-qm (ii) under non-competitive condition with probe [CuI(Fs)2]3− (composition: [Cu]tot = 30 μM, [Fs]tot = 70 μM). (b) Quantification of Cu(I) binding affinity with probe [CuI(Fz)2]3− (composition: [Cu]tot = 50 μM, [Fz]tot = 300 μM) under competitive conditions for APP E2 (ii) with non-competitive control of APP E2-qm (i). The best curve fitting of the experimental data set (i) to eqn (6) (shown in solid trace) provided a log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD = −11.9 for the Cu(I) binding affinity of the M1 site in APP E2. All experiments were conducted in Mops buffer (50 mM, pH 7.4) with added reductants (1.0 mM NH2OH and 0.5 mM Asc). KD = −11.9 for the Cu(I) binding affinity of the M1 site in APP E2. All experiments were conducted in Mops buffer (50 mM, pH 7.4) with added reductants (1.0 mM NH2OH and 0.5 mM Asc). | ||
Systematic testing showed that the APP E2/heparin complex competed for Cu(I) effectively with ligand Fz according to eqn (4) but that the complex with APP E2-qm competed only weakly under the same conditions (Fig. 6b(i)vs.Fig. 6b(ii)). This is consistent with the highest affinity being associated with the M1 site. The experimental data in Fig. 6b(i) fitted satisfactorily to eqn (6) to yield log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD = −11.9 for the M1 site at pH 7.4.
KD = −11.9 for the M1 site at pH 7.4.
Ligand Bca competed more strongly for Cu(I) with the APP E2/heparin complex and equivalent experiments with Bca estimated log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD = −12.2 for the M1 site (Fig. 7a(i)). New data acquired from a 1
KD = −12.2 for the M1 site (Fig. 7a(i)). New data acquired from a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 dilution of each analytical solution (Fig. 7a(ii)) provided an identical estimate of the log
1 dilution of each analytical solution (Fig. 7a(ii)) provided an identical estimate of the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD value. Note that the dilution did not simply halve the absorbance values (solid black versus dotted red dashed traces) but induced a lower concentration of [CuI(Bca)2]3− and a higher concentration of [Cu(I)–E2] to maintain the relationship of eqn (5) (as required by Le Chatelier's principle). In fact, the derived Cu(I) affinity for the E2/heparin complex was not altered within the experimental error (log
KD value. Note that the dilution did not simply halve the absorbance values (solid black versus dotted red dashed traces) but induced a lower concentration of [CuI(Bca)2]3− and a higher concentration of [Cu(I)–E2] to maintain the relationship of eqn (5) (as required by Le Chatelier's principle). In fact, the derived Cu(I) affinity for the E2/heparin complex was not altered within the experimental error (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD = −12.1 ± 0.2, see Table 2) upon systematic variation of the concentration of each competing component and/or upon swapping competing ligands with close affinities (i.e., Fz for Bca).
KD = −12.1 ± 0.2, see Table 2) upon systematic variation of the concentration of each competing component and/or upon swapping competing ligands with close affinities (i.e., Fz for Bca).
 | ||
Fig. 7 Quantification of Cu(I) binding affinities of APP E2 with probe [CuI(Bca)2]3− (compositions: [Cu]tot = 36 μM, [Bca]tot = 82 μM) in the presence (a) or absence (b) of heparin H3393 (1.0 equiv. relative to E2). The experimental data set (ii) in empty circles were obtained by a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 dilution of each solution for the data set (i). The best curve fittings of the experimental data sets (i and ii) to eqn (6) (shown in two solid traces) derived a consistent log 1 dilution of each solution for the data set (i). The best curve fittings of the experimental data sets (i and ii) to eqn (6) (shown in two solid traces) derived a consistent log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD = −12.2 for the highest Cu(I) binding affinity for E2/heparin complex in (a), but inconsistent log KD = −12.2 for the highest Cu(I) binding affinity for E2/heparin complex in (a), but inconsistent log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD (=−13.4 and −13.1, respectively) for APP E2 domain in (b). The dashed traces in (ii) are the simple 1 KD (=−13.4 and −13.1, respectively) for APP E2 domain in (b). The dashed traces in (ii) are the simple 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 dilution curve of data sets (i). All experiments were conducted in Mops buffer (50 mM, pH 7.4) with added reductant (1.0 mM NH2OH). 1 dilution curve of data sets (i). All experiments were conducted in Mops buffer (50 mM, pH 7.4) with added reductant (1.0 mM NH2OH). | ||
| Ligand (L) | Components for Cu(I) competition (μM) | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD for CuI–E2a KD for CuI–E2a |
|||
|---|---|---|---|---|---|
| [Cu]tot | [L]tot | [E2]tot | − heparin | + heparin | |
| a Determined in the absence and presence of one equivalent of heparin (relative to APP E2) and calculated viaeqn (6). b The protein affinity is too weak to be defined under the given experimental conditions. | |||||
| Bcs | 40 | 100 | 50 | −14.8 | n/ab |
| 150 | −14.5 | n/ab | |||
| Bca | 36 | 82 | 10 | −13.0 | −12.2 |
| 50 | −13.4 | −12.1 | |||
| 18 | 41 | 5 | −12.8 | −12.2 | |
| 25 | −13.1 | −12.2 | |||
| 7.2 | 16.6 | 4 | −11.5 | −11.9 | |
| 12 | −11.9 | −12.0 | |||
| Fz | 50 | 300 | 20 | −11.7 | −12.0 |
| 42.5 | −11.0 | −11.9 | |||
This confirms that the His-rich M1 site in E2/heparin provides a single Cu(I) binding site of picomolar affinity. A structural study of metal ion binding to the M1 site (via soaking of apo-E2 crystals) documented CuII(His)4 (Fig. 2c) and ZnII(His)3 sites.25 A further mutation study demonstrated that stable binding of Cu(II) by the M1 site requires coordination by all four His ligands, but only a sub-set of 2–3 for Zn(II).36 As heparin itself shows no detectable affinity for Cu(I), it is unlikely that the glycosaminoglycan is directly involved in the Cu(I) binding.
However, in the absence of heparin, equivalent experiments and data analysis generated misleading log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD values. These were scattered over four orders of magnitude when the concentrations of the species in eqn (4) were varied and/or the competing ligand was swapped (Table 2). The scattering is symptomatic of the formation of ternary complexes, according to eqn (7). This disobeys the assumptions of eqn (4) and invalidates the estimations. Note that a 1
KD values. These were scattered over four orders of magnitude when the concentrations of the species in eqn (4) were varied and/or the competing ligand was swapped (Table 2). The scattering is symptomatic of the formation of ternary complexes, according to eqn (7). This disobeys the assumptions of eqn (4) and invalidates the estimations. Note that a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 dilution of the test solutions of Fig. 7b(i) simply halved the absorbance of each solution without shifting the binding equilibrium, as required by eqn (4) (compare behaviour of Fig. 7a(i and ii)). These observations are consistent with the presence of ternary complexes: note that the equilibrium of eqn (7) is unaffected by dilution.
1 dilution of the test solutions of Fig. 7b(i) simply halved the absorbance of each solution without shifting the binding equilibrium, as required by eqn (4) (compare behaviour of Fig. 7a(i and ii)). These observations are consistent with the presence of ternary complexes: note that the equilibrium of eqn (7) is unaffected by dilution.
The above experiments demonstrate that: (i) an effective ligand competition between a protein P and a ligand L for a metal ion M described by eqn (4) must satisfy eqn (6) with consistent estimates of the constants KDβ2 upon variation of the concentrations of each competing component and/or upon change of the competing ligands of overlapping affinities; (ii) dilution of a set of competing solutions provides a simple diagnostic evaluation of whether the equilibrium of eqn (4) holds for the system; (iii) in the presence of heparin, the M1 site in APP E2 possesses picomolar affinity for Cu(I).
Isolation and characterization of the stable E2–Cu(I)–Bca ternary complex
Formation of the ternary complex(es) involving ligand Bca was investigated systematically. A series of solutions were prepared in Mops buffer (50 mM; pH 7.4; NaCl, 100 mM) containing ligand Bca (80 μM) plus APP E2 or a control protein (APP E2-qm, APP D2 or Atox1; each 30 μM). Cu(I) solution (CuIISO4, 30 μM, NH2OH, 1.0 mM) and/or heparin (30 μM) were added selectively. The details are listed in Table 3.| Expt | [Cu(II)] (μM) | [Bca] (μM) | [NH2OH] (mM) | [heparin] (μM) | Protein (30 μM) | Ternary complexb |
|---|---|---|---|---|---|---|
| a Conducted in Mops buffer (50 mM, pH 7.4, 100 mM NaCl). b Detected by co-elution of respective protein and ligand Bca from a desalting gel filtration column. | ||||||
| 1 | — | 80 | 1.0 | — | E2 | No |
| 2 | 30 | 80 | — | — | E2 | No |
| 3 | 30 | 80 | 1.0 | — | E2 | Yes |
| 4 | 30 | 80 | 1.0 | 30 | E2 | No |
| 5 | 30 | 80 | 1.0 | — | E2-qm | No |
| 6 | 30 | 80 | 1.0 | — | D2 | No |
| 7 | 30 | 80 | 1.0 | — | Atox1 | No |
The possibility of stable complex formation in each case was tested by separation of the reaction mixture on a simple desalting column (see inset in Fig. 8). Fraction I contained protein and/or protein complex(es) with molar masses >6 kDa whereas fraction II contained components of small molecules with molar masses <6 kDa only. Each fraction was characterized by comparing its solution spectrum against controls of each added purified protein, the ligand Bca and a solution containing [CuI(Bca)2]3− anions (Fig. 8 and Fig. S5, S6, ESI†). The results demonstrated: (i) ligand Bca alone cannot form a detectable complex with APP E2, nor in the presence of Cu(II) (Fig. 8a and Table 3, expts 1 and 2); (ii) in the presence of Cu(I), ligand Bca formed a stable (ternary) complex with APP E2 that can be isolated (Fig. 8c and d; expt 3), but that was disrupted by heparin (Fig. 8a and b; expt 4); (iii) no Bca-protein complex was detected with control proteins APP E2-qm (disabled M1 site), APP D2 or Atox1 under the same conditions as (ii) (Fig. 8a and b; expts 5–7). The results suggest that the E2-Bca complex detected and isolated in expt 3 is specific for the APP E2 protein and is mediated by Cu(I) binding to the M1 site in the protein. We have demonstrated that all three control proteins bind Cu(I) with different affinities: log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD ∼ −10 for both APP E2-qm and APP D2 (see Fig. 6 and ref. 26) and log
KD ∼ −10 for both APP E2-qm and APP D2 (see Fig. 6 and ref. 26) and log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD = −17.4 for Atox1 (see ref. 4).
KD = −17.4 for Atox1 (see ref. 4).
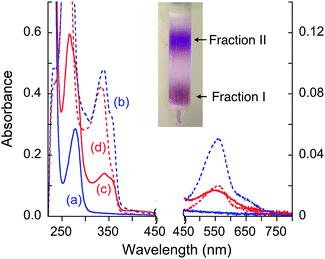 | ||
| Fig. 8 Solution spectra of protein fraction I (in solid line) and non-protein fraction II (in dashed line) separated by a desalting gel-filtration column (see inset) from the expts listed in Table 3: (a and b) fraction I (a) from expts 1, 2, 4–7 and fraction II (b) from expts 4–6; (c and d) fraction I (c) and fraction II (d) from expt 3. Absorbance intensity at λ < 450 nm and λ > 450 nm is shown on the left and right vertical axes, respectively. The spectrum (a) is indistinguishable from the spectrum of each purified protein sample alone. | ||
The isolated E2–Cu(I)–Bca complex was chromophoric and characterized by a full solution spectrum shown in Fig. 8c. Fingerprints for the presence of Cu(I), Bca and protein are seen by absorbance at 560, 335 and 280 nm, respectively, although that at 280 nm for APP E2 is masked by the intense absorbance of the Bca ligand (Fig. S4, ESI†). On the other hand, the absorbance intensity at 560 nm for non-protein fraction II from expt 3 was considerably weaker than those of the equivalent fraction from expt 4 (compare Fig. 8dversusFig. 8b), consistent with the observation that Cu(I) was trapped by APP E2 as a ternary complex in expt 3 only. All these observations support the above model of formation of a Cu(I)-mediated ternary complex.
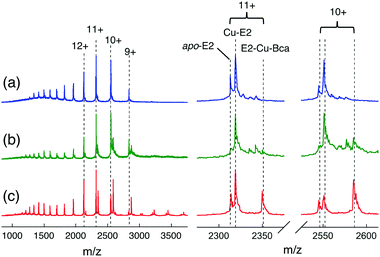
Further support and characterization are provided by native ESI-MS analysis (Fig. 9 and Table S3, ESI†). Addition of one equiv. of CuIISO4 into apo-E2 in volatile ammonium acetate buffer (10 mM, pH ∼ 7) allowed detection of both a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 Cu–E2 complex and apo-E2 by native ESI-MS (Fig. 9a and Table S3, ESI†). Addition of Bca (2 equiv.) led to detection of new protein components at low relative abundances (Fig. 9b). Further addition of 5 equiv. of reductant ascorbate (Asc) into the latter solution increased dramatically the relative abundance of one peak in each of the charge states (Fig. 9c). The deconvoluted masses corresponding to those peaks was 25
1 Cu–E2 complex and apo-E2 by native ESI-MS (Fig. 9a and Table S3, ESI†). Addition of Bca (2 equiv.) led to detection of new protein components at low relative abundances (Fig. 9b). Further addition of 5 equiv. of reductant ascorbate (Asc) into the latter solution increased dramatically the relative abundance of one peak in each of the charge states (Fig. 9c). The deconvoluted masses corresponding to those peaks was 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 833 (±3) Da, consistent with the theoretical mass of 25
833 (±3) Da, consistent with the theoretical mass of 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 834 Da for the ternary complex E2
834 Da for the ternary complex E2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Cu(I)
Cu(I)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Bca = 1
Bca = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. These experiments identify the stoichiometry of the stable ternary complex as E2–Cu(I)–Bca and confirm the existence of eqn (7).
1. These experiments identify the stoichiometry of the stable ternary complex as E2–Cu(I)–Bca and confirm the existence of eqn (7).
Discussion
Nature of the APP E2 ternary complexes and their biological implications
This work demonstrated that Cu(I) mediates formation of stable ternary complexes between the APP E2 domain and a set of bidentate ligands (Bcs, Bca, Fz and Fs; Fig. 1). A systematic study of the representative ligand Bca showed that: (i) formation of such ternary complexes is specific for the APP E2 domain and is dependent on Cu(I), but not Cu(II), binding to the M1 site in E2; (ii) the composition of the ternary complex with ligand Bca is E2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Cu(I)
Cu(I)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Bca = 1
Bca = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 as, presumably, is the case for the other ligands; (iii) the ternary complexes are disrupted readily by heparin, a partner that interacts physiologically with APP.
1 as, presumably, is the case for the other ligands; (iii) the ternary complexes are disrupted readily by heparin, a partner that interacts physiologically with APP.
The APP E2 domain features a tetra-His M1 metal binding site (Fig. 2c) which, in conjunction with several nearby basic residues, forms a positively charged surface patch.25 APP E2 and homologues bind heparins (a class of negatively charged polysaccharide molecules of varying length) via interaction with these basic residues.32–35 Metal binding to the M1 site enhances the binding affinity of APP E2 for heparin.36 These previous observations may help to understand the specificity of APP E2 in forming stable Cu(I)-mediated ternary complexes with the Cu(I) probe ligands that can be disrupted by heparin. Each ligand (Fig. 1) carries two negative charges and may interact electrostatically with the positively charged residues surrounding the M1 site. A shared coordination of Cu(I) between the M1 site and the probe ligand L may enhance such interactions to allow formation of a stable 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ternary complex. There are several lines of evidence supporting this model:
1 ternary complex. There are several lines of evidence supporting this model:
(i) Control proteins such as APP D2, Atox1 and many other Cu(I) binding proteins bind Cu(I) with different affinities, but they do not form ternary complexes with the Cu(I) probe ligands of Fig. 1 (expts 6 and 7, Table 3). In fact, APP E2 is the first example of a protein forming a stable Cu(I)-mediated ternary complex.
(ii) Variant APP E2-qm lacks a functional M1 site but can still bind two equiv. of Cu(I) weakly. However, such binding does not promote formation of a ternary complex (Table 3, expt 5), demonstrating that Cu(I) binding to the M1 site is essential.
(iii) A previous study demonstrated that the M1 site can also bind Cu(II) and Zn(II), but the stable binding of Cu(II) is conditional upon the presence of an intact M1 site with all four His sidechains acting as ligands (Fig. 2c).36 In contrast, stable binding of Zn(II) does not require an intact M1 site and exhibits a need for 2–3 His ligands from the M1 site.36 Cu(I) has the same d10 electronic configuration as Zn(II) and is even more flexible with its coordination number (2–5) and geometry. Consequently, while stable Cu(II) binding by the M1 site cannot be shared by an external ligand and thus cannot promote ternary complex formation, the flexibility of Cu(I) binding to the M1 site apparently allows sharing of the Cu(I) coordination by an external ligand L and formation of the stable ternary complexes.
(iv) Heparins constitute a group of highly negatively charged molecules of varying sizes that have been demonstrated to interact with and bind to E2 and E2-like proteins via interaction with a group of basic residues located close to the M1 site. Heparin appears to disrupt weak interactions between E2 and ligand molecules L. This apparently allows Cu(I) to bind to the M1 site with picomolar affinity.
Formation of the Cu(I)-mediated ternary complexes observed in this work may be an anomaly specific to the in vitro experiments. However, the APP E2 domain and, in particular, the region involving the M1 site has been demonstrated to be an important site of interaction with both metal ions and other components of the extracellular matrix.22,23 Accumulating evidence suggests that these interactions are important for the physiological functions of APP and its homologues APLP1 and APLP2.22,23,42 This work demonstrates that APP E2 is able to interact, via formation of ternary complexes, with each of the four Cu(I) ligands shown in Fig. 1 whose affinities for Cu(I) span a range over six orders of magnitude (Table 1).
It is interesting to speculate that similar metal-bridged interactions involving biological co-ligand(s) may exist in vivo, and may be dependent on specific metal ions, such as Cu(I). Furthermore, this work suggests that ternary interactions could be regulated by biological co-ligand(s) and by heparin: these ligands could promote copper transfer to and/or from the M1 site, thereby facilitating diverse physiological functions.23,42 Such interactions may be significant in modulating the functions of APP at synapses where high concentrations of copper are reportedly released.43–45 In addition, APP and the HSPG glypican-1 (Gpc-1) co-localise in intracellular compartments46 and Cu-APP complexes are thought to play a role in the degradation of Gpc-1 heparan sulfate chains in endosomes.47 The interesting interplay between Cu(I)-binding, heparin-binding and ternary complex formation could be important factors influencing APP function and Alzheimer disease pathogenesis – a possibility that remains to be explored in future research.
A lesson for conditions of reliable quantification of metal-binding affinity by ligand competition
Metal binding affinities in proteins frequently reach nanomolar concentration or below and can only be assessed reliably via ligand competition at the present time, regardless of the detection readouts (absorbance, fluorescence, NMR, EPR, isothermal titration calorimetry (ITC) or enzyme activity).1,48 Reliable quantification requires an effective competition (as described by eqn (2)) that must satisfy the metal speciation analysis at equilibrium by eqn (3) or, more specifically, by eqn (4)–(6) for the Cu(I) probes employed in this work. Although the transfer of metal ions between the competing partners is usually mediated via formation of transient ternary complexes, this study documented that the formation of stable ternary complexes violates the conditions of eqn (2)–(6) and invalidates subsequent affinity determination.A set of useful diagnostic experiments was documented for detection of the formation of stable ternary complexes. These include:-
(i) Close examination of the fingerprints of the selected detection probe (including but not limited to the solution spectrum) to ensure that they are not altered by the ligand competition (e.g., Fig. 3, 4 and Fig. S3, S4, ESI†);
(ii) For an absorbance probe, conducting control metal ion titrations of the probe ligand in the absence and presence of the target protein. Inconsistent metal-probe complex recovery at the saturating metal titration point indicates the possibility of formation of stable ternary complex(es) (e.g., Fig. 5);
(iii) Confirmation of the binding equilibrium of eqn (2) by variation of the concentrations of each individual competing component and/or by observation of a simple ‘dilution effect’. Affinity data invariant with these changes indicates negligible (if any) ternary effects (e.g., Fig. 6, 7 and Table 2).
(iv) Employment of alternate competing ligands with overlapping affinity ranges. Invariant estimates documents reliability, provided that a unified affinity scale is used (see Table 2).
It is anticipated that formation of ternary complexes may alter the spectroscopic properties of a given probe. However, this may not be obvious in certain cases such as those shown in Fig. 4 and Fig. S4 (ESI†) for ligand L = Bca, Bcs. Then the experiments of (ii) and (iii) above become important. Dilution controls are a simple but effective mean of verifying the integrity of the equilibrium for a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 competition such as eqn (4). Dilution with buffer will shift the binding equilibrium to a new position, without affecting determination of the metal affinity. Such controls have been conducted for characterization of many Cu(I)-binding protein targets previously and confirm the reliability of these data.16,26,49–51
2 competition such as eqn (4). Dilution with buffer will shift the binding equilibrium to a new position, without affecting determination of the metal affinity. Such controls have been conducted for characterization of many Cu(I)-binding protein targets previously and confirm the reliability of these data.16,26,49–51
Where possible, affinity estimates should also be conducted with at least two independent competing standards with overlapping affinity ranges. This is possible when both probes impose appropriate free metal buffering conditions for a given target.16 This is demonstrated for the APP E2/heparin complex in this work: in the presence of heparin, a consistent Cu(I) KD of picomolar affinity was determined for the M1 site in E2 using two independent probe ligands Fz and Bca (Table 2). The Cu(I) affinity of ligand Fs is too weak while that of Bcs is too strong for this purpose (Fig. 6a and Fig. S4, ESI†).
Conclusions
This work documents the formation of stable Cu(I)-mediated ternary complexes between the APP E2 protein domain and each of four Cu(I)-responsive probe ligands (Fs, Fz, Bca and Bcs). This is the first reported example of a stable ternary complex involving these commonly used probe ligands and provides a convenient ‘negative control’ since unreliable KD values are extracted under the conditions of ternary complex formation. Several simple diagnostic control experiments are recommended to probe the presence of stable ternary complexes in metal-binding competition experiments. Utilizing these criteria, we demonstrated that ternary complexes are precluded when APP E2 is bound to its biological ligand heparin. Competition experiments provided reliable estimates of a picomolar affinity for the His-rich M1 site in APP E2/heparin complex. This work allows new insights into a potential regulation of APP functions regulated by copper binding and heparin interaction.Abbreviations
| APP D2 | Amyloid precursor protein D2 domain |
| APP E2 | Amyloid precursor protein E2 domain |
| APLP1 | Amyloid precursor-like protein 1 |
| APLP2 | Amyloid precursor-like protein 2 |
| Asc | Ascorbate |
| Bca | Bicinchoninic anion |
| Bcs | Bathocupröine disulfonate |
| eq | Equation |
| equiv. | Equivalent(s) |
| ESI-MS | Electrospray ionization mass spectrometry |
| Fs | Ferene S |
| Fz | Ferrozine |
| GAG | Glycosaminoglycan |
| HSPGs | Heparan sulfate proteoglycans |
| His | Histidine |
| Mops | 3-(N-Morpholino)propanesulfonic acid |
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was supported by funds from the Australian Research Council Grant DP130100728. Additional financial support for TRY was generously provided by the Norma Hilda Schuster (née Swift) Scholarship Fund and the Albert Shimmins Fund. We thank Associate Professor Tara Pukala (University of Adelaide) for assistance with native ESI-MS experiments. Additional ESI-MS analysis was conducted at the Bio21 Mass Spectrometry and Proteomics Facility. We thank Professor Roberto Cappai (University of Melbourne) for kindly donating the Pichia pastoris strain expressing APP D2.References
- Z. Xiao and A. G. Wedd, The challenges of determining metal-protein affinities, Nat. Prod. Rep., 2010, 27, 768–789 RSC.
- A. W. Foster, S. J. Dainty, C. J. Patterson, E. Pohl, H. Blackburn, C. Wilson, C. R. Hess, J. C. Rutherford, L. Quaranta, A. Corran and N. J. Robinson, A chemical potentiator of copper-accumulation used to investigate the iron-regulons of Saccharomyces cerevisiae, Mol. Microbiol., 2014, 93, 317–330 CrossRef CAS PubMed.
- L. A. Finney and T. V. O'Halloran, Transition metal speciation in the cell: insights from the chemistry of metal ion receptors, Science, 2003, 300, 931–936 CrossRef CAS PubMed.
- Z. Xiao, J. Brose, S. Schimo, S. M. Ackland, S. La Fontaine and A. G. Wedd, Unification of the copper(I) binding affinities of the metallo-chaperones Atx1, Atox1 and related proteins: detection probes and affinity standards, J. Biol. Chem., 2011, 286, 11047–11055 CrossRef CAS PubMed.
- P. Bagchi, M. T. Morgan, J. Bacsa and C. J. Fahrni, Robust Affinity Standards for Cu(I) Biochemistry, J. Am. Chem. Soc., 2013, 135, 18549–18559 CrossRef CAS PubMed.
- B.-E. Kim, T. Nevitt and D. J. Thiele, Mechanisms for copper acquisition, distribution and regulation, Nat. Chem. Biol., 2008, 4, 176–185 CrossRef CAS PubMed.
- S. Lutsenko, Human copper homeostasis: a network of interconnected pathways, Curr. Opin. Chem. Biol., 2010, 14, 211–217 CrossRef CAS PubMed.
- R. A. Festa and D. J. Thiele, Copper: An essential metal in biology, Curr. Biol., 2011, 21, R877–R883 CrossRef CAS PubMed.
- P. C. Bull, G. R. Thomas, J. M. Rommens, J. R. Forbes and D. W. Cox, The Wilson disease gene is a putative copper transporting P-type ATPase similar to the Menkes gene, Nat. Genet., 1993, 5, 327–337 CrossRef CAS PubMed.
- E. Gaggelli, H. Kozlowski, D. Valensin and G. Valensin, Copper homeostasis and neurodegenerative disorders (Alzheimer's, prion, and Parkinson's diseases and amyotrophic lateral sclerosis), Chem. Rev., 2006, 106, 1995–2044 CrossRef CAS PubMed.
- P. S. Donnelly, Z. Xiao and A. G. Wedd, Copper and Alzheimer's disease, Curr. Opin. Chem. Biol., 2007, 11, 128–133 CrossRef CAS PubMed.
- Z. Xiao, F. Loughlin, G. N. George, G. J. Howlett and A. G. Wedd, C-terminal domain of the membrane copper transporter Ctr1 from Saccharomyces cerevisiae binds four Cu(I) ions as a cuprous-thiolate polynuclear cluster: sub-femtomolar Cu(I) affinity of three proteins involved in copper trafficking, J. Am. Chem. Soc., 2004, 126, 3081–3090 CrossRef CAS PubMed.
- L. X. Chong, M. R. Ash, M. J. Maher, M. G. Hinds, Z. Xiao and A. G. Wedd, Unprecedented binding cooperativity between Cu(I) and Cu(II) in the copper resistance protein CopK from Cupriavidus metallidurans CH34: implications from structural studies by NMR spectroscopy and X-ray crystallography, J. Am. Chem. Soc., 2009, 131, 3549–3564 CrossRef CAS PubMed.
- Y. Fu, H. C. Tsui, K. E. Bruce, L. T. Sham, K. A. Higgins, J. P. Lisher, K. M. Kazmierczak, M. J. Maroney, C. E. Dann, 3rd, M. E. Winkler and D. P. Giedroc, A new structural paradigm in copper resistance in Streptococcus pneumoniae, Nat. Chem. Biol., 2013, 9, 177–183 CrossRef CAS PubMed.
- A. Badarau and C. Dennison, Copper Trafficking Mechanism of CXXC-Containing Domains: Insight from the pH-Dependence of Their Cu(I) Affinities, J. Am. Chem. Soc., 2011, 133, 2983–2988 CrossRef CAS PubMed.
- Z. Xiao, L. Gottschlich, R. van der Meulen, S. R. Udagedara and A. G. Wedd, Evaluation of quantitative probes for weaker Cu(I) binding sites completes a set of four capable of detecting Cu(I) affinities from nanomolar to attomolar, Metallomics, 2013, 5, 501–513 RSC.
- G. D. Ciccotosto, D. J. Tew, S. C. Drew, D. G. Smith, T. Johanssen, V. Lal, T. L. Lau, K. Perez, C. C. Curtain, J. D. Wade, F. Separovic, C. L. Masters, J. P. Smith, K. J. Barnham and R. Cappai, Stereospecific interactions are necessary for Alzheimer disease amyloid-beta toxicity, Neurobiol. Aging, 2011, 32, 235–248 CrossRef CAS PubMed.
- I. Coburger, S. O. Dahms, D. Roeser, K. H. Guhrs, P. Hortschansky and M. E. Than, Analysis of the overall structure of the multi-domain amyloid precursor protein (APP), PLoS One, 2013, 8, e81926 Search PubMed.
- C. Russo, V. Venezia, E. Repetto, M. Nizzari, E. Violani, P. Carlo and G. Schettini, The amyloid precursor protein and its network of interacting proteins: physiological and pathological implications, Brain Res. Brain Res. Rev., 2005, 48, 257–264 CrossRef CAS PubMed.
- R. Cappai, B. Elise Needham and G. D. Ciccotosto, in Abeta Peptide and Alzheimer's Disease: Celebrating a Century of Research, ed. C. J. Barrow and D. H. Small, Springer London, London, 2007, pp. 37–51 DOI:10.1007/978-1-84628-440-3_3.
- I. W. Hamley, The Amyloid Beta Peptide: A Chemist's Perspective. Role in Alzheimer's and Fibrillization, Chem. Rev., 2012, 112, 5147–5192 CrossRef CAS PubMed.
- K. Wild, A. August, C. U. Pietrzik and S. Kins, Structure and Synaptic Function of Metal Binding to the Amyloid Precursor Protein and its Proteolytic Fragments, Front. Mol. Neurosci., 2017, 10, 21 Search PubMed.
- L. J. Sosa, A. Cáceres, S. Dupraz, M. Oksdath, S. Quiroga and A. Lorenzo, The physiological role of the amyloid precursor protein as an adhesion molecule in the developing nervous system, J. Neurochem., 2017, 143, 11–29 CrossRef CAS PubMed.
- G. K. Kong, J. J. Adams, H. H. Harris, J. F. Boas, C. C. Curtain, D. Galatis, C. L. Masters, K. J. Barnham, W. J. McKinstry, R. Cappai and M. W. Parker, Structural Studies of the Alzheimer's Amyloid Precursor Protein Copper-binding Domain Reveal How it Binds Copper Ions, J. Mol. Biol., 2007, 367, 148–161 CrossRef CAS PubMed.
- S. O. Dahms, I. Konnig, D. Roeser, K. H. Guhrs, M. C. Mayer, D. Kaden, G. Multhaup and M. E. Than, Metal binding dictates conformation and function of the amyloid precursor protein (APP) E2 domain, J. Mol. Biol., 2012, 416, 438–452 CrossRef CAS PubMed.
- S. L. Leong, T. R. Young, K. J. Barnham, A. G. Wedd, M. G. Hinds, Z. Xiao and R. Cappai, Quantification of Copper Binding to Amyloid Precursor Protein Domain 2 and its Caenorhabditis elegans Ortholog. Implications for Biological Function, Metallomics, 2014, 6, 105–116 RSC.
- M. E. Rice, Ascorbate regulation and its neuroprotective role in the brain, Trends Neurosci., 2000, 23, 209–216 CrossRef CAS PubMed.
- D. H. Small, V. Nurcombe, R. Moir, S. Michaelson, D. Monard, K. Beyreuther and C. L. Masters, Association and release of the amyloid protein precursor of Alzheimer's disease from chick brain extracellular matrix, J. Neurosci., 1992, 12, 4143–4150 CAS.
- G. Multhaup, Identification and regulation of the high affinity binding site of the Alzheimer's disease amyloid protein precursor (APP) to glycosaminoglycans, Biochimie, 1994, 76, 304–311 CrossRef CAS PubMed.
- S. Hoefgen, I. Coburger, D. Roeser, Y. Schaub, S. O. Dahms and M. E. Than, Heparin induced dimerization of APP is primarily mediated by E1 and regulated by its acidic domain, J. Struct. Biol., 2014, 187, 30–37 CrossRef CAS PubMed.
- S. S. Mok, G. Sberna, D. Heffernan, R. Cappai, D. Galatis, H. J. Clarris, W. H. Sawyer, K. Beyreuther, C. L. Masters and D. H. Small, Expression and analysis of heparin-binding regions of the amyloid precursor protein of Alzheimer's disease, FEBS Lett., 1997, 415, 303–307 CrossRef CAS PubMed.
- J. T. Hoopes, X. Liu, X. Xu, B. Demeler, E. Folta-Stogniew, C. Li and Y. Ha, Structural characterization of the E2 domain of APL-1, a Caenorhabditis elegans homolog of human amyloid precursor protein, and its heparin binding site, J. Biol. Chem., 2010, 285, 2165–2173 CrossRef CAS PubMed.
- Y. Xue, S. Lee and Y. Ha, Crystal structure of amyloid precursor-like protein 1 and heparin complex suggests a dual role of heparin in E2 dimerization, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 16229–16234 CrossRef CAS PubMed.
- Y. Xue, S. Lee, Y. Wang and Y. Ha, Crystal structure of the E2 domain of amyloid precursor protein-like protein 1 in complex with sucrose octasulfate, J. Biol. Chem., 2011, 286, 29748–29757 CrossRef CAS PubMed.
- S. O. Dahms, M. C. Mayer, D. Roeser, G. Multhaup and M. E. Than, Interaction of the amyloid precursor protein-like protein 1 (APLP1) E2 domain with heparan sulfate involves two distinct binding modes, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2015, 71, 494–504 CAS.
- C. Dienemann, I. Coburger, A. Mehmedbasic, O. M. Andersen and M. E. Than, Mutants of Metal Binding Site M1 in APP E2 Show Metal Specific Differences in Binding of Heparin but Not of sorLA, Biochemistry, 2015, 54, 2490–2499 CrossRef CAS PubMed.
- S. O. Dahms, S. Hoefgen, D. Roeser, B. Schlott, K.-H. Gührs and M. E. Than, Structure and biochemical analysis of the heparin-induced E1 dimer of the amyloid precursor protein, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 5381–5386 CrossRef CAS PubMed.
- S. Hoefgen, S. O. Dahms, K. Oertwig and M. E. Than, The Amyloid Precursor Protein Shows a pH-Dependent Conformational Switch in Its E1 Domain, J. Mol. Biol., 2015, 427, 433–442 CrossRef CAS PubMed.
- Z. Xiao, P. S. Donnelly, M. Zimmermann and A. G. Wedd, Transfer of Copper between Bis(thiosemicarbazone) Ligands and Intracellular Copper-Binding Proteins. Insights into Mechanisms of Copper Uptake and Hypoxia Selectivity, Inorg. Chem., 2008, 47, 4338–4347 CrossRef CAS PubMed.
- M. Johnson, A. T. Coulton, M. A. Geeves and D. P. Mulvihill, Targeted Amino-Terminal Acetylation of Recombinant Proteins in E. coli, PLoS One, 2010, 5, e15801 CAS.
- G. K. Kong, D. Galatis, K. J. Barnham, G. Polekhina, J. J. Adams, C. L. Masters, R. Cappai, M. W. Parker and W. J. McKinstry, Crystallization and preliminary crystallographic studies of the copper-binding domain of the amyloid precursor protein of Alzheimer's disease, Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun., 2005, 61, 93–95 CrossRef CAS PubMed.
- U. C. Muller, T. Deller and M. Korte, Not just amyloid: physiological functions of the amyloid precursor protein family, Nat. Rev. Neurosci., 2017, 18, 281–298 CrossRef PubMed.
- J. Kardos, I. Kovacs, F. Hajos, M. Kalman and M. Simonyi, Nerve endings from rat brain tissue release copper upon depolarization. A possible role in regulating neuronal excitability, Neurosci. Lett., 1989, 103, 139–144 CrossRef CAS PubMed.
- M. L. Schlief and J. D. Gitlin, Copper homeostasis in the CNS: a novel link between the NMDA receptor and copper homeostasis in the hippocampus, Mol. Neurobiol., 2006, 33, 81–90 CrossRef CAS PubMed.
- M. Kawahara, M. Kato-Negishi and K. Tanaka, Cross talk between neurometals and amyloidogenic proteins at the synapse and the pathogenesis of neurodegenerative diseases, Metallomics, 2017, 9, 619–633 RSC.
- R. Cappai, F. Cheng, G. D. Ciccotosto, B. E. Needham, C. L. Masters, G. Multhaup, L. A. Fransson and K. Mani, The amyloid precursor protein (APP) of Alzheimer disease and its paralog, APLP2, modulate the Cu/Zn-Nitric Oxide-catalyzed degradation of glypican-1 heparan sulfate in vivo, J. Biol. Chem., 2005, 280, 13913–13920 CrossRef CAS PubMed.
- F. Cheng, L. A. Fransson and K. Mani, Cytochromeb561, copper, beta-cleaved amyloid precursor protein and niemann-pick C1 protein are involved in ascorbate-induced release and membrane penetration of heparan sulfate from endosomal S-nitrosylated glypican-1, Exp. Cell Res., 2017, 360, 171–179 CrossRef CAS PubMed.
- D. E. Wilcox, Isothermal titration calorimetry of metal ions binding to proteins: An overview of recent studies, Inorg. Chim. Acta, 2008, 361, 857–867 CrossRef CAS.
- J. Brose, S. La Fontaine, A. G. Wedd and Z. Xiao, Redox Sulfur Chemistry of the Copper Chaperone Atox1 is Regulated by the Enzyme Glutaredoxin 1, the Reduction Potential of the Glutathione Couple GSSG/2GSH and the Availability of Cu(I), Metallomics, 2014, 6, 793–808 RSC.
- T. R. Young, A. Kirchner, A. G. Wedd and Z. Xiao, An Integrated Study of the Affinities of the Aβ16 Peptide for Cu(I) and Cu(II): Implications for the Catalytic Production of Reactive Oxygen Species, Metallomics, 2014, 6, 505–517 RSC.
- C. J. K. Wijekoon, A. A. Ukuwela, A. G. Wedd and Z. Xiao, Evaluation of employing poly-lysine tags versus poly-histidine tags for purification and characterization of recombinant copper-binding proteins, J. Inorg. Biochem., 2016, 162, 286–294 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Expression vector modification (Table S1), ESI-MS data (Tables S2 and S3), protein characterization (Fig. S1 and S2); solution spectra (Fig. S3–S5), ternary complex separation (Fig. S6). See DOI: 10.1039/c7mt00291b |
| ‡ It has been suggested that long-chain heparins, such as the preparation used in this study (H3393, 19 kDa), may form dimeric (E2)2–heparin complexes. However, detailed studies by Hoefgen et al. indicated that this was due to two E2 domains independently binding at distant ends of the same (long) heparin chain, rather than stabilising a dimeric E2–E2 interface (see ref. 30). Accordingly, in the quantitative copper-binding analysis presented here, each E2 molecule is treated as an independent Cu(I)-binding moiety. As the exact molecular nature of the structure has not been elucidated, the complex is referred to as ‘E2/heparin’ herein. |
| This journal is © The Royal Society of Chemistry 2018 |