Open Access Article
Open Access ArticleCandida rugosa lipase covalently immobilized on facilely-synthesized carbon nitride nanosheets as a novel biocatalyst†
Ya Li‡
a,
Zhijun Ruan‡ b,
Mingming Zhenga,
Qianchun Denga,
Shan Zhanga,
Chang Zhenga,
Hu Tang
b,
Mingming Zhenga,
Qianchun Denga,
Shan Zhanga,
Chang Zhenga,
Hu Tang a,
Fenghong Huang*a and
Jie Shi
a,
Fenghong Huang*a and
Jie Shi *a
*a
aHubei Key Laboratory of Lipid Chemistry and Nutrition, Oil Crops and Lipids Process Technology National & Local Joint Engineering Laboratory, Key Laboratory of Oilseeds Processing, Ministry of Agriculture, Oil Crops Research Institute, Chinese Academy of Agricultural Sciences, Wuhan 430062, China. E-mail: shijie@caas.cn
bHubei Key Laboratory of Processing and Application of Catalytic Materials, College of Chemistry and Chemical Engineering, Huanggang Normal University, Huanggang 438000, China
First published on 17th April 2018
Abstract
The immobilization of lipase on solid supports provides a significant improvement to the stability and reusability of lipase. During immobilization, the restricted surface area and inferior separation capacity of matrix materials are crucial for obtaining high-quality immobilized lipase. Carbon nitride nanosheets (C3N4-NS) as a type of two-dimensional nanomaterial have attracted various attentions for their prominent 2D planar nanostructure, characteristic surface area, thermostability and biocompatibility. Herein, we report a rational design and fabrication of immobilized Candida rugosa lipase based on carbon nitride nanosheets (C3N4-NS) as the matrix. The synthetic C3N4-NS are characterized by transmission electron microscopy, Brunauer–Emmett–Teller gas sorptometry measurement, X-ray powder diffraction, Fourier transform infrared spectroscopy and thermogravimetric analysis. These results show that C3N4-NS possess an as-expected two-dimensional nanostructure with a large surface area of 74.374 m2 g−1. In addition, we chose glutaraldehyde-assisted covalent attachment to combine C3N4-NS and Candida rugosa lipase (CRL) via amino groups at the margins of C3N4-NS. The as-constructed immobilized lipase (C3N4-NS@CRL) exhibits satisfactory enzyme-loading (44.76 mg g−1), pH-flexibility, thermostability (after 180 min at 50 °C, 67% of the initial activity remained) and recyclability (after 10 runs, 72% of the initial activity remained). When compared with the free CRL, all experimental data indicate that C3N4-NS@CRL exhibited improved stability and enhanced practicability. To our knowledge, this is the first report of the application of carbon nitride nanosheets to enzyme immobilization.
1. Introduction
Due to their mild catalytic conditions, high catalytic efficiency, wide substrate specificity and fewer side effects in contrast with chemical catalysts, enzymes have rapidly attracted increasing attention from researchers and have been generally applied in extensive areas such as biocatalysis, pharmaceuticals, environmental protection, the food industry and biosensors.1 Noticeably, lipase (triacylglycerol ester hydrolase, E.C. 3.1.1.3), as a major player in the area of enzymes, is remarkably excellent in the versatile reactions it catalyses, such as hydrolysis, esterification, transesterification, alcoholysis, ammolysis and C–C bond formation.2 However, taking commercial utility into consideration, free lipase suffers from problems of low operational stability, difficult reusability and high cost.3 Immobilization of lipase on suitable matrix materials via physical adsorption or covalent attachment has led it to show superior performance in enzyme activity, acid–base resistance properties, thermostability and recyclability when compared to the free form of lipase and provided glorious new trains of thought to break through these intractable dilemmas. Aimed at obtaining greater usefulness and better acclimation, appropriate supporting materials are becoming crucial to the immobilization of lipase. Although many sorts of carrier materials ranging from inorganics4 to organics5 have been applied to immobilized lipase, outstanding strategies to design and fabricate immobilized lipase with high activity and stability are still in urgent demand.Two-dimensional nanomaterials have offered great promise in the area of immobilized enzymes as carrier materials due to their unique nanostructure and attractive properties. It is clear to see that 2D nanomaterials could provide a large surface area to capture plentiful amounts of protein, with controllable surface functionalization to match multiple immobilization methods and a 2D nanostructure to guarantee enough space for mass transfer during catalytic processes.6 In recent years, graphene and graphene oxide (GO), shining stars in the 2D family, have exhibited outstanding performances as carrier materials and have been used to build various novel biocatalysts.7–9 However, there still exist several stumbling blocks to the use of graphene and GO, such as their complex, tedious and time-consuming synthesis processes, and their high cost as commercial products, and these impose restrictions on the actual application value of immobilized enzymes based on them.
Recently, carbon nitride (C3N4), a two-dimensional layered structure, as a new type of metal-free conjugated semiconductor, has attracted a great deal of attention and has been widely applied in the field of photocatalysis research.10 Compared with bulk samples, carbon nitride nanosheets (C3N4-NS), the 2D nanostructure of carbon nitride with fewer layers, possess a better 2D planar microstructure, higher surface area and abundance of active sites, which reveals huge potential as a promising anchored material in the area of enzymes.11 Firstly, the 2D nanostructure of C3N4-NS could ensure efficient mass transfer during the biocatalysis process, while the surface area of C3N4-NS may assure adequate enzyme loading in the same way as other 2D nanomaterials.12 Secondly, numerous active chemical sites are exposed to the surface of C3N4-NS.13 The amino groups at the margins of the nanosheets are conducive to forming more stable biocatalysts via a covalent attachment procedure with the enzyme, accompanied by improved recovery efficiency. In addition, the synthesis processes of C3N4-NS are simple, and it is easy to achieve large-scale production. The nitrogen-rich precursors chosen to fabricate the C3N4-NS are cheap and available, for example cyanamide, dicyandiamide, melamine and urea.10 In the light of practical applications, C3N4-NS seem to be more competitive for low cost. Besides, C3N4-NS have been shown to have favorable biocompatibility, thermostability, chemical resistance stability and environmental friendliness in a series of literature reports.14 Hence, C3N4-NS could work in harmony with various kinds of enzyme when extended to the area of immobilized enzymes. Shen et al.15 first selected mesoporous carbon nitride as the anchored material, realized the immobilization of Candida rugosa lipase via physical absorption and obtained an improved thermal stability of free CRL. However, after their research, related literature has rarely been reported continuously on immobilized enzymes based on C3N4, in particular C3N4-NS.
Herein, we designed and facilely manufactured carbon nitride nanosheets (C3N4-NS) with a satisfactory 2D plane nanostructure and abundant residual amino groups as a novel anchored material. We selected Candida rugosa lipase (CRL) as the proof-of-concept model and assessed the potential applicability of C3N4-NS as an anchored material in immobilized lipase. On one hand, the 2D plane nanostructure of C3N4-NS guarantees enzymatic reactions operating in a sufficient space and accelerates mass transfer efficiency. On the other hand, abundant residual amino groups make it easy to adopt a chemical cross-linking approach to bond the enzyme and anchored matrix together, offering a stable configuration and avoiding the leaching of the enzyme. The whole rationale design is briefly illustrated in Scheme 1 and explained as follows. Large-scale carbon nitride nanosheets were successfully prepared via a top-down approach with dicyanodiamine as the nitrogen-rich precursor under a high-temperature calcination process. In addition, Candida rugosa lipase (CRL) was firmly immobilized on carbon nitride nanosheets via covalent attachments assisted by glutaraldehyde molecules. Experimental results indicated that the resultant immobilized lipase (C3N4-NS@CRL) revealed an enhanced enzyme-loading (44.76 mg g−1), pH-flexibility (pH 4.0–8.0), thermostability (180 min at 50 °C) and recyclability (after 10 runs, 72% of the initial activity remained). Consequently, our work could not only prove that C3N4-NS is an effective carrier for immobilized lipase, but also may provide a new method to construct biocatalysts with improved catalytic efficiency and convenient reuse. To the best of our knowledge, this paper is the first report of the application of 2D-nanostructured carbon nitride nanosheets in the area of immobilized enzymes.
2. Materials and methods
2.1. Materials
Dicyandiamide (DCDA, 99%), p-nitrophenylpalmitate (p-NPP, Aldrich, 99%), and Candida rugosa lipase (CRL, lyophilized powder, Type VII, 700 U per mg solid) were purchased from Sigma-Aldrich (St. Louis, USA). Absolute ethanol (EtOH), glutaraldehyde solution (25%), disodium hydrogen phosphate dodecahydrate (Na2HPO4·12H2O), and sodium dihydrogen phosphate dihydrate (NaH2PO4·2H2O) were supplied by Sinopharm Chemical Reagent Co., Ltd (Shanghai, China). An Enhanced (BCA) Protein Assay Kit was obtained from Shanghai Labaide Biotechnology Co., Ltd (Shanghai, China). Ultrapure water was obtained with Milli-Q apparatus (Millipore, Bedford, MA, USA). All chemicals were of analytical reagent grade and used as received without any further purification.2.2. Characterization
The morphologies of obtained samples were characterized using a transmission electron microscope (Tecnai G2 F30, FEI, USA) operated on 300 kV. The crystal phases of samples were measured using a Bruker D8 Advance X-ray diffractometer (Bruker, Germany) with a 2θ range from 10° to 80° with Cu Kα irradiation. The nitrogen sorption isotherms were measured with an Autosorb-iQ2 (Quantachrome, USA) analyzer. X-ray photoelectron spectroscopy (XPS) analysis was conducted with a Quantera II (ULVAC-PHI, Japan) X-ray photoelectron spectrometer equipped with an Al Kα X-ray radiation source. FT-IR spectra were characterized on a Bruker Tensor 27 FT-IR spectrometer (Thermoelectron, USA) with the KBr pellet technique. The thermal stabilities of C3N4-NS and C3N4-NS@CRL were studied with a thermogravimetric (TGA) analyzer (TAQ50, Netzsch, Germany) at a heating rate of 5 °C min−1 in a N2 atmosphere. The UV-vis absorption spectra were recorded using a DU 800 UV-vis spectrometer (Beckman Coulter, Japan).2.3. Preparation of graphitic carbon nitride nanosheets (C3N4-NS)
Graphitic carbon nitride nanosheets (C3N4-NS) were prepared with reference to literature methods with moderate modifications.16 Dicyandiamide (6 g) was placed into an alumina crucible (50 mL) with a lid, then transferred into a tube furnace, heated at a rate of 2.3 °C min−1 up to 550 °C and held at 550 °C for 4 h in a N2 atmosphere before being cooled down naturally. Then the yellow solid in the alumina crucible was collected and ground into a powder with an agate mortar. Next, the obtained yellow powder was placed into a porcelain crucible (50 mL), heated at a rate of 5 °C min−1 up to 500 °C and maintained at this temperature for 2 h in a N2 atmosphere before being cooled down naturally in the tube furnace. The finally-obtained pale yellow powder was graphitic carbon nitride nanosheets (C3N4-NS).2.4. Immobilization of Candida rugosa lipase on C3N4-NS (C3N4-NS@CRL)
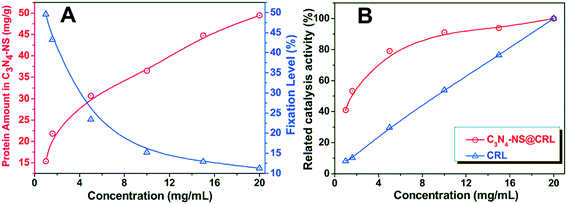
The immobilization of Candida rugosa lipase (CRL) on C3N4-NS was performed by the cross-linking method assisted by glutaraldehyde according to literature reports.17 79 mL PBS buffer (0.1 M, pH 7.0) and 25 mL of glutaraldehyde solution (25%, w/v) were successively added to 3.22 g C3N4-NS in a 250 mL Erlenmeyer flask with sonication treatment. Then, the mixture was incubated at 65 °C in a shaker operating at 160 rpm for 12 h. Subsequently, the activated carrier was separated and repeatedly washed with distilled water 3 times via vacuum filtration. Finally, the product was dried in a vacuum oven at 25 °C overnight, and the material was named G-C3N4-NS.4 g CRL was fully dissolved in 200 mL 0.1 M PBS (pH 7.0) with mild shaking at 37 °C. After the obtained CRL solution was centrifuged, the supernatant was collected and retained. The obtained G-C3N4-NS was dispersed in the lipase solution, and then incubated at 37 °C in a shaker operating at 160 rpm for 12 h. The immobilized lipase (C3N4-NS@CRL) was collected by refrigerated centrifugation, washed with 0.1 M PBS (pH 7.0) 3 times, freeze-dried overnight and stored in a refrigerator at 4 °C. The influences of initial concentration of the CRL solution on the efficiency of protein loading were explored by bringing carriers (30 mg) into contact with 1–20 mg mL−1 of CRL solutions with different initial concentrations at a fixed temperature of 37 °C and 0.1 M pH 7.0 PBS for 6 h.
The protein concentration of C3N4-NS@CRL was determined by the Bradford method, using bovine serum albumin as a standard.18 The amount of immobilized lipase was calculated by detecting the amount of un-immobilized lipase. Calculated from the experimental results, the free CRL solution contained 0.062 mg of protein per mL.
2.5. Enzymatic activity assay
The activities of the free and immobilized lipases were measured by the hydrolysis of p-NPP in a buffer solution at 37 °C for 5 min at 160 rpm. The concentration of the hydrolysis product p-nitrophenol (p-NP) was measured using a spectrophotometric method on an ultraviolet-visible spectrophotometer and the value of absorbance at 410 nm was collected. 30 mg of immobilized lipase (or 100 mg of free lipase) was added to a tube that contained 1 mL of 0.5% p-nitrophenylpalmitate ethanol solution (w/v) and 1 mL of 0.1 M PBS (pH 7.0), then incubated at 160 rpm at 37 °C for 5 min. The reaction was terminated by adding 2 mL of 25 g L−1 Na2CO3 solution. After refrigerated centrifuging at 5000 rpm for 10 min, 1 mL of the supernatant was taken and diluted with distilled water, then the value of absorbance at 410 nm was detected on an ultraviolet-visible spectrophotometer. The molar absorption coefficient (ε = 410 nm) was 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 519 M−1 cm−1 under the same conditions as above according to the Beer–Lambert law. One unit of activity (U) was defined as the amount of enzyme that hydrolyzes 1 μmol of p-nitrophenylpalmitate per minute under the conditions described previously.
519 M−1 cm−1 under the same conditions as above according to the Beer–Lambert law. One unit of activity (U) was defined as the amount of enzyme that hydrolyzes 1 μmol of p-nitrophenylpalmitate per minute under the conditions described previously.
The activities obtained from other conditions were defined as relative activities, and we assumed the maximum activity value of the immobilized and free lipase under optimal conditions to be 100%. The effect of temperature on the activities of free and immobilized lipases was investigated at pH 7.0 at different temperatures (30, 40, 50, 60, 70, and 80 °C) for 1 h. The effect of pH on the activities of free and immobilized lipases was investigated at 37 °C under a variety of pHs (3.0, 4.0, 5.0, 6.0, 7.0, and 8.0) for 1 h. Each treatment was repeated three times.
2.6. Thermal stability of C3N4-NS@CRL
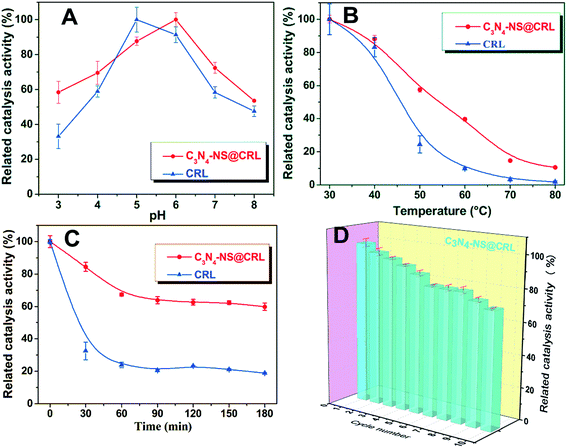
The thermal stability of the immobilized lipase was studied by incubating the biocatalyst in 0.1 M PBS (pH 7.0) from 0 min to 180 min at 50 °C. Periodically, samples were withdrawn every 30 min, and their residual activities were assayed by the hydrolysis of p-NPP as described above. Residual activity was expressed as a percentage of initial activity (hydrolytic activity before thermal incubation).2.7. Reusability of C3N4-NS @CRL
The reusability of the immobilized lipase was determined by the following procedure: 30 mg of immobilized lipase was used for the hydrolysis of p-NPP ethanol solution in 0.1 M pH 7.0 phosphate buffer for 5 min at 37 °C under mild stirring. After each batch reaction, the immobilized lipase was collected by refrigerated centrifugation, washed three times with absolute ethanol, dried with a nitrogen purge and reused for the next cycle. The lipase activity was calculated as in the above description.3. Results and discussions
3.1. Characterization
In order to prove that the designed immobilization was performed successfully, we investigated the morphologies of C3N4-NS and C3N4-NS@CRL by transmission electron microscopy (TEM). Generally, the bulk C3N4 showed a layered structure with dicyandiamide as the precursor via a thermal condensation process. After the thermal exfoliation step, we obtained the 2D planar graphene-like carbon nitride nanosheets (C3N4-NS), which exhibited silk-like plicated slices with a micron-scale planar size as seen in the TEM image (Fig. 1A). After immobilization of Candida rugosa lipase on C3N4-NS assisted by a cross-linking agent, the silk-like C3N4-NS surface appeared to contain some blocky materials as shown in Fig. 1B, which were considered to be the agglomerate shape of CRL consistent with previous reports, and proved that CRL was bonded with C3N4-NS.9 This point could be confirmed by the nitrogen adsorption–desorption isotherm observations of C3N4-NS and C3N4-NS@CRL in Fig. 1C. According to the IUPAC classification, both of the two adsorption isotherms were not reversible and were mainly attributed to a type IV isotherm with hysteresis loops. The C3N4-NS displayed a large BET surface area of 74.4 m2 g−1; while after immobilization of free CRL, the BET surface area of C3N4-NS@CRL was only 19.6 m2 g−1. The decrease in the surface area of C3N4-NS@CRL compared to the parent C3N4-NS was due to the blockage of carbon nitride nanosheets with CRL lipase after immobilization. In addition, we examined the critical lattice structures of all of the prepared nanomaterials during the immobilization process using X-ray diffraction (XRD). As shown in Fig. 1D, all of the four samples revealed two similar characteristic diffraction angles at 13° and 27°. The small peak (2θ = 13°) was attributed to the (100) in-plane repeat units of tri-s-triazine. Meanwhile, the later dominant peak (2θ = 27°) originated from the (002) interlayer diffraction of graphite-like layered structures.19 The slight weakening of the characteristic peaks of C3N4-NS compared to the C3N4 samples suggested that the interlayer stacking pattern of C3N4 and the planar size of C3N4 layers were weakened during the exfoliation process. Meanwhile, the characteristic diffraction angles of glutaraldehyde-modified C3N4-NS (G-C3N4-NS) and C3N4-NS@CRL exhibited no obvious difference to C3N4-NS, indicating that C3N4-NS retained its specific crystal texture and could ensure the mass transfer on the surface of C3N4-NS@CRL during catalytic reactions.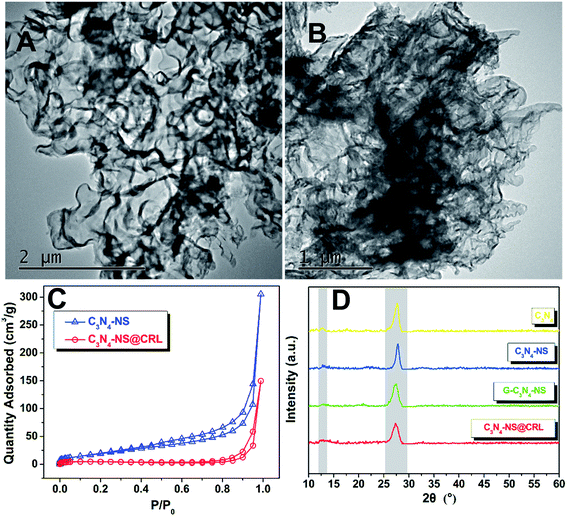 | ||
| Fig. 1 TEM images of C3N4-NS (A) and C3N4-NS@CRL (B); (C) nitrogen adsorption–desorption isotherms of C3N4-NS and C3N4-NS@CRL; (D) the powder XRD patterns of C3N4, C3N4-NS, G-C3N4-NS and C3N4-NS@CRL. | ||
In addition, we figured out another clue by X-ray photoelectron spectroscopy (XPS) to verify the existence of covalent bonding via checking the surface compositions and the valence states of those obtained samples. According to the survey XPS spectra, all of the obtained samples contained C 1s (287 eV), N 1s (397 eV) and O 1s (531 eV) elements (Fig. 2A). The amount of oxygen in C3N4-NS, glutaraldehyde-modified C3N4-NS (G-C3N4-NS) and C3N4-NS@CRL was 3.4%, 2.1% and 15.3%, respectively. The negligible O 1s peaks of C3N4-NS and G-C3N4-NS could be ascribed to the tiny amount of O2 adsorbed on the surface of the two products. Nevertheless, due to the CRL containing more oxygen and O2 adsorbed on the surface of C3N4-NS, the O 1s peak at 531 eV of C3N4-NS@CRL became much stronger, which could also illustrate that CRL was favourably anchored to the surface of C3N4-NS. All of the high-resolution N 1s spectra of the C3N4-NS could be deconvoluted into two peaks (Fig. 2B). The main peak at 397.3 eV corresponded to sp2 hybridized N atoms bonded with carbon atoms (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–C) in tri-s-triazine rings. The peak at 399.1 eV was attributed to the bridging N atoms bonded to C atoms in the form of amino groups ((C)2–NH) connected with structural defects or N–(C)3 groups in the aromatic cycles.19 The above results indicated that C3N4-NS consisted of the heptazine heterocyclic ring unit. As we expected, for C3N4-NS modified by glutaraldehyde or equipped with CRL, the 399.1 eV peak in C3N4-NS was slightly shifted to weaker bonding energy regions, 398.8 eV in G-C3N4-NS and 398.6 eV in C3N4-NS@CRL, which could be attributed to the amide bonds (–CO–NH–) generated from the cross-linking procedures. It could be found that C 1s of C3N4-NS could be separated into two peaks at 283.6 and 286.8 eV, indicating two types of carbon bonding in the C3N4-NS (Fig. 2C). Among them, the binding energy peak at 283.6 eV was ascribed to characteristic sp2 C–C and N–C
N–C) in tri-s-triazine rings. The peak at 399.1 eV was attributed to the bridging N atoms bonded to C atoms in the form of amino groups ((C)2–NH) connected with structural defects or N–(C)3 groups in the aromatic cycles.19 The above results indicated that C3N4-NS consisted of the heptazine heterocyclic ring unit. As we expected, for C3N4-NS modified by glutaraldehyde or equipped with CRL, the 399.1 eV peak in C3N4-NS was slightly shifted to weaker bonding energy regions, 398.8 eV in G-C3N4-NS and 398.6 eV in C3N4-NS@CRL, which could be attributed to the amide bonds (–CO–NH–) generated from the cross-linking procedures. It could be found that C 1s of C3N4-NS could be separated into two peaks at 283.6 and 286.8 eV, indicating two types of carbon bonding in the C3N4-NS (Fig. 2C). Among them, the binding energy peak at 283.6 eV was ascribed to characteristic sp2 C–C and N–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N bonds, and the binding energy peak at 286.8 eV was attributed to C–OR and O–C
N bonds, and the binding energy peak at 286.8 eV was attributed to C–OR and O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bonds.19 After C3N4-NS was modified by glutaraldehyde or equipped with CRL, a new peak at 285 eV appeared, which could be attributed to sp2 C
O bonds.19 After C3N4-NS was modified by glutaraldehyde or equipped with CRL, a new peak at 285 eV appeared, which could be attributed to sp2 C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bonds in the aldehyde group of glutaraldehyde in G-C3N4-NS and amide bonds in C3N4-NS@CRL. All of the results from the high-resolution XPS analysis demonstrated that CRL was successfully linked to C3N4-NS through a covalent method.
O bonds in the aldehyde group of glutaraldehyde in G-C3N4-NS and amide bonds in C3N4-NS@CRL. All of the results from the high-resolution XPS analysis demonstrated that CRL was successfully linked to C3N4-NS through a covalent method.
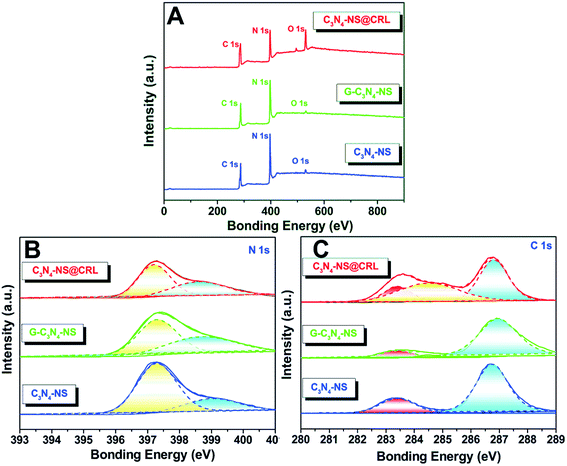 | ||
| Fig. 2 XPS spectra (A), high-resolution N 1s (B) and high-resolution C 1s (C) of C3N4-NS@CRL, G-C3N4-NS and C3N4-NS. | ||
The FT-IR spectra of C3N4-NS@CRL, CRL, G-C3N4-NS and C3N4-NS were shown in Fig. 3A. C3N4-NS, G-C3N4-NS and C3N4-NS@CRL displayed the similar characteristic absorption bands of C3N4 networks. The broad peaks between 3600 and 3000 cm−1 were attributed to –NH2 stretching of the aromatic ring. Meanwhile, the bands of 1635, 1574, 1545, 1462 and 1402 cm−1 were assigned to characteristic stretching vibrations of the tri-s-triazine derived repeating units. The peaks at 1319 and 1244 cm−1 corresponded to stretching vibrations of aromatic C–N connected units of N–(C)3, C–NH–C or C–NH2. The sharp band of 811 cm−1 represented the out-of-plane bending vibration characteristic of heptazine rings.19 Although the characteristic absorption peaks of free CRL were highly overlapped with the absorption peaks of C3N4-NS, peaks could also be revealed due to CRL and C3N4-NS in the curve of C3N4-NS@CRL. These results suggest that the whole immobilization process could make sure that the basic structure of carbon nitride nanosheets was intact. In addition, we determined the free NH2 content of C3N4-NS via a fluorescamine method and the average value was 1.7 nmol free NH2 groups per 1 mg C3N4-NS (Fig. S2†).
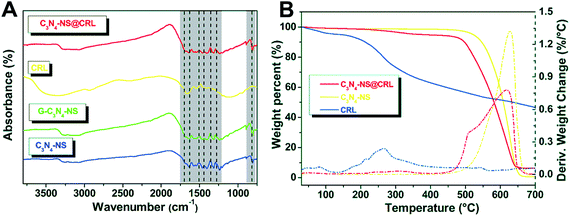 | ||
| Fig. 3 FT-IR spectra (A) of C3N4-NS@CRL, CRL, G-C3N4-NS and C3N4-NS; (B) TGA analysis of C3N4-NS, C3N4-NS@CRL and CRL. | ||
The thermal behaviour of C3N4-NS, CRL and C3N4-NS@CRL was studied by TGA analysis in a nitrogen atmosphere. In Fig. 3B, a sharp decrease in the C3N4-NS weight occurred around 600 °C,10 which could be attributed to the pyrolysis and decomposition of the C3N4-NS polymer fragments. In addition, the TGA curve of CRL showed two major weight loss steps at 80 °C and 260 °C. The weight loss onset around 80 °C indicated the evaporation of absorbed water, while the decomposition at 260 °C could be accredited to denaturation of the CRL structure,9 compared with the TGA curve of free CRL. It is worth noting that both C3N4-NS and C3N4-NS@CRL showed no obvious weight loss (less than 4%) from 80 °C to 300 °C, which could prove the enhanced thermostability of C3N4-NS@CRL when CRL was immobilized on the C3N4-NS.
3.2. Assay of biocatalyst activity
To investigate the application of C3N4-NS in the area of biocatalysis, we utilized it to immobilize Candida rugosa lipase (CRL) by covalent binding coupled with glutaraldehyde and evaluated the enzyme activity depending on the hydrolysis of p-nitrophenylpalmitate (p-NPP). Due to the multipoint interactions between lipase, glutaraldehyde and tri-s-triazine rings on the surface of C3N4-NS, we speculate that those interactions may promote exposure to the catalytic domain of lipase molecules through conformational changes of the open lid domain, thus partly enhancing the enzymatic activity of the hydrolysis of substrate molecules at active sites.As shown in Fig. 5B, C3N4-NS@CRL exhibited better relative enzyme activity in the range from 30 °C to 50 °C (57% of the initial activity at 50 °C). Due to denaturation, the relative activity of the free CRL decreased rapidly when there was an increase in temperature (10% of the initial activity at 60 °C). In contrast, the C3N4-NS@CRL was more heat-resistant than the free CRL at higher temperatures. This may be explained by the formation of chemical bonds during the cross-linking reaction which could effectively restrict the conformation distortion or damage during periods of elevated temperature.
Under the optimal conditions, the kinetic parameters of the free CRL and C3N4-NS@CRL were tested and are listed in Table S1†. The Km value of the C3N4-NS@CRL was 84 times higher than that of the free CRL, indicating that the substrate’s affinity capacity for the C3N4-NS@CRL was stronger compared to the free CRL. In addition, the specific activity was defined as the initial reaction rate divided by the amount of protein. Based on the Vm values of free CRL and C3N4-NS@CRL, the specific activities were 0.37 U mg−1 and 141.7 U mg−1.
4. Conclusion
In summary, we designed and constructed a novel anchored material for an immobilized enzyme. Benefiting from its expanded surface area, increased number of active sites and 2D nanostructure, C3N4-NS was demonstrated to be an excellent carrier for the immobilization of Candida rugosa lipase. Experimental results indicated that the obtained immobilized lipase (C3N4-NS@CRL) demonstrated enhanced enzyme-loading (44.76 mg g−1), pH-flexibility (pH 4.0–8.0), thermostability (after 180 min at 50 °C, 67% of the initial activity remained) and recyclability (after 10 runs, 72% of the initial activity remained). To the best of our knowledge, it is the first time that the application of carbon nitride nanosheets to the area of immobilized enzymes has been reported. Finally, our present strategy could be generalized to construct other immobilized enzyme combinations through the use of other templates.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We are grateful to the National Science Foundation of China (no. 31501423, 31701560 and 31671820), the Young Elite Scientists Sponsorship Program by CAST (2017QNRC001), the Director Fund of Oil Crops Research Institute of CAAS (no. 1610172015006) and the Agricultural Science and Technology Innovation Project of Chinese Academy of Agricultural Sciences (CAAS-ASTIP-2013-OCRI) for financial support.References
- S. A. Ansari and Q. Husain, Biotechnol. Adv., 2012, 30, 512–523 CrossRef CAS PubMed.
- S. Benjamin and A. A. Pandey, Yeast, 1998, 14, 1069–1087 CrossRef CAS PubMed.
- R. Sharma, Y. Chisti and U. C. Banerjeea, Biotechnol. Adv., 2001, 19, 627–662 CrossRef CAS PubMed.
- I. V. Pavlidis, T. Tsoufis, A. Enotiadis, D. Gournis and H. Stamatis, Adv. Eng. Mater., 2010, 12, 179–183 CrossRef.
- J. F. Shi, X. L. Wang, S. H. Zhang, L. Tang and Z. Y. Jiang, J. Mater. Chem. B, 2016, 4, 2654–2661 RSC.
- B. Luo, G. Liu and L. Z. Wang, Nanoscale, 2016, 8, 6904–6920 RSC.
- I. V. Pavlidis, M. Patila, U. T. Bornscheuer, D. Gournis and H. Stamatis, Trends Biotechnol., 2014, 32, 312–320 CrossRef CAS PubMed.
- S. Hermanova, M. Zarevucka, D. Bousa, M. Pumera and Z. Sofer, Nanoscale, 2015, 7, 5852–5858 RSC.
- S. Zhang, J. Shi, Q. C. Deng, M. M. Zheng, C. Y. Wan, C. Zheng, Y. Li and F. H. Huang, Molecules, 2017, 22, 1205–1220 CrossRef PubMed.
- Y. Wang, X. C. Wang and M. Antonietti, Angew. Chem., Int. Ed., 2012, 51, 68–89 CrossRef CAS PubMed.
- A. W. Wang, C. D. Wang, L. Fu, W. Wong-Ng and Y. C. Lan, Nano-Micro Lett., 2017, 9, 47–68 CrossRef.
- C. L. Tan, X. H. Cao, X. J. Wu, Q. Y. He, J. Yang, X. Zhang, J. Z. Chen, W. Zhao, S. K. Han, G. H. Nam, M. Sindoro and H. Zhang, Chem. Rev., 2017, 117, 6225–6331 CrossRef CAS PubMed.
- X. D. Zhang, X. Xie, H. Wang, J. J. Zhang, B. C. Pan and Y. Xie, J. Am. Chem. Soc., 2013, 135, 18–21 CrossRef CAS PubMed.
- L. C. Chen and J. B. Song, Adv. Funct. Mater., 2017, 27, 1702695 CrossRef.
- W. Z. Shen, L. W. Ren, H. Zhou, S. C. Zhang and W. B. Fan, J. Mater. Chem., 2011, 21, 3890 RSC.
- P. Niu, L. L. Zhang, G. Liu and H. M. Cheng, Adv. Funct. Mater., 2012, 22, 4763–4770 CrossRef CAS.
- Y. L. Fan, F. Su, K. Li, C. X. Ke and Y. J. Yan, Sci. Rep., 2017, 7, 45643 CrossRef PubMed.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS PubMed.
- H. Xu, J. Yan, X. J. She, L. Xu, J. X. Xia, Y. Xu, Y. G. Song, L. Y. Huang and H. M. Li, Nanoscale, 2014, 6, 1406–1415 RSC.
- T. L. d. Albuquerque, N. Rueda, J. C. S. dos Santos, O. Barbosa, C. Ortiz, B. Binay, E. Özdemir, L. R. B. Gonçalves and R. Fernandez-Lafuente, Process Biochem., 2016, 51, 865–874 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available: Kinetic parameters for the free CRL and C3N4-NS@CRL, effect of different organic solvents on the relative activity of the C3N4-NS@CRL, fluorescence intensity of the fluorescamine method plotted as a function of increasing amounts of amino groups of fresh APTES. See DOI: 10.1039/c8ra00536b |
| ‡ These authors equally contributed to this study. |
| This journal is © The Royal Society of Chemistry 2018 |