Open Access Article
Open Access ArticleSelection of aptamers for AMACR detection from DNA libraries with different primers†
Deng-Kai Yang ab,
Chia-Fu Choub and
Lin-Chi Chen
ab,
Chia-Fu Choub and
Lin-Chi Chen *a
*a
aDepartment of Bio-Industrial Mechatronics Engineering, National Taiwan University, No. 1 Roosevelt Road Section 4, Taipei, Taiwan 10617, R. O. C. E-mail: chenlinchi@ntu.edu.tw; Fax: +886-2-2362-7620; Tel: +886-2-3366-5343
bInstitute of Physics, Academia Sinica, 128 Academia Rd. Sec. 2, Nangang District, Taipei, Taiwan 11574, R. O. C
First published on 23rd May 2018
Abstract
In this work, we investigated aptamer selection against alpha-methylacyl-CoA racemase (AMACR) with three DNA libraries bearing different primers. Because of increased selection diversity, aptamers with varied structural properties, such as pre-folded, induced-folding, high and low primer-dependent conformations, were discovered. From the selection results, a dimeric aptamer was constructed and capable of detecting over-expression of AMACR in prostate cancer cell lines. In summary, this study demonstrates a library-based approach to obtain aptamers with different binding properties and provides distinct aptamers for flexible design of AMACR detection.
Introduction
Aptamers are synthetic oligonucleotides or peptides that can bind to targets with high specificity and affinity, and they have been widely studied as antibody surrogates for diagnostic1–3 and therapeutic4–6 applications since the 1990s. Compared with antibodies, aptamers have a number of advantages, such as low molecular weight, high chemical stability, high compatibility with various chemical modifications, and so on.7,8 In addition, aptamers are generated by a combinatorial chemistry-based in vitro ligand selection process called systematic evolution of ligands by exponential enrichment (SELEX),9,10 which has a considerably lower cost than that of monoclonal antibody production with the hybridoma technique or phage display. Although aptamers have become as an emerging molecular strategy for both biomarker detection and drug delivery system developments, a typical SELEX approach is not efficient to generate aptamer variants with distinct binding properties (e.g., different target binding epitopes or different conformations). Thus, there are both needs and interests to study an effective approach to generate distinct aptamers for a single target, since this will allow to construct a multivalent aptamer resembling a multivalent antibody for improving targeting efficiency and also allow flexible molecular sensing design. Because aptamers are selected from the library of SELEX, a rational library design is considered to play a crucial role in the discovery of distinct aptamers.Taking a nucleic acid aptamer selection as an example, lengthening the random sequence region is the easiest way to increase the diversity of the library (diversity = 4N, N is the number of bases of the random sequence region), but the method is limited due to the low yields of lengthier sequences in chemical synthesis11 and poor PCR efficiency. Moreover, compared with proteins having 20 different standard amino acids, the diversity of DNA aptamers is also constrained by the composition of only four different nucleotides.11 Thus, many researchers have been endeavouring to break through the diversity limit of the nucleic acid library. To bridge the diversity gap, Gold and his team used various unnatural nucleotides modified with protein-like side chains, such as benzyl (Bn), iso-butyl (iBu), R-2-hydroxypropyl (Thr) and so on, in libraries for aptamer screening.12 Their method remarkably increased not only the diversity but also the chances to find aptamers with high affinity and specificity for target molecules. Kimoto et al. reported another advancement of nucleic acid library design.13 They devised a random library containing the four natural nucleotides and an unnatural nucleotide with hydrophobic base 7-(2-thienyl)imidazo[4,5-b]pyridine (Ds). This library design increased the chemical and structural diversity of the library and demonstrated the role of hydrophobic modification in improving the affinities of aptamers for their targets. Also, an unnatural nucleic acid, called xeno nucleic acid (XNA), was reported.14 The deoxyribose sugar of nucleotides was substituted by an unnatural sugar moiety, such as arabinose, threose, cyclohexane and so on. Holliger and his team successfully in vitro replicated XNA with specially mutated polymerases.15 They also introduced XNA in a SELEX library and generated XNA-containing aptamers.16 Besides, some researchers also attempted to find a method that could enhance both the sequence diversity and structural complexity of a SELEX library.11,17
In principle, the fixed regions (i.e., primers) of the library sequences also affect the selection diversity through a structural effect. For instance, some research groups introduced predetermined stem-loop or other defined motifs into the library sequences to generate aptamer for different purposes, such as self-reporting or structure-switching aptamers.18–20 These studies implied that the fixed regions of the library sequences could lead the evolution direction of aptamers. This is because that the fixed regions, which play roles of priming sites for PCR amplification of the selected DNA ligands, can influence the conformations of SELEX library sequences through base pairing with the random sequence region. Moreover, the primer regions are known to potentially involve in target binding and to lead to positive or negative effects on aptamer discovery.21–23 Therefore, post-SELEX optimization is often required to determine the primer-independent, minimal core sequence of an as-selected aptamer. This is especially needed for developing an effective aptasensor probe or multivalent aptamer construct for specific biomarker detection.
In this work, we investigate the primer effect on the SELEX process for the discovery of aptamers for cancer biomarker detection. Considering that prostate cancer is one of the most frequently diagnosed cancers24 and is the first leading cause of new cancer cases and the second leading cause of cancer deaths in males in the United States in 2013,25 we chose alpha-methylacyl-CoA racemase (AMACR, also known as P504S in cancer literature) as the SELEX target. It is because that AMACR has been proven to be highly expressed in prostate cancer cells and has become a novel protein biomarker for prostate cancer diagnosis.26–29 For enhancing the chance of discovery of distinct anti-AMACR aptamers, we performed SELEX with three DNA libraries (Library A,23 Library B,30 and Library C31) bearing different primers. The three different libraries were used to select aptamers against AMACR in parallel. The single-bead SELEX method was adopted in this study32 (shown in Scheme 1). The real-time quantitative PCR (qPCR), circular dichroism (CD) and enzyme-linked aptamer assay (ELAA) were used to monitor the selection process and to evaluate the target binding properties.
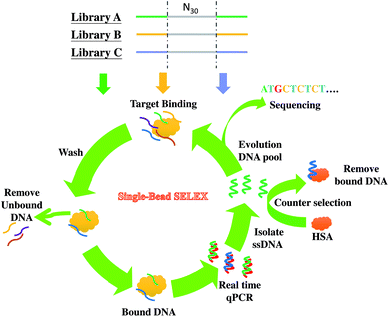 | ||
| Scheme 1 Selection of anti-AMACR aptamers from three DNA libraries (N30) with different primers by single-bead SELEX. | ||
Experimental section
Preparation of protein-coated glass beads
Epoxide-functionalized glass beads (EGBs) with 500 microns diameter were used as solid supports for protein immobilization, and AMACR-coated EGBs were prepared according to our previous report.32 A similar condition was adopted to prepare the HSA-coated EGBs (300 mg EGBs incubated in 1 mL 3% HSA solution), for the counter selection and a control experiment in the SELEX process. Before SELEX, the AMACR-coated and HSA-coated EGBs were blocked with glycine solution (ca. 75 mg glycine dissolved in 1 mL of selection buffer, 50 mM Tris–HCl, 100 mM NaCl, 4 mM KCl, 2.5 mM MgCl2 and 1 mM CaCl2, pH8) at room temperature for 2 hours. Then, the beads were washed with the selection buffer and stored at 4 °C before use.SELEX with three different DNA libraries
Three kinds of DNA libraries (A: 69 mer, B: 72 mer, and C: 61 mer) were used in parallel to screen AMACR aptamers. The SELEX process is illustrated in Scheme 1. The libraries all included 30 randomized nucleotides (N30) flanked by two primer binding sequences for PCR, and the sequences are listed in ESI Table S1.† The SELEX process was referred to our previous research,32 but the pH values of selection and washing buffer were changed to pH 8 to decrease non-specific interactions. Besides, the NaCl concentration of the wash buffer was replaced by 100 mM NaCl for the 1st and 2nd SELEX rounds, 150 mM NaCl for the 3rd and 4th rounds, and 200 mM NaCl for the 5th round. All the DNA sequences bound on protein-coated beads were amplified and simultaneously quantified using a Qiagen q-PCR machine (Rotor-Gene®Q). Moreover, qPCR analyses from the non-specific DNA binding on HSA were done for comparison. After several rounds of SELEX, the winning pools from the three libraries were harvested, cloned, and sequenced.Sequence alignment and secondary structure prediction
After SELEX, 92 clones (28 clones from Library A, 33 clones from Library B, and 31 clones from Library C) were prepared and sequenced. ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/) was used to align these DNA sequences. From the sequence alignment analysis, two or three groups with higher alignment scores were identified, and one of each group was randomly picked up as the representative aptamer candidates. The RNAstructure Version 5.6 (http://rna.urmc.rochester.edu/index.html) was used to predict the secondary structures of the aptamer candidates.Circular dichroism characterization
The CD spectra were recorded at 4 °C in a 0.1 cm path length cuvette. The parameters were set as follows: sensitivity 100 mdeg, data pitch 1 nm, response time 1 second, bandwidth 1 nm, and scanning mode continuous scanning with a scanning speed of 50 nm min−1. The reported CD spectra were averaged over three scans, from wavelengths of 320 to 200 nm. To observe the similarity of sequence compositions of Library A, Library B, and Library C. The differential CD melting curves of the random sequence in each library were measured by recording the molar ellipticity value at 275 nm along with an increasing temperature, a temperature gradient from 20 to 95 °C at a rate of 1 °C min−1.Validation of the anti-AMACR aptamer candidates
Enzyme-linked aptamer assays (ELAAs) with standard black opaque 96-well polystyrene microtiter plates were carried out for the assessment of the affinity and specificity of the selected AMACR aptamers. Serially diluted concentrations of aptamers were tested with a constant protein coating concentration (0.2 μM) for the affinity measurement, and 0.5 μM aptamers were used to detect serially diluted protein coating concentrations for the specificity measurement. For protein coating, AMACR diluted in PBS buffer (pH 7.4) was added to the wells of the plates (100 μL per well) and incubated at 4 °C overnight. After incubation, the wells were washed three times with 200 μL per well of selection buffer containing 0.05% Tween 20. For blocking, each AMACR-coated well was added with 200 μL of 3 wt% BSA in the selection buffer containing 0.05% Tween 20 and was then incubated at 37 °C for 2 hours. Afterward, the wells were washed as previously described. Before protein assays, biotin-labeled aptamer sequences were suspended in the reaction buffer (50 mM Tris–HCl, 100 mM NaCl, 4 mM KCl, 2.5 mM MgCl2 and 1 mM CaCl2, pH 7.4), heated to 95 °C for 5 min, and gradually cooled to 25 °C. Then each protein-coated well was added with 100 μL of aptamer solution (in the reaction buffer) and was incubated at room temperature for 1 hours followed by the same wash procedure. After that, each well was added with two drops (ca. 100 μL) of an alkaline phosphatase-conjugated streptavidin (Strep-AP) solution (0.003 mg mL−1) (Abcam) and incubated at room temperature for 30 min followed by the washes. For fluorescent signal development, each well as added with 100 μL of AttoPhos® AP fluorescent substrate solution (Promega) and incubated in the dark for at least 15 min. Finally, the fluorescent signal of each well was read by a fluorescent microplate reader (excitation at 435 nm and emission at 555 nm).AMACR detection in cell lysates
Two groups of cell lysate samples (total protein extracts) from (1) Escherichia coli (competent cells) with or without AMACR gene recombination and (2) prostate cancer cell line lysates (LNCaP and PC3) were detected by aptamers with the ELAA method. For sample preparations, each total protein sample was diluted to 100 ng μL−1 in the selection buffer (pH 8), transferred to the microtiter plate (50 μL per well), and then incubated at 4 °C overnight before the ELAA assay.Results and discussion
Primer effects on the structural composition of a N30 library
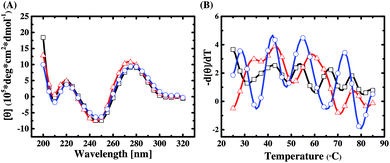
The base composition and melting temperature of primer regions in a library sequence are the two important factors to determine their interaction with the random region. In this work, a DNA secondary structure prediction tool, RNAstructure Version 5.6, is used to predict the conformation of the primer regions of the three libraries. We find that the primer regions of Library A and Library B both tend to form a stem-loop structure on both sides (refer to ESI Fig. S6†), and the predicted free energies are −4.0 (kcal mol−1) and −0.9 (kcal mol−1), respectively. By contrast, the primer region of Library C can help the library sequences fold into hairpin structures (refer to ESI Fig. S6†), and its predicted free energy is −0.1 (kcal mol−1). This implies that primer regions can potentially affect the structural composition of a SELEX library and thus influence on the selection diversity. To prove this implication, we perform CD experiments on the sequence pools of the libraries before SELEX.From CD spectral characterization of the library sequences of Library A, Library B, and Library C, as shown in Fig. 1(A), we observe that the CD spectra of the library sequences all have two positive bands around 220 and 275 nm and two negative bands around 210 and 245 nm. This data suggests that the structures of library sequences belong to a subtype of B-form secondary conformation.33 Taking a closer look, we see that the positive band around 275 nm of Library B has a blue shift by about 3–4 nm as compared to those of Libraries A and C. While the CD spectra measured at 4 °C seem to suggest no obvious primer effect on the structural composition of a library sequence pool, the differential CD melting data (Fig. 1(B)) unequivocally provides the evidence how different primers result in different structural compositions of SELEX libraries, despite having the same N30 random region.
A differential CD melting data can precisely determine the melting point (Tm) of a double-helix or a folded single-strand nucleic acid with a defined sequence and structure. Since a SELEX library contains a pool of 1015 random sequences, we expect to observe a Tm pattern or profile of a library using the differential CD melting analysis. As can be seen in Fig. 1(B), the Tm patterns of Library A, Library B, and Library C are obviously different while all of them show a periodic wave-like melting patterns. Compared with Fig. 1(A), we can infer that all three libraries tend to result in an N30 random sequence pool of B-form conformation at low temperature; however, a different primer contributes different thermodynamic stability and folding tendency for each sequence in a library at the elevated temperature. In addition, it can be seen in Fig. 1(B) that the wave-pattern amplitude of the melting curve of Library C is larger than those of Library A and Library B. It suggests that the library sequences in Library C have higher proportions of folded or more rigid conformations with similar Tm values, especially when compared to Library A. Therefore, it is evident that the primer regions play an important role in the structural composition and diversity of a SELEX library, as predicted. That is why the discovery of distinct aptamers for biomarker detection is more likely achieved with multiple libraries with different primers than with a single library in a typical SELEX practice, which is to be elucidated in the next section.
Evolution of AMACR-binding ligands from different libraries
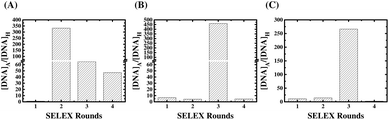
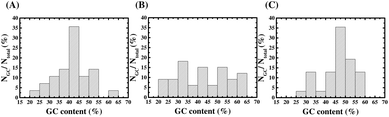
The results of comparative qPCR quantification for the specific ssDNA ligands bound to the AMACR-coated beads with respect to the non-specific ssDNA molecules bound to the HSA-coated beads from SELEX rounds 1 to 4 of the Library A, Library B, and Library C are compared in the ESI Fig. S1, S2 and S3,† respectively. The ratios of specific binding DNA ligands to non-specific DNA molecules are plotted against the SELEX round number in Fig. 2 to serve as the measure of ligand evolution for each library. The result of SELEX from Library A is shown in Fig. 2(A). It can be observed that the differential qPCR analysis of the DNA ligand pool from the 2nd round gives the highest ratio of specific binding ligands to non-specific molecules. In addition, it can also be found that the ratio decreases but maintains a certain value in the 3rd and 4th rounds. Unlike Library A, Library B and Library C both attains the maximal ratios in the 3rd round (refer to Fig. 2(B) and (C), respectively), and both libraries show under-selection in rounds 1 and 2 and over-selection in round 4, which yields no aptamers. It is obvious that the primer effect also exists for the rate of aptamer evolution since the structural composition of the selection library is significantly affected by the primer regions (refer to Fig. 1(B)). To avoid the “over-selection” problem, the ssDNA pool from the 2nd round of Library A and the pools from the 3rd round of Library B and Library C were regarded as the selected winning pools for subsequent cloning, sequencing, and analysis to yield aptamer candidates.After SELEX, we sequenced 28, 33 and 31 clones of the aptamer candidates from Library A, Library B, and Library C, respectively. The GC contents of the aptamer candidates are listed in ESI Table S2 to S4,† and the GC content distributions are plotted as a histogram in Fig. 3. In Fig. 3(A), the result from Library A shows that there is more than 80% of the DNA sequences with a GC content below 50%, and almost half of the sequences have a GC content ranging between 35 and 45%. In Fig. 3(B), the result from Library B shows that there is 64% of the DNA sequences with a GC content below 50%, but it does not show a clear normal distribution of the GC content. In Fig. 3(C), the result from Library C shows that there is more than 65% of the DNA sequences with a GC content below 50%, and almost half of the sequences have a GC content ranging between 45 and 55%. From the results, it can be observed that the GC content distributions of aptamer candidates from these three libraries are entirely different to each other. This is consistent with the presumption of selection of distinct aptamers from multiple libraries with different primers.
The aptamer sequences and their structural characteristics
The 92 sequences from the three libraries are analyzed by ClustalW2. From the selected sequences of each library, two pairs of aptamer candidates with the highest two alignment scores are picked out and listed in ESI Table S5.† Then six candidates (A24, A38, B4, B38, C14, and C20) are chosen from each pair for the following study. The secondary structures of the chosen aptamers are predicted by RNAstructure Version 5.6 and plotted in ESI Fig. S4,† which features multiple stem-loop structures and intramolecular variants of the B-form structure. The CD spectra of the six aptamers, which were dissolved in the selection buffer, are shown in Fig. 4 (curves with red hollow triangle). As can be seen, the CD spectra of the aptamers all have two positive bands around 220 and 275 nm and two negative bands around 210 and 245 nm. This data proves that the aptamer structures belong to a subtype of B-form secondary conformation,33 which is consistent with in silico structural prediction in ESI Fig. S4.†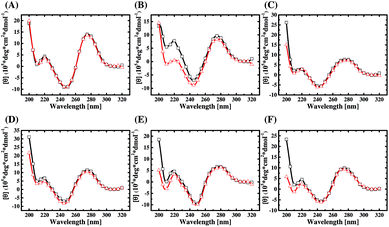 | ||
| Fig. 4 The CD spectra for (A) A24, (B) A38, (C) B4, (D) B38, (E) C14, and (F) C20 with (black line, □) and without (red line, △) AMACR. | ||
Also, the CD spectra of the six aptamers mixed with an equal concentration of AMACR in the selection buffer are compared in Fig. 4 (curves with black hollow square). It can be found that the peak height of the positive band around 220 nm in the CD spectra of A38, B38, C14, and C20 increases after the aptamers are mixed with the target protein. Although the CD data cannot explain what exact structures are formed for aptamers, they still elucidate that AMACR induces conformation changes of the aptamers. Thus, these aptamers have a potential to be used as structurally active sensing elements for the development of beacon-like aptamer sensors. Unlike the above data of the four sequences, the CD spectra of A24 and B4 do not exhibit a clear structural change after mixing with AMACR. This result means that these two sequences do not have the property of target-induced conformational change and are supposed to be pre-folded AMACR aptamers.
AMACR binding and detection characteristics of the aptamers from different libraries
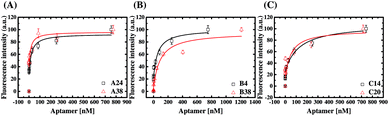
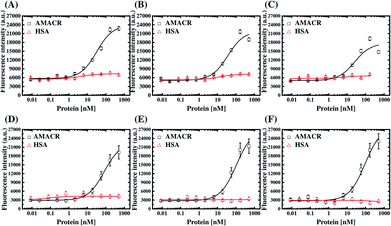
We performed fluorescent ELAA experiments to assess the binding affinity and specificity of the aptamers. Fig. 5 shows the results of the equilibrium AMACR total binding assays using the fluorescent ELAA for the six aptamers. From this data, the apparent dissociation constant (KD) for the binding events between AMACR and the aptamers are estimated to be in the range from 14 to 128 nM (A24 = 21.8 ± 13.6 nM and A38 = 14.6 ± 14.1 nM; B4 = 57.7 ± 29.4 nM and B38 = 92.1 ± 33.2 nM; C14 = 52.5 ± 17.5 nM and C20 = 128 ± 97.8 nM). The results show that A24 and B4 aptamers can recognize AMACR, which further suggests that A24 and B4 aptamers are pre-folded and do not change their confirmations after binding to AMACR (refer to Fig. 4). Moreover, it can be observed that the KD values of the aptamers from Library A are both smaller than that of the aptamers from the other two libraries, even when the aptamer candidates were generated using the same protocol.The six aptamers were also assayed with different amounts of AMACR and HSA, which were coated on 96-well microtiter plates at different coating concentrations (5 × 10−7 M to 8.5 × 10−12 M), and the results are shown in Fig. 6. All of the aptamers show ELAA responses depend on AMACR concentration and can attain nM range detection. Yet, it can be seen that A24 and A38 aptamers both have significant responses to a higher HSA concentration. It means that the A24 and A38 aptamers would have less promising detection limits for AMACR in un-purified samples with the presence of interfering non-target proteins like HSA, serum or cell lysate. In contrast, B4, B38, C14, and C20 aptamers maintain baseline ELAA responses at high concentrations of HSA. Accordingly, the aptamers from Library A exhibit less-promising specificity for AMACR recognition despite with remarkable binding affinity. By contrast, the aptamers from Library B and Library C are more specific for AMACR binding. The above data demonstrates that the library's primer design affects the affinity and specificity of the selected aptamers and proves the advantage of multiple libraries with different primers for discovery of more specific aptamers.
“High primer-dependent” vs. “low primer-dependent” aptamers
The above results have demonstrated that the primer design plays a pivotal role in the ligand evolution progress of a DNA library in SELEX. Further evidence is provided here to show that the binding properties of as-selected, full-length aptamers are affected by their primer regions. In this experiment, the forward and reverse primers of these six aptamers were removed to result in the truncated aptamers composed of merely selected N30 sequences, and fluorescent ELAA assays were also used to assess the binding affinity of the truncated aptamers. The results are shown in Fig. S5,† and the KD values for the binding events between AMACR and the truncated aptamers are estimated to be in the range from 150 to 869 nM (A24 = 477 ± 170 nM and A38 = 813 ± 405 nM; B4 = 869 ± 215 nM and B38 = 173 ± 91.8 nM; C14 = 150 ± 55.8 nM and C20 = 206 ± 36.4 nM). Fig. 7 shows comparisons of the binding characteristics of three pairs of as-selected (full-length) and primer-truncated aptamers: A38, B4, and C14. From the results, we find that the dissociation constants of aptamers all increase after removing the primers of the aptamers. In other words, the primer regions of the full-length aptamers participate in the binding of AMACR and reinforce the binding strength. In particular, the primer removal leads to a large decrease of affinities for A24, A38, and B4 aptamers. In comparison, a minor reduction in the binding affinity is observed for B38, C14, and C20 aptamers. An interesting example is C14, which shows no obvious binding property change after primer truncation (Fig. 7(C)).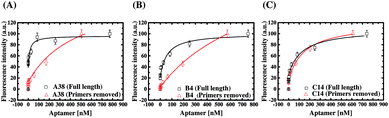 | ||
| Fig. 7 Comparisons of the binding characteristics of the as-selected and primer-truncated aptamers using the fluorescent ELAA method. (A) A38, (B) B4, and (C) C14. | ||
Furthermore, we use the website tool, RNAstructure Version 5.6, to predict the secondary structures of the six aptamers and the other 86 candidates (refer to ESI Table S2 to S4†). In Fig. S6,† we choose A38, B4, and C14 as examples, and compare their secondary structures of primer regions with those of Library A, Library B, and Library C, respectively. As the same as the A38 sequence shown in Fig. S6(A),† we find self-folding structures of the 5′-primer regions (with lower thermal stability) in 25 of the other 27 aptamer sequences (ca. 96.2%) from Library A are deconstructed, and some of the regions tend to hybridize to the N30 regions. On the other hand, the 3′-primer regions (with higher thermal stability) in the all other 27 aptamer sequences from Library A maintain their stem-loop conformation. The similar results are found in 30 of the 33 aptamers (ca. 90.9%) from Library B, but most of the sequences have a structural feature that the 5′- and 3′-primer regions are both deconstructed as the same as the B4 structure shown in Fig. S6(B).† Interestingly, the partial forward (5′-ACGG-3′) and reverse (5′-CCGT-3′) priming sites of the 16 of the 31 aptamer sequences (ca. 51.6%) from Library C are found to hybridize to each other and let the sequences form closed-loop structures. Therefore, the primer structures of Library C prevent the primer regions of aptamers to hybridize with the middle N30 region to participate in target recognition. (refer to Fig. S6(C)†). This could explain why we consider that C14 is rather a low primer-dependent aptamer while A38 and B4 are high primer-dependent. Yet, it should be noted that this is just a presumed comment from the observed data, and more experiments are required to determine whether that secondary-structure primers would make the post-SELEX optimization easier.
For sure, it can be deduced that the primer design affects aptamer evolution through the extent of primer-N30 region hybridization, which then causes a primer-dependent library structural composition and selection diversity. Meanwhile, the above results suggest that, with a proper use of primer sequence design, the effect of primer sequence on target recognition can be minimized, and post-SELEX truncation can be possibly as simple as directly removing the primer regions after cloning and sequencing (e.g., Library C).
Dimeric aptamer construct for AMACR detection
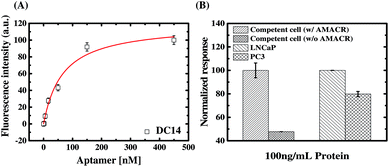
From the SELEX with libraries having different primers, we have identified a number of aptamers with distinct structural characteristics and AMACR binding properties. To demonstrate the advantage of the present SELEX library strategy, we select aptamer C14 for developing a specific bivalent molecular probe for detection of AMACR over-expression in prostate cancer cell lines. As proven above, C14 is a low primer-dependent aptamer, so direct truncation of its primer regions does not apparently affect its binding affinity and specificity to AMACR (see Fig. 7(C)). In addition, an aptamer is not like the antibody, which has more than one binding valence to capture antigen targets. Such multivalent binding property might lead better binding energy between antibodies and antigens even if the energy in per combining site is low.34 Accordingly, we developed a dimeric aptamer probe (DC14) by directly connecting two truncated C14 aptamers. As shown in Fig. 8(A), the apparent KD of DC14 with AMACR is estimated to be 66.6 ± 17.0 nM. From the result, it can be observed that the KD of the dimeric aptamer is about 2.3-fold higher than the monomeric C14 aptamer (150 ± 55.8 nM). Then we used DC14 as an ELAA probe to detect AMACR in cell lysates (total protein extracts) of E. coli with and without recombinant AMACR gene and two prostate cancer cell lines, and the results are shown in Fig. 8(B). As can be seen, DC14 can efficiently distinguish the cell lysates from the E. coli cells with and without inserted AMACR gene. For a demonstration of detecting AMACR overexpression in cancer cells. The ELAA detection results of cell lysates from LNCaP and PC3 prostate cancer cell lines are compared in Fig. 8(B), too. It is found that the detection result of DC14 is consistent with the reported data that AMACR expression of LNCaP is higher than PC3.35 In other words, DC14 can successfully detect the expression amounts of AMACR in different prostate cancer cell lines and has a potential to be used for prostate cancer diagnosis and prognosis monitoring.From above study, it is further confirmed that C14 is a low primer-dependent aptamer as deduced from Fig. 7(C). It is because that the binding strength to AMACR is not decreased but enhanced after we directly connect two truncated C14 aptamers to form a dimeric aptamer DC14. The result means that the truncated aptamer C14 can form a stable AMACR-sensing structure without primers. In other words, this experiment data implies that a low primer-dependent aptamer can be easily reconstructed as a high-affinity, high-specificity multimeric aptamer probe by directly omitting the primer regions even when its original monomeric affinity is not promising. In addition, it supports our presumption of multiple-library SELEX for enhancing discovery of distinct aptamers for flexible biomarker detection design.
Conclusions
From SELEX with library sequences bearing different primers, this study successfully discovers specific aptamers with distinct structural and binding properties (pre-folded vs. target-induced folding aptamers; high vs. low primer-dependent aptamers) for detection of the emerging cancer biomarker AMACR. Also, we demonstrate that the primer regions of library sequences exert pivotal influences on not only the structural composition and diversity of SELEX library but also the ligand evolution rate and both affinity and specificity of the selected aptamers. Comparison between the results of secondary structure prediction, CD experiments, and aptamer binding data, we prove that the primer regions affect SELEX through the base-pairing interaction between the primers and the randomized regions. By comparing the SELEX results from three different DNA libraries, it suggests that the aptamers from Library C show no obvious decreases in binding strength and specificity after primer removal. From this finding, we construct a dimeric aptamer probe (DC14) by directly connecting two monomeric, primer-truncated C14 aptamers for the detection of AMACR expression in recombinant E. coli cells and prostate cancer cell lines. The success of DC14 encourages the discovery of the aptamer by simply truncating the two fixed primer regions, since the affinity can be enhanced by folds via constructing a dimeric probe by a tandem repeat of the primer-truncated aptamer. Although some attempts were devoted to developing primer-free SELEX,22,36 common SELEX practices using a library with a central random sequence flanked with two fixed primer regions for PCR amplification are still wildly utilized for aptamer discovery. Through this study, we conclude that parallel or simultaneous SELEX with multiple libraries can increase the selection diversity from the structural aspect and enhance the likelihood of discovering different aptamers for a single target. Also, we suggest the researchers of this field to bring attention to the primer design, which has to be taken into account, in an aptamer screening process.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We gratefully acknowledge Prof. Chun-Hua Hsu (Department of Agricultural Chemistry, National Taiwan University) for providing the AMACR sample and cell lysates from E. coli with or without AMACR gene recombination for testing and the facility for conducting the CD experiments. We also acknowledge Ms Yu-En Lee (Department of Bio-Industrial Mechatronics Engineering, National Taiwan University) for providing the prostate cancer cell lysates. This work was sponsored by the Academia Sinica of the Republic of China (Taiwan) with the grant numbers AS-103-TP-A01 & AS-106-TP-A03, and Ministry of Science and Technology of the Republic of China (Taiwan) with the grant numbers MOST 101-2221-E-002-174-MY3 and 106-2119-M-001-005.Notes and references
- W. Zhou, P. J. Huang, J. Ding and J. Liu, Analyst, 2014, 139, 2627–2640 RSC.
- M. Jarczewska, L. Gorski and E. Malinowska, Anal. Methods, 2016, 8, 3861–3877 RSC.
- H. G. Sun, W. H. Tan and Y. L. Zu, Analyst, 2016, 141, 403–415 RSC.
- J. H. Zhou and J. Rossi, Nat. Rev. Drug Discovery, 2017, 16, 181–202 CrossRef PubMed.
- H. T. Ma, J. P. Liu, M. M. Ali, M. A. I. Mahmood, L. Labanieh, M. R. Lu, S. M. Iqbal, Q. Zhang, W. A. Zhao and Y. Wan, Chem. Soc. Rev., 2015, 44, 1240–1256 RSC.
- W. H. Tan, H. Wang, Y. Chen, X. B. Zhang, H. Z. Zhu, C. Y. Yang, R. H. Yang and C. Liu, Trends Biotechnol., 2011, 29, 634–640 CrossRef PubMed.
- M. Ilgu and M. Nilsen-Hamilton, Analyst, 2016, 141, 1551–1568 RSC.
- M. Yuce, N. Ullah and H. Budak, Analyst, 2015, 140, 5379–5399 RSC.
- C. Tuerk and L. Gold, Science, 1990, 249, 505–510 Search PubMed.
- A. D. Ellington and J. W. Szostak, Nature, 1990, 346, 818–822 CrossRef PubMed.
- T. K. Sharma, J. G. Bruno and A. Dhiman, Biotechnol. Adv., 2017, 35, 275–301 CrossRef PubMed.
- J. C. Rohloff, A. D. Gelinas, T. C. Jarvis, U. A. Ochsner, D. J. Schneider, L. Gold and N. Janjic, Mol. Ther.--Nucleic Acids, 2014, 3, e201 CrossRef PubMed.
- M. Kimoto, R. Yamashige, K. Matsunaga, S. Yokoyama and I. Hirao, Nat. Biotechnol., 2013, 31, 453–457 CrossRef PubMed.
- P. Herdewijn and P. Marliere, Chem. Biodiversity, 2009, 6, 791–808 Search PubMed.
- V. B. Pinheiro, A. I. Taylor, C. Cozens, M. Abramov, M. Renders, S. Zhang, J. C. Chaput, J. Wengel, S. Y. Peak-Chew, S. H. McLaughlin, P. Herdewijn and P. Holliger, Science, 2012, 336, 341–344 CrossRef PubMed.
- I. Alves Ferreira-Bravo, C. Cozens, P. Holliger and J. J. DeStefano, Nucleic Acids Res., 2015, 43, 9587–9599 Search PubMed.
- M. Darmostuk, S. Rimpelova, H. Gbelcova and T. Ruml, Biotechnol. Adv., 2015, 33, 1141–1161 CrossRef PubMed.
- S. S. Oh, K. Plakos, X. Lou, Y. Xiao and H. T. Soh, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 14053–14058 CrossRef PubMed.
- R. Nutiu and Y. Li, Angew. Chem., Int. Ed. Engl., 2005, 44, 1061–1065 CrossRef PubMed.
- S. S. Oh, K. Plakos, Y. Xiao, M. Eisenstein and H. T. Soh, ACS Nano, 2013, 7, 9675–9683 CrossRef PubMed.
- Z. Huang and J. W. Szostak, RNA, 2003, 9, 1456–1463 CrossRef PubMed.
- W. Pan, P. Xin, S. Patrick, S. Dean, C. Keating and G. Clawson, J. Visualized Exp., 2010, 41, 2039 Search PubMed.
- X. Li, W. Zhang, L. Liu, Z. Zhu, G. Ouyang, Y. An, C. Zhao and C. J. Yang, Anal. Chem., 2014, 86, 6596–6603 CrossRef PubMed.
- P. D. Baade, D. R. Youlden and L. J. Krnjacki, Mol. Nutr. Food Res., 2009, 53, 171–184 Search PubMed.
- R. Siegel, D. Naishadham and A. Jemal, Ca-Cancer J. Clin., 2013, 63, 11–30 CrossRef PubMed.
- Z. Jiang, B. A. Woda, K. L. Rock, Y. Xu, L. Savas, A. Khan, G. Pihan, F. Cai, J. S. Babcook, P. Rathanaswami, S. G. Reed, J. Xu and G. R. Fanger, Am. J. Surg. Pathol., 2001, 25, 1397–1404 CrossRef PubMed.
- M. A. Rubin, M. Zhou, S. M. Dhanasekaran, S. Varambally, T. R. Barrette, M. G. Sanda, K. J. Pienta, D. Ghosh and A. M. Chinnaiyan, JAMA, J. Am. Med. Assoc., 2002, 287, 1662–1670 CrossRef PubMed.
- J. Luo, S. Zha, W. R. Gage, T. A. Dunn, J. L. Hicks, C. J. Bennett, C. N. Ewing, E. A. Platz, S. Ferdinandusse, R. J. Wanders, J. M. Trent, W. B. Isaacs and A. M. De Marzo, Cancer Res., 2002, 62, 2220–2226 Search PubMed.
- M. Zhou, A. M. Chinnaiyan, C. G. Kleer, P. C. Lucas and M. A. Rubin, Am. J. Surg. Pathol., 2002, 26, 926–931 CrossRef PubMed.
- M. M. Masud, M. Kuwahara, H. Ozaki and H. Sawai, Bioorg. Med. Chem., 2004, 12, 1111–1120 CrossRef PubMed.
- Y. C. Chang, C. Y. Yang, R. L. Sun, Y. F. Cheng, W. C. Kao and P. C. Yang, Sci. Rep., 2013, 3, 1863 CrossRef PubMed.
- D. K. Yang, L. C. Chen, M. Y. Lee, C. H. Hsu and C. S. Chen, Biosens. Bioelectron., 2014, 62, 106–112 CrossRef PubMed.
- J. Kypr, I. Kejnovska, D. Renciuk and M. Vorlickova, Nucleic Acids Res., 2009, 37, 1713–1725 CrossRef PubMed.
- O. Makela, E. Ruoslaht and I. J. T. Seppala, Immunochemistry, 1970, 7, 917–932 CrossRef PubMed.
- B. A. Wilson, H. Wang, B. A. Nacev, R. C. Mease, J. O. Liu, M. G. Pomper and W. B. Isaacs, Mol. Cancer Ther., 2011, 10, 825–838 CrossRef PubMed.
- W. H. Pan, P. Xin and G. A. Clawson, BioTechniques, 2008, 44, 351–360 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Materials, equipment and software, differential qPCR analyses, list of the three library sequences and related primers, list of all the sequenced aptamer candidates, predicted secondary structures of testing aptamers are detailed in ESI Material. See DOI: 10.1039/c8ra01808a |
| This journal is © The Royal Society of Chemistry 2018 |