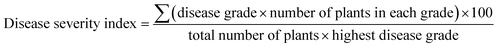Open Access Article
Open Access ArticleDiscovery of novel inhibitors of RNA silencing suppressor P19 based on virtual screening†
Fan
Hu‡
 ab,
Rong
Lei‡
ab,
Rong
Lei‡
 b,
Yu-Fang
Deng
ab,
Jun
Wang
b,
Gui-Fen
Li
b,
Chao-Nan
Wang
ab,
Zhi-Hong
Li
a and
Shui-Fang
Zhu
*ab
b,
Yu-Fang
Deng
ab,
Jun
Wang
b,
Gui-Fen
Li
b,
Chao-Nan
Wang
ab,
Zhi-Hong
Li
a and
Shui-Fang
Zhu
*ab
aCollege of Plant Protection, China Agricultural University, Beijing, 100193, China. E-mail: zhusf@caiq.gov.cn
bInstitute of Plant Quarantine, Chinese Academy of Inspection and Quarantine, Beijing, 100029, China
First published on 16th March 2018
Abstract
To control plant viruses, viral RNA silencing suppressors are important drug targets due to their key roles in interfering antiviral RNA silencing. Here we have presented a strategy, combining virtual and experimental screening, to discover the inhibitors of viral suppressor. By docking 157![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 026 compounds from a natural product library into P19 model, eighteen candidate compounds were selected. Candidates VS2, VS12, VS14 and VS15 displayed strong binding ability to P19 in the surface plasmon resonance imaging assay with KD values of 136.2, 111.6, 81.2 and 124.5 nM, respectively. Then the inhibition activities of these inhibitors on the association between P19 and siRNA were also affirmed by electrophoretic mobility shift assay. Moreover, the antiviral effects on plants showed that compounds VS14 and VS15 both exhibited antiviral activities against Tomato bushy stunt virus (TBSV) in vivo with inhibition rates of 32.35% and 16.61% in 11 dpi, respectively. This strategy would be a powerful tool for the discovery of novel antiviral agents and provide new insights into the control of plant viruses.
026 compounds from a natural product library into P19 model, eighteen candidate compounds were selected. Candidates VS2, VS12, VS14 and VS15 displayed strong binding ability to P19 in the surface plasmon resonance imaging assay with KD values of 136.2, 111.6, 81.2 and 124.5 nM, respectively. Then the inhibition activities of these inhibitors on the association between P19 and siRNA were also affirmed by electrophoretic mobility shift assay. Moreover, the antiviral effects on plants showed that compounds VS14 and VS15 both exhibited antiviral activities against Tomato bushy stunt virus (TBSV) in vivo with inhibition rates of 32.35% and 16.61% in 11 dpi, respectively. This strategy would be a powerful tool for the discovery of novel antiviral agents and provide new insights into the control of plant viruses.
Introduction
Plant viruses cause huge losses in agriculture worldwide. To control viruses in crops, many approaches have been tried including plant genetic resistance and chemical control.1–4 However, most current antivirals based on induction of plant resistance are prophylactic with little therapeutic effect,5,6 highlighting the need for the development of novel inhibitors against important targets based on a main antiviral mechanism. In recent years, RNA silencing has been identified as a major mechanism against viruses in plant and animal kingdoms.7,8 Trigged by viral double stranded RNA (dsRNA), RNA silencing causes a sequence-specific shut down of the expression of genes to limit damage owing to viral infection. As a counter defensive strategy, most viruses encode RNA silencing suppressor (RSS) proteins to counteract the antiviral silencing.9,10 These competitions can be seemed as an arm race between viruses and their hosts (Fig. 1). Interestingly, previous studies showed that deletion of viral RSS actually reduced viral RNA accumulation and alleviated systemic symptoms in plants.11–13 Thus, it raises the possibility that plants can finally win the host-pathogen arm race with the help of the chemical viral RSS inhibitors.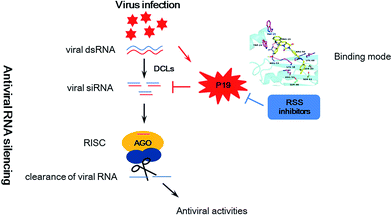 | ||
| Fig. 1 Schematic representation of antiviral RNA silencing in plants, counter-defense by viral silencing suppressor P19 and counter–counter defense by RSS inhibitors. | ||
In a previous study, a strategy was introduced to screen inhibitors of RSSs and gained several chemicals that can inhibit suppressors both in vitro and in vivo.14 The siRNA was immobilized on chips through streptavidin–biotin interaction and RSS was incubated with candidate chemicals before analyzed by SPR method. Furthermore, the inhibition activities against RSS–siRNA complex in vitro and in vivo of the chemicals screened by SPR were also confirmed by EMSA and protoplast-based assay, respectively. The highlight of this study is that the selected chemicals can efficiently suppress RSS–siRNA interactions, not just bind to RSSs without affecting their counter-PTGS ability. Similarly, a fluorescence-detection assay was presented to screen small-molecule inhibitors of RSS P19.15 The P19 with His tag was immobilized on 96-well Ni2+-NTA plates and the siRNA with fluorophore was captured by P19 after incubation, thus the chemicals can be screened by determining the fluorescence intensity per well. Additionally, based on the presence of thiosulfonate functional groups in both identified inhibitors, they also found that covalent modification of cysteine residues in P19 can lead to a reduction of binding activity to siRNA of P19. However, the traditional methods for developing drugs, such as random synthesis and screening and analogue synthesis, are not only expensive but also time-consuming. In recent years, the combination of computer aided drug design (CADD) and experimental high throughput screening (HTS) has become an effective method in lead compounds discovery and development.16–18 One common used approach in CADD is structure-based virtual screening, which enables docking of numerous compounds into a certain bio-molecular target and then evaluates in a speedy and inexpensive manner. Moreover, this molecular docking can also be used to view the binding modes between compounds and target, even further analyze the possible mechanisms.
Among viral RSS proteins, a 19 kDa protein (P19) of Tombusviruses has been widely studied.19 P19 can bind and sequester small interfering RNAs (siRNA), and subsequently inhibit the silencing pathway by preventing these siRNAs incorporation into the RNA-induced silencing complex (RISC) (Fig. 1). Also, sequestration of siRNAs has been seemed as a common strategy for many RSSs to suppress the silencing pathway.20–22 Therefore, inhibition of interactions between siRNAs and RSSs can enhance host RNA silencing resistance directly and then attenuates virus disease severity. In addition, the reports of crystal structures of P19–siRNA complex demonstrate how P19 bind to siRNA and make this RSS an attractive target for structure-based virtual screening of novel inhibitors.23,24
The aim of this work is to discover new antiviral chemicals for management of plants even after viral infection. First of all, we used RSS P19 as the molecular target to screen a natural product library containing 157![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 026 compounds. Then we analyzed the selected candidate compounds by surface plasmon resonance imaging (SPRi). The inhibition activities of these candidates on the association between P19 and siRNA were also tested by electrophoretic mobility shift assay (EMSA). Finally, we identified four strong inhibitors against P19 and examined their antiviral activities against TBSV in plants. Taken together, we have introduced a strategy to screen novel inhibitors of RSS by combined virtual screening and experimental HTS, and obtained several antiviral agents.
026 compounds. Then we analyzed the selected candidate compounds by surface plasmon resonance imaging (SPRi). The inhibition activities of these candidates on the association between P19 and siRNA were also tested by electrophoretic mobility shift assay (EMSA). Finally, we identified four strong inhibitors against P19 and examined their antiviral activities against TBSV in plants. Taken together, we have introduced a strategy to screen novel inhibitors of RSS by combined virtual screening and experimental HTS, and obtained several antiviral agents.
Experimental
Molecular docking
The crystal structure of P19 (PDB ID:4JGN) used in this study was prepared by the following steps: all water and solvent molecules were removed from structure in Pymol,25 then hydrogen atoms and Gasteiger charges were added using AutoDock Tools.26 The modified structure was used to define the binding pocket according to PocketPicker,27 siRNA binding sites and the analyzed key residues (Fig. 2B). After that, the structure was submitted for docking in AutoDock Vina (version:1.1.2) against a natural product database containing 157![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 026 small molecules. (3D structures prepared by Guo et al.28). The results were scored by AutoDock Vina and the top 200 ranked compounds were chosen for further binding mode analysis.
026 small molecules. (3D structures prepared by Guo et al.28). The results were scored by AutoDock Vina and the top 200 ranked compounds were chosen for further binding mode analysis.
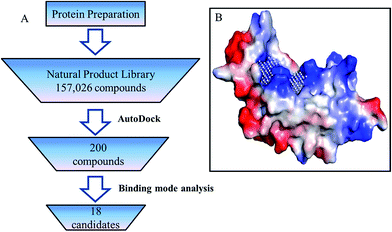 | ||
| Fig. 2 (A) Virtual screen scheme to retrieve inhibitors of RSS P19. (B) Binding sites predication of RSS P19. | ||
Protein expression and purification
The RSS P19 protein was expressed in Escherichia coli strain BL21 (DE3) pLysS by use of the pet-28a expression system. Briefly, the P19 sequence was cloned at Nde I and Not I sites into the expression vector pET-28a(+), which resulted in P19 C-terminal fusion with a His-tag. (Primers were shown in Table S2.†) After transformation of Escherichia coli, a pre-culture of 5 mL volume was grown at 37 °C overnight, and added to 500 mL Luria-Bertani (LB) medium and grown to an OD600 of 0.6–0.8. Then, expression of P19 was induced by the addition of 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG). To avoid misfolding, the temperature of the culture was reduced to 25 °C. After 8 h, the expression bacterial cultures were harvested and proteins were purified using Qiagen Ni-NTA Agarose according to the manufacturer's instruction.Preparation of small molecule microarrays for SPRi
The photo-cross-linked (PCL) surface sensor chip was prepared as described previously.29,30 Compounds dissolved in DMSO (10 mM) were spotted in multiplex using a Genetix QArray spotter (produced 300 μM features). The chip was dried to evaporate DMSO under vacuum and then exposed to UV irradiation 2.4 J cm−2 (365 nm) in a UV chamber (Amersham Life Science). Subsequently the chip was washed using DMF, ethanol and ultrapure water for 15 min respectively.The prepared PCL chip was placed in an SPR imaging instrument (PlexArray, USA). After washing with running buffer (PBS, pH 7.2) for 10 min, the RSS proteins diluted in PBS (pH 7.2) were injected at a flow rate of 2 μL s−1 with both association and dissociation time of 300 s.
Electrophoretic mobility shift assay
The P19-siRNA inhibition activity of compounds was tested by chemiluminescent EMSA Kit (Beyotime, China). (siRNA sequences were shown in Table S2.†) Viral suppressor P19 and a candidate compound were incubated in 18 μL binding buffer for 25 min at room temperature before adding 2 μL siRNA. After incubation for another 25 min, 2.2 μL loading buffer was added to the binding reaction. Then the mixture was analyzed by electrophoresis in 6.0% polyacrylamide gels at 110 V for 1 h in 0.5× TBE running buffer. The Protein–siRNA complexes in the gel were transferred to a nylon membrane at 380 mA for 45 min, and then the nylon membrane was crosslinked by UV-light instrument and visualized according to manufacturer's instruction.Evaluation of antiviral activity of inhibitors against TBSV in vivo
Tobacco plants (Nicotiana benthamiana) were grown in greenhouse on a cycle of 14 h light at 24 °C and 10 h dark at 20 °C. Approximately 0.2 g fresh TBSV systemically infected leaf tissue was homogenized with kieselguhr in 20 mL distilled-water to prepare the virus inoculum. Then the leaves of tobacco were rub-inoculated with the sap. The chemicals dissolved in DMSO were diluted in water to the concentration of 150 mg L−1 and 0.05% pesticide synergist was added which can help enhance the spreadability and penetration of chemicals. The same compositions except chemical were also added to solution used for control group. Subsequently, the prepared solutions were atomizing sprayed on the entire inoculated plants within 1 hour post inoculation and again at 1, 3 days post inoculation (dpi). The antiviral activities of inhibitors were evaluated by disease severity index and quantitative PCR.Plant disease severity was graded by using a 0–4 rating scale. 0 = no disease symptom, 1 = slight symptoms on inoculated leaves, 2 = vein clearing on one systemic leave, 3 = vein clearing on two or more upper leaves, 4 = the whole plant necrosis. And disease severity index was calculated by the following formula:
Total RNAs were extracted from three individual inoculated leaves at different time points after inoculation. Then the virus replication levels and gene expression levels were measured by quantitative real-time PCR analysis and the primers were shown in Table S2.† Relative quantitation of the target gene expression level was performed using the comparative Ct method. Each experiment was analyzed with three technical replications. Elongation factor 1α (EF-1α) gene was used as an internal control.
Results
Molecular docking
By comparing with the available crystal structures of P19-siRNA, 4JGN, a complete short loop structure in N-terminal near siRNA binding sites and diffracted to 1.86 Å resolution without any mutations, was selected to screen potential inhibitors.Based on the studies about P19–siRNA interaction,23,24,31,32 13 key residues were selected and divided into four groups: (A) TRP17, TRP20, LYS38, LYS49; (B) ARG50, ARG53, CYS88; (C) PRO15, ARG93, SER102; (D) SER40, SER91, SER98. Among these, the key residues in group A can interact with siRNA directly, the mutations of these residues would result in decrease of the lethal necrosis phenotype in viral-infected plants. While the residues in group B are important in keeping structural stable of P19, thus affect the P19–siRNA interaction indirectly. The residues in group C and D are located in the siRNA binding domain of P19 and have interactions with siRNA. But substitutions of these residues can not affect P19 severely. In addition, the residues in group C are conserved among P19 family whereas residues in group D are not. Thus, the importance order of the residues group is: A ≥ B > C > D.
Virtual screening was performed using the following steps (Fig. 2A). All 157![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 026 compounds in the library were docked into the binding pocket (Fig. 2B) of prepared P19 model using AutoDock Vina (version: 1.1.2).26 Docking calculation was performed by AutoDock Vina.26 Briefly, the scoring function, which combines certain advantages of empirical and knowledge-based scoring functions, consists of several main terms including hydrophobic, hydrogen bonding and steric interactions. Based on the calculated free energy of binding, the top 200 ranked compounds were selected. Then the binding models of these 200 compounds were manually checked according to the following criteria: (1) forming at least one pi–pi interaction or stacking interaction with TRP17 or TRP20, (2) forming at least two hydrogen bonds with residues in group A and B, (3) well occupy the defined cavity of P19, (4) availability. Finally, 18 compounds were selected for experimental screening (for details, see the ESI†). As shown in Table S1,† the binding affinity calculated for all these 18 ligands binding with P19 range from −8.7 to −10.6 kcal mol−1.
026 compounds in the library were docked into the binding pocket (Fig. 2B) of prepared P19 model using AutoDock Vina (version: 1.1.2).26 Docking calculation was performed by AutoDock Vina.26 Briefly, the scoring function, which combines certain advantages of empirical and knowledge-based scoring functions, consists of several main terms including hydrophobic, hydrogen bonding and steric interactions. Based on the calculated free energy of binding, the top 200 ranked compounds were selected. Then the binding models of these 200 compounds were manually checked according to the following criteria: (1) forming at least one pi–pi interaction or stacking interaction with TRP17 or TRP20, (2) forming at least two hydrogen bonds with residues in group A and B, (3) well occupy the defined cavity of P19, (4) availability. Finally, 18 compounds were selected for experimental screening (for details, see the ESI†). As shown in Table S1,† the binding affinity calculated for all these 18 ligands binding with P19 range from −8.7 to −10.6 kcal mol−1.
Experimental screening
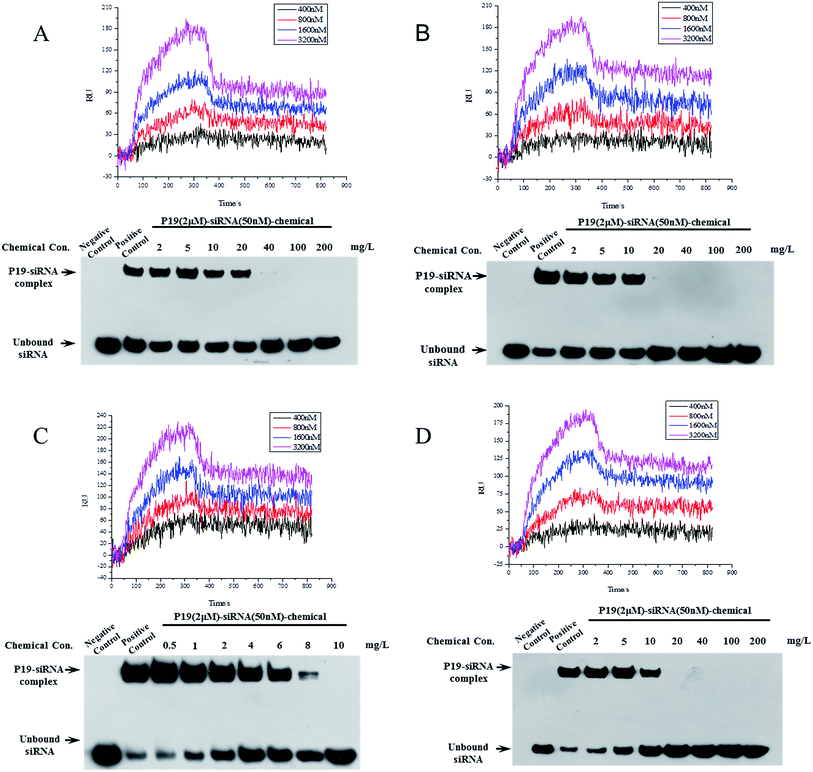
Eighteen candidate compounds were purchased from ChemDiv and the binding ability to P19 were tested using a SPRi method. The result of protein P19 (19 kDa) purification was shown in Fig. S1.† All compounds were dissolved in DMSO (10 mM) and spotted in a PCL chip. DMSO and rapamycin were spotted as control and FKBP12 was used to test the PCL chip (Fig. S2†). DMSO and surface background were taken as negative control to subtract noise signal. Different concentrations of P19 diluted in PBS were used as analyte. Among the candidates, four compounds showed relatively high binding resonance unit (RU) values were considered to have potential P19-binding activity. Detailed kinetic parameters of these four candidates including association rate constants (Ka), dissociation rate constants (Kd) and equilibrium dissociation constant (KD) were calculated and presented in Table 1. KD values were calculated by the equation: KD = Kd/Ka, and the smaller KD values represent higher binding affinity. The smallest KD value was 81.2 nM for compound VS14. Then these four compounds were selected to test their inhibition of association between P19 and siRNA by Electrophoretic mobility shift assay (EMSA) (Fig. 3).| Compound | K a (1/Ms) | K d (1/s) | K D (nM) |
|---|---|---|---|
| VS2 | 2.46 × 103 | 3.35 × 10−4 | 136.2 |
| VS12 | 1.72 × 103 | 1.92 × 10−4 | 111.6 |
| VS14 | 3.56 × 103 | 2.89 × 10−4 | 81.2 |
| VS15 | 2.49 × 103 | 3.1 × 10−4 | 124.5 |
In EMSA experiment, different concentrations of chemicals and constant amount of P19 (2 μM) were used to test if these inhibitors are dose-dependent. As shown in Fig. 3, negative control represented mixture running buffer only contained 50 nM siRNA whereas mixture buffer in lane positive control contained 50 nM siRNA and 2 μM P19. In lane positive control, the measured P19–siRNA complex revealed that P19 was able to sequester siRNA. The following lanes represented increasing concentrations of chemicals were added in mixture buffers contained constant amounts of siRNA and P19, which resulted in corresponding reduced P19–siRNA complexes. As shown in Fig. 3, the molecular ratio of P19–siRNA complex and siRNA was 3.62![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for VS14 in positive lane, as the concentration of added chemical increased, the ratio reduced correspondingly. The ratio were 3.03, 0.99, 0.34 for the added VS14 concentration of 1, 4, 8 mg L−1 respectively and the ratio was close to zero in the concentration lane of 10 mg L−1. For VS15, the molecular ratio of P19–siRNA complex and siRNA was 2.93 in positive lane while the ratio were 2.76, 1.75, 0.67 for the added chemical concentration of 2, 5, 10 mg L−1 respectively. For VS12, the molecular ratio was 1.74 in positive lane while the ratio were 1.08, 0.9, 0.83 for the added chemical concentration of 2, 5, 10 mg L−1 respectively, and in the concentration lane of 20 mg L−1 the ratio was approximately 0.01. For VS2, the molecular ratio was 0.56 in positive lane while the ratio were 0.7, 0.59, 0.53 for the added chemical concentration of 2, 10, 20 mg L−1 respectively, and in the concentration lane of 40 mg L−1 the ratio was approximately 0.01. These results indicate that these candidates could inhibit the binding between P19 and siRNA in a dose-dependent manner. All these four compounds suppressed P19–siRNA interaction with concentration values less than 100 mg L−1. Among which, VS14 completely suppressed P19–siRNA interactions at 10 mg L−1. Both VS12 and VS15 showed strong inhibitions against P19–siRNA complex at 20 mg L−1.
1 for VS14 in positive lane, as the concentration of added chemical increased, the ratio reduced correspondingly. The ratio were 3.03, 0.99, 0.34 for the added VS14 concentration of 1, 4, 8 mg L−1 respectively and the ratio was close to zero in the concentration lane of 10 mg L−1. For VS15, the molecular ratio of P19–siRNA complex and siRNA was 2.93 in positive lane while the ratio were 2.76, 1.75, 0.67 for the added chemical concentration of 2, 5, 10 mg L−1 respectively. For VS12, the molecular ratio was 1.74 in positive lane while the ratio were 1.08, 0.9, 0.83 for the added chemical concentration of 2, 5, 10 mg L−1 respectively, and in the concentration lane of 20 mg L−1 the ratio was approximately 0.01. For VS2, the molecular ratio was 0.56 in positive lane while the ratio were 0.7, 0.59, 0.53 for the added chemical concentration of 2, 10, 20 mg L−1 respectively, and in the concentration lane of 40 mg L−1 the ratio was approximately 0.01. These results indicate that these candidates could inhibit the binding between P19 and siRNA in a dose-dependent manner. All these four compounds suppressed P19–siRNA interaction with concentration values less than 100 mg L−1. Among which, VS14 completely suppressed P19–siRNA interactions at 10 mg L−1. Both VS12 and VS15 showed strong inhibitions against P19–siRNA complex at 20 mg L−1.
Binding modes of inhibitors
Based on the dock conformation, the binding modes of all 4 chemicals with P19 were shown in Fig. 4. Markedly, all of the hits have several hydrogen bonds interactions with residues of P19. Moreover, they all have interactions with Trp20 or Trp17, which are the key residues in P19 for siRNA binding.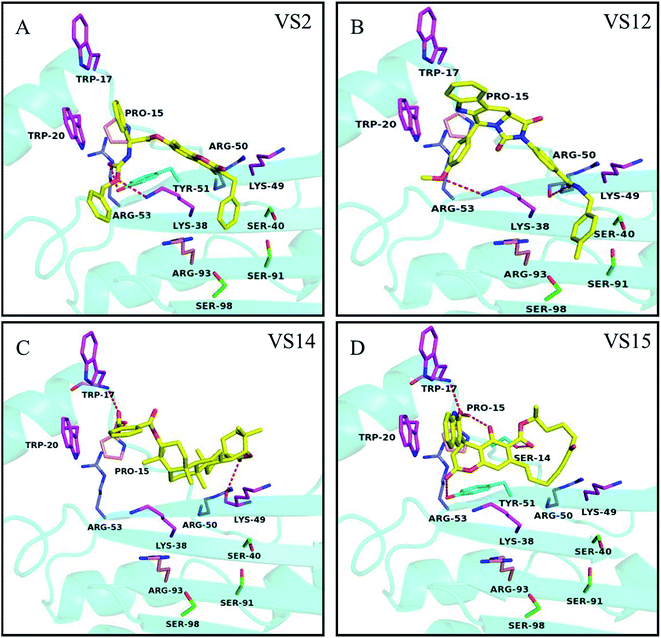 | ||
| Fig. 4 Potential binding modes of inhibitors (A) VS2, (B) VS12, (C) VS14 and (D) VS15 docked to the protein structures P19 (4JGN). Compounds are shown as yellow sticks. The key residues in group A, B, C and D are displayed as magenta, purple, pink and green, respectively. Hydrogen bonding interactions are represented as orange dotted lines. | ||
In the binding of VS2 to P19, the oxygen of carbonyl and the other oxygen of ester in VS2 create hydrogen bonds with residues Tyr51 and Lys38, respectively (Fig. 4A). Notably, the pi–pi interaction between the benzene ring and residue Trp20 was supposed to be the major interaction. And the similar pi–pi interaction between residue Trp20 and aromatic ring can be also viewed in VS15. In addition, different oxygen atoms of carbonyl in VS15 form four respective hydrogen bonds with Trp17, Pro15, Ser14 and Tyr51 (Fig. 4D). The inhibition activity of VS12 is similar to that of VS15, but the binding patterns are somehow different. VS12 have both pi–pi and stacking interactions with residue Trp20. Besides, the other two hydrogen bonds between VS12 and P19 are the oxygen of carbonyl with residue Arg50 and the oxygen of anisole with residue Lys38 (Fig. 4B). The activities of VS12 and VS15 were 2 fold increment compared to that of VS2. Both of them have one pi–pi interaction and at least three other interactions with P19. It seemed that more interactions with P19 can increase the chemicals' activity significantly. Among these chemicals, VS14 showed the highest activity of inhibition. Three hydrogen bonds interacting with the residues Lys49, Pro15 and Trp17 were observed. Also, the aromatic ring can form pi–pi interaction with Trp20 (Fig. 4C). The high activity of VS14 may attribute to that it has interactions with three critical residues in analyzed key residues group A.
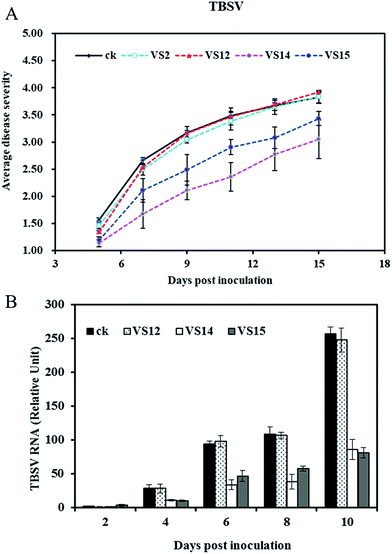
Antiviral effects of inhibitors in vivo
The antiviral activities of inhibitors against TBSV in vivo were evaluated via disease severity index calculation and quantitative PCR method. The average disease severity with treatment by VS2, VS12, VS14, VS15 are shown in Fig. 5A. Obviously, VS14 and VS15 were effective in reducing disease symptom whereas VS2 and VS12 showed relatively lower efficacy. After 9 dpi, the plants in control group displayed necrosis while treatment of VS14 and VS15 significantly decreased this lethal necrosis phenotype. The disease severity indexes in 11 dpi are listed in Table 2. Among these inhibitors, VS14 exhibited the best antiviral activity against TBSV with the inhibition rate of 32.35%. The inhibition rates of VS15, VS2 and VS12 were 16.61%, 3.31% and 2.04% respectively (inhibition rate > 0). It should be noted that most viral-infected plants in water control group showed lethal necrosis whereas some of that treated with VS14 and VS15 just showed slight symptoms in 13 dpi.| Compound | Disease severity index | Inhibition rate (%) |
|---|---|---|
| Control (water) | 87.17 ± 2.02 | — |
| VS2 | 84.60 ± 4.10 | 3.31 ± 4.68 |
| VS12 | 86.71 ± 3.94 | 2.04 ± 2.89 |
| VS14 | 58.97 ± 6.60 | 32.35 ± 7.30 |
| VS15 | 72.69 ± 3.53 | 16.61 ± 3.37 |
The viral accumulation levels at different days post inoculation were measured by quantitative RT-PCR. As shown in Fig. 5B, both treatments with VS14 and VS15 resulted in less virus accumulation in inoculated leaves than that of water control. Viral-infected plants in group water control and VS12 had a similar variation trend in viral RNA levels. These results are consistent with disease severity visual observed. (The viral accumulation levels of treatment with VS2 was not determined because of their similar symptom developments with control group and VS12). Taken together, all these results indicate that inhibitors VS14 and VS15 are effective in reducing viral accumulation levels in the inoculated leaves and attenuating system symptoms.
Discussion
Since RNA silencing has been seemed as an important antiviral defense mechanism, various viral silencing suppressors have been identified consistently. The crystal structures of some suppressors also have been characterized, such as 2b and P19.23,24,33 Both the crystal structures of 2b and P19 elucidate the binding modes between dsRNA and proteins. And this dsRNA binding ability is speculated to be a common strategy to counter antiviral silencing for RSSs.20–22 Tomato aspermy virus (TAV) 2b forms a homodimer with two long α-helices linked by a short linker. This homodimer has a pair of hook-like structures to bind to the siRNA duplex at its major groove.33 P19 also acts as a dimer to sequester siRNA duplex by providing stacking interactions between two conserved tryptophan residues from α helix and terminal bases of siRNA, and by forming β sheet concave binding surface to interact with the phosphates and sugar 2′-hydroxyls of siRNA.23 Previous studies showed some critical residues in dsRNA-binding ability of P19. For instance, mutations on the two tryptophan residues W39 and W42 obviously attenuate the lethal necrosis phenotype in virus-infected plants, as these two residues are the critical amino acids for P19 to recognize terminals of dsRNA.23 This reduced silencing suppression ability of P19 mutations was also confirmed by agro-infiltration. Two other mutations in residues K60 and K71, which are involved in salt bridges to phosphate backbone of RNA, also reduce the necrosis phenotype in hosts.23 A few residues were not selected, although they may also be responsible for inhibition, their location far from the siRNA-binding sites makes them less likely to interact with candidate inhibitors in this study (e.g. K67, Q107). Then the chosen analyzed key residues were used to define the binding pocket for molecular docking and be divided into four groups according to their importance in the interaction between P19 and siRNA. These four residue-groups would be crucial criteria in the binding mode checking. By analyzing the available X-ray structures of P19–siRNA complex, we found that one file (PDB ID 4JGN) has a relatively complete structure in N-terminal loop that near the RNA-binding domain. Moreover, the 4JGN complex has a high-resolution at 1.86 Å which is also important for virtual screening. The corresponding key residues in 4JGN to that of 1RPU complex are listed in Table S3.†Applying a similar screen procedure described before,28 we screened a natural product database and purchased 18 selected compounds for experimental validation by using a SPRi method. The combined small molecule microarray (SMM) and SPRi method was developed to screen lead compounds against drug targets recently.34 In comparison with the SPR method applying in the previous study,14 siRNA was not used in the primary experiment because the selected 18 candidate compounds are more specific to RSS-siRNA interaction and it would be more convenient and cost-effective to observe the direct interactions between protein and compounds. Another advantage of SPRi is high-throughput, allowing the parallel evaluation of hundreds or thousands of compounds simultaneously. After this label-free experimental screening of candidates, four possible inhibitors were obtained.
Suppressors inhibit RNA silencing by targeting different steps, such as dicing, RISC assembly, secondary siRNA production. It has been suggested that P19 can bind to and sequester viral siRNAs, and thus to evade host RNA silencing defense in infected plants. To test whether these inhibitors actually work through this pathway, the inhibition activities of these inhibitors on the association between P19 and siRNA were confirmed by EMSA. In the preliminary EMSA experiment, we used 1 nM dsRNA and different concentrations of P19 in the binding reaction, but the processed nylon membrane cannot show any signal. Then we increased the concentrations of dsRNA and P19 gradually, at last we found that 50 nM dsRNA and 2 μM P19 (approximately 1000 ng) could be appropriate in this test. As shown in Fig. 3, when increasing amount of compounds were added to the binding reactions which have a constant concentration of P19 and dsRNA, the detected complexes of P19–siRNA correspondingly reduced. These results suggest the selected inhibitors can inhibit the binding between P19 and siRNA in a concentration-dependent manner. Furthermore, we tested the inhibition activities of previously reported RSS inhibitors croconic acid and 1,4-benzoquinon,14,35 and found that both compounds showed complete inhibition in concentration of 200 mg L−1 (Fig. S3†) Obviously, the inhibitors screened by our strategy have stronger inhibition activity than the tested known inhibitors. This can be attributed to the high activity and more specific function way of inhibitors selected by virtual screening.
The possible interaction mechanism was further analyzed according to the potential binding modes of identified inhibitors. As mentioned above, one of the important criteria in manual analysis was to check if the candidates have interactions with the key residues TRP17 or TRP20 of P19 (refer to W39 and W42 in 1RPU, respectively). Among these inhibitors, VS14, which has pi–pi interaction with Trp20 and hydrogen bond interaction with TRP17, exhibited strong inhibition activity. But inhibitor VS15 which also has interactions with TRP17 and TRP20 showed relatively weaker activity. Several candidates which have interactions with TRP17 and TRP20 were selected in manual checking, but only two of them can inhibit P19 in vitro. It seemed that dual interactions with this two critical residues viewed in the binding mode may not be a key factor affecting compounds' inhibition activity. We have to point out that although criteria applied in manual checking step increase success rate of virtual screening notably, it still have some limitations. For instance, all selected candidates in our study have at least one pi–pi interaction or stacking interaction with TRP17 or TRP20, and have a relatively similar mechanism of action although they have different scaffolds. Unlike P19 inhibitors identified previously,15 all compounds selected in our study do not have thio-sulfonate functional groups which are reactive toward cysteine residues. Hence our candidates would not affect P19 activity through covalent modification of cysteine residues.
Among the identified inhibitors, VS14 and VS15 displayed relatively strong activities in attenuating viral symptoms and reducing viral accumulation. Comparing the chemical structures and inhibition effects of VS14, VS15, VS12 and VS2, we deduced that the key factor might be the ester group in VS14, which can be hydrolyzed in plant cells by the endogenous plant esterase, releasing phthalic acid and triptolide-like compounds. Phthalic acid is a kind of benzoic acid, which might play an important role in the precursor of salicylic acid, which is a key endogenous component of local and systemic disease resistance in plants.36 Triptolide possesses antitumor properties, which is an inhibitor of RNA polymerase I and II-dependent transcription leading predominantly to down-regulation of short-lived mRNA.37 VS15 contains a macrocyclic lactone structure, which have antibiotic activity.38 As for V12 and VS2, the amide structure may not facilitate the antiviral effect. However, the antiviral effects of these inhibitors in plant were not as good as predicted. Therefore, the dosage amount, dosage form and the spray times may need more optimization. Inspection of RSS-dsRNA crystal structures provides new insights in discovering RSS inhibitors and analyzing their functional mechanism. Considering that many RSSs show a common pattern to bind siRNA,20–22 it raises the possibility that inhibitors of P19 can also suppress other RSSs. That means P19 inhibitors identified in this study may have inhibition activity on other RSSs and can be used to treat many different plant viruses. Moreover, to manage viruses based on the mechanism of RNA silencing probably would be a focus of future research. In medicine, a recent study has introduced an RNAi-based high-throughput method to screen inhibitors against X protein which is the silencing suppressor of hepatitis B virus and the identified inhibitor can efficiently suppress hepatitis B virus replication in culture medium.39 In agriculture, several interesting studies have been conducted on topical application of dsRNA for plant virus control.40,41 Spraying dsRNA, which is the elicitor of RNA silencing, on viral-infected plants can lead to degradation of homologous viral RNA. However, limitations still exist (e.g. cost and instability). Therefore, combined utilization of dsRNA and RSS inhibitors would be a better choice.
Conclusions
In summary, we have developed a strategy to discover novel antiviral agents targeted RNA silencing suppressors. With the combination of virtual screen and bio-assays, several inhibitors were identified. The success rate of about 22% (4 out of 18 virtual screened small molecules) and the strong inhibition effectiveness both verify this strategy. Furthermore, this strategy may be also applicable for animal viruses which encode suppressors to counter PTGS. This study would provide new insights on the development of pesticides and help resolving the problems faced by genetically resistant crops.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was supported by the National Key Research and Development Program (2016YFC1200605).References
- L. C. Galvez, J. Banerjee, H. Pinar and A. Mitra, Plant Sci., 2014, 228, 11–25 CrossRef CAS.
- S. Whitham, S. P. Dinesh-Kumar, D. Choi, R. Hehl, C. Corr and B. Baker, Cell, 1994, 78, 1101–1115 CrossRef CAS.
- H. Takahashi, J. Miller, Y. Nozaki, Sukamto, M. Takeda, J. Shah, S. Hase, M. Ikegami, Y. Ehara and S. P. Dinesh-Kumar, Plant J., 2002, 32, 655–667 CrossRef CAS.
- F. Faoro and F. Gozzo, Plant Sci., 2015, 234, 1–13 CrossRef CAS.
- C. E. Wong, R. A. J. Carson and J. P. Carr, Mol. Plant-Microbe Interact., 2002, 15, 75–81 CrossRef CAS.
- L. Friedrich, K. Lawton, W. Ruess, P. Masner, N. Specker, M. G. Rella, B. Meier, S. Dincher, T. Staub, S. Uknes, J.-P. Métraux, H. Kessmann and J. Ryals, Plant J., 1996, 10, 61–70 CrossRef CAS.
- D. Baulcombe, Nature, 2004, 431, 356–363 CrossRef CAS PubMed.
- S. W. Ding, Nat. Rev. Immunol., 2010, 10, 632–644 CrossRef CAS.
- W. X. Li and S. W. Ding, Curr. Opin. Biotechnol., 2001, 12, 150–154 CrossRef CAS.
- J. Burgyan and Z. Havelda, Trends Plant Sci., 2011, 16, 265–272 CrossRef CAS.
- A. J. Soards, A. M. Murphy, P. Palukaitis and J. P. Carr, Mol. Plant-Microbe Interact., 2002, 15, 647–653 CrossRef CAS PubMed.
- H. Ziebell, T. Payne, J. O. Berry, J. A. Walsh and J. P. Carr, J. Gen. Virol., 2007, 88, 2862–2871 CrossRef CAS PubMed.
- G. Szittya, A. Molnár, D. Silhavy, C. Hornyik and J. Burgyán, Plant Cell, 2002, 14, 359–372 CrossRef CAS.
- H. Shimura, T. Fukagawa, A. Meguro, H. Yamada, M. Oh-Hira, S. Sano and C. Masuta, FEBS Lett., 2008, 582, 4047–4052 CrossRef CAS.
- S. M. Sagan, R. Koukiekolo, E. Rodgers, N. K. Goto and J. P. Pezacki, Angew. Chem., 2007, 119, 2051–2055 CrossRef.
- D. L. Ma, D. S. Chan and C. H. Leung, Chem. Soc. Rev., 2013, 42, 2130–2141 RSC.
- G.-H. Ma, Y. Ye, D. Zhang, X. Xu, P. Si, J.-L. Peng, Y.-L. Xiao, R.-Y. Cao, Y.-L. Yin, J. Chen, L.-X. Zhao, Y. Zhou, W. Zhong, H. Liu, X.-M. Luo, L.-L. Chen and X. Shen, Eur. J. Med. Chem., 2016, 124, 981–991 CrossRef CAS PubMed.
- T. T. Yao, S. W. Fang, Z. S. Li, D. X. Xiao, J. L. Cheng, H. Z. Ying, Y. J. Du, J. H. Zhao and X. W. Dong, J. Agric. Food Chem., 2017, 65, 3204–3211 CrossRef CAS.
- W. Qiu, J.-W. Park and H. B. Scholthof, Mol. Plant-Microbe Interact., 2002, 15, 269–280 CrossRef CAS.
- F. Li and S. W. Ding, Annu. Rev. Microbiol., 2006, 60, 503–531 CrossRef CAS.
- Z. Mérai, Z. Kerényi, S. Kertész, M. Magna, L. Lakatos and D. Silhavy, J. Virol., 2006, 80, 5747–5756 CrossRef.
- L. Lakatos, T. Csorba, V. Pantaleo, E. J. Chapman, J. C. Carrington, Y. P. Liu, V. V. Dolja, L. F. Calvino, J. J. López-Moya and J. Burgyán, EMBO J., 2006, 25, 2768–2780 CrossRef CAS.
- J. M. Vargason, G. Szittya, J. Burgyán and T. M. T. Hall, Cell, 2003, 115, 799–811 CrossRef CAS PubMed.
- K. Ye, L. Malinina and D. J. Patel, Nature, 2003, 426, 874–878 CrossRef CAS PubMed.
- Schrödinger, LLC, http://www.pymol.org/, 2010.
- O. Trott and A. J. Olson, J. Comput. Chem., 2010, 31, 455–461 CAS.
- M. Weisel, E. Proschak and G. Schneider, Chem. Cent. J., 2007, 1, 7 CrossRef PubMed.
- R. Guo, Y. Zhang, X. Li, X. Song, D. Li and Y. Zhao, J. Mol. Model., 2016, 22, 135 CrossRef.
- N. Kanoh, M. Kyo, K. Inamori, A. Ando, A. Asami, A. Nakao and H. Osada, Anal. Chem., 2006, 78, 2226–2230 CrossRef CAS PubMed.
- V. Singh and A. Nand, RSC Adv., 2015, 5, 87259–87265 RSC.
- J. Cheng, R. Koukiekolo, K. Kieliszkiewicz, S. M. Sagan and J. P. Pezacki, Biochim. Biophys. Acta, 2009, 1794, 1197–1203 CrossRef CAS PubMed.
- Y.-C. Hsieh, R. T. Omarov and H. B. Scholthof, J. Virol., 2009, 83, 2188–2200 CrossRef CAS.
- H. Y. Chen, J. Yang, C. Lin and Y. A. Yuan, EMBO Rep., 2008, 9, 754–760 CrossRef CAS PubMed.
- V. Singh, K. Singh, A. Nand, H. Dai, J. Wang, L. Zhang, A. Merino and J. Zhu, Arabian J. Chem., 2015 DOI:10.1016/j.arabjc.2014.12.020.
- S. Sano, T. Fukagawa, H. Yamada, C. Masuta and H. Shimura, U.S. Pat., 8,680,158, 2014.
- J. Chong, M.-A. Pierrel, R. Atanassova, D. Werck-Reichhart, B. Fritig and P. Saindrenan, Plant Physiol., 2001, 125, 318–328 CrossRef CAS.
- S. Vispé, L. DeVries, L. Créancier, J. Besse, S. Bréand, D. J. Hobson, J. Q. Svejstrup, J.-P. Annereau, D. Cussac, C. Dumontet, N. Guilbaud, J.-M. Barret and C. Bailly, Mol. Cancer Ther., 2009, 8, 2780–2790 CrossRef PubMed.
- X. Li, S. Vanner, W. Wang, Y. Li, V. A. Gallardo and N. A. Magarvey, J. Antibiot., 2013, 66, 443 CrossRef CAS.
- S. Ghosh, A. Kaushik, S. Khurana, A. Varshney, A. K. Singh, P. Dahiya, J. K. Thakur, S. K. Sarin, D. Gupta, P. Malhotra, S. K. Mukherjee and R. K. Bhatnagar, J. Biol. Chem., 2017, 292, 12577–12588 CrossRef CAS PubMed.
- D. Gan, J. Zhang, H. Jiang, T. Jiang, S. Zhu and B. Cheng, Plant Cell Rep., 2010, 29, 1261–1268 CrossRef CAS PubMed.
- N. Mitter, E. A. Worrall, K. E. Robinson, P. Li, R. G. Jain, C. Taochy, S. J. Fletcher, B. J. Carroll, G. Q. Lu and Z. P. Xu, Nat. Plants, 2017, 3, 16207 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra01311j |
| ‡ These authors contributed equally to this study. |
| This journal is © The Royal Society of Chemistry 2018 |