Open Access Article
Open Access ArticleUser-friendly point-of-care detection of influenza A (H1N1) virus using light guide in three-dimensional photonic crystal†
Nuree Lee‡
a,
Cong Wang‡ a and
Jungyul Park
a and
Jungyul Park *ab
*ab
aDepartment of Mechanical Engineering, Sogang University, 35 Baekbeom-ro (Shinsu-dong), Mapo-gu, Seoul 04107, Korea. E-mail: sortpark@sogang.ac.kr
bInterdisciplinary Program of Integrated Biotechnology, Sogang University, 35 Baekbeom-ro (Shinsu-dong), Mapo-gu, Seoul 04107, Korea
First published on 22nd June 2018
Abstract
This paper proposes a user-friendly and highly sensitive detection method for influenza A (H1N1) virus using the cooperation of quantum dot (Qdot)-aptamer beacons and light guide in a three-dimensional (3D) photonic crystal (PC). For easy use, we present a fluorescent probe-based ‘OFF–ON’ detection protocol. First, a mixture composed of Qdot-aptamer beacons and dark quencher-labeled guard DNA (G-DNA) was prepared. It initially quenched the fluorescent signals to significantly low intensity, i.e., ‘OFF’ state. Then, the influenza A (H1N1) virus preferentially bound to the aptamer and G-DNA was released, so that the fluorescent signal was restored and biosensor turned to ‘ON’ state. The restored fluorescence signal changed quantitatively according to the concentration of the target influenza A (H1N1) virus. Owing to the light guide of the 3D nanoporous PC structure, we achieved high sensitivity with ultra-low limit of detection (LOD) of 138 pg mL−1 and high selectivity over other species of influenza A virus and biomolecules. Additionally, with the benefit of enhanced output fluorescent signals, the target virus could be easily detected with a low-cost and portable home-made setup (total cost of only 20 US dollars) and a built-in camera in a smartphone.
Introduction
In recent years, the threat of influenza pandemics has massively increased and many countries have drawn up epidemic prevention plans following the World Health Organization (WHO) guidelines. The propagator of this epidemic disease, i.e., influenza virus, is a single-stranded and negative-infected RNA virus of the orthomyxoviridae strain that causes regular seasonal epidemics in birds, humans, and other mammals. The worldwide prevalent influenza virus that infects humans has three genera identified by antigenic differences in their nucleoproteins and matrix proteins, i.e., types A, B, and C. Among them, the influenza virus of type A, also called influenza A virus, has the greatest genetic diversity and infects the widest range of host species.1 In particular, a novel swine-origin influenza virus (S-OIV) H1N1, a subtype of the influenza A virus, was isolated from humans in Mexico and the United States in April 2009. Subsequently, the WHO declared an H1N1 pandemic, which was marked the first global pandemic since the 1968 Hong King flu. Recent research shows that the infectivity of S-OIV is more efficient and fatal than seasonal influenza viruses.2,3 It was reported by WHO that more than 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 138 deaths were caused by the influenza A (H1N1) virus in more than 214 countries. Followed by a series of rapid responses for prevention and control, the detection and identification of the influenza A (H1N1) virus at an early stage can effectively inhibit the spread of the pandemic influenza. Classical methods for detecting influenza viruses, such as haemagglutination (HA) assay,4 haemagglutination inhibition (HI) assay5 and enzyme-linked immunosorbent assay (ELISA),6,7 require long periods of time (3–10 days), large amounts of target virus, special instruments, and well-trained operators. Though the reverse transcriptase polymerase chain reaction (RT-PCR) can significantly improve the assay speed and sensitivity, it is still complicated, expensive and time-consuming (1–2 days).8 To address the abovementioned drawbacks, point-of-care (POC) diagnostic method can be an effective and simple approach to detect the influenza A (H1N1) virus in a user-friendly way instantly or in a very short time even by an unskilled user.9,10 The available commercial POC methods are mainly based on immunoassays that detect influenza viral antigen and are used for screening influenza A virus within 5–30 minutes. However, low sensitivity and false negative results by POC methods are the main concerns that remain to be improved.
138 deaths were caused by the influenza A (H1N1) virus in more than 214 countries. Followed by a series of rapid responses for prevention and control, the detection and identification of the influenza A (H1N1) virus at an early stage can effectively inhibit the spread of the pandemic influenza. Classical methods for detecting influenza viruses, such as haemagglutination (HA) assay,4 haemagglutination inhibition (HI) assay5 and enzyme-linked immunosorbent assay (ELISA),6,7 require long periods of time (3–10 days), large amounts of target virus, special instruments, and well-trained operators. Though the reverse transcriptase polymerase chain reaction (RT-PCR) can significantly improve the assay speed and sensitivity, it is still complicated, expensive and time-consuming (1–2 days).8 To address the abovementioned drawbacks, point-of-care (POC) diagnostic method can be an effective and simple approach to detect the influenza A (H1N1) virus in a user-friendly way instantly or in a very short time even by an unskilled user.9,10 The available commercial POC methods are mainly based on immunoassays that detect influenza viral antigen and are used for screening influenza A virus within 5–30 minutes. However, low sensitivity and false negative results by POC methods are the main concerns that remain to be improved.
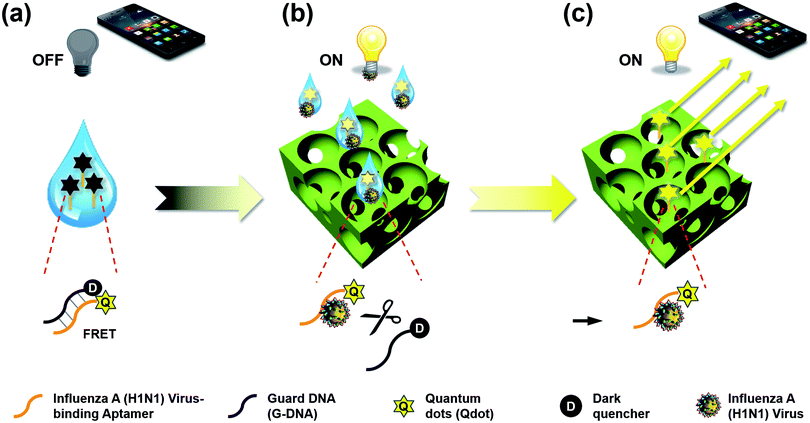
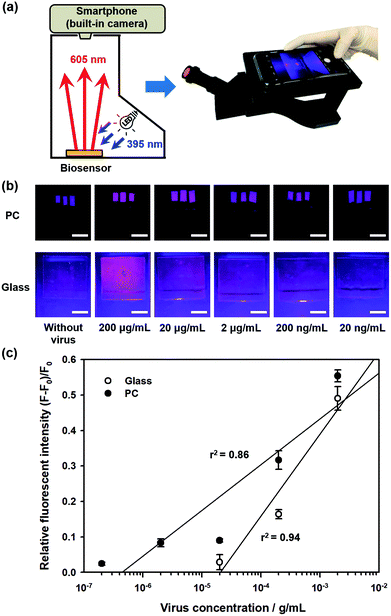
In this study, we propose a direct detection method for the influenza A (H1N1) virus using the cooperation of quantum dot (Qdot)-aptamer beacons and emission light guide for signal enhancement based on 3D photonic crystals (PC). Also, we developed a portable setup using a smartphone camera for easy diagnosis without limit of location. Fig. 1 shows the proposed detection principle using Qdot-aptamer conjugates11 and 3D PC.12,13 For user-friendliness, we applied a fluorescence-based ‘OFF–ON’ detection strategy. First, in a pre-prepared solution for influenza A (H1N1) virus detection, the energy from the Qdot is transferred to a dark quencher conjugated with guard DNA(G-DNA), through fluorescence resonance energy transfer (FRET).14 Hence, the fluorescence emission signals from the Qdot are reduced to an extremely low level, i.e., “OFF” state (Fig. 1a). Then, in the presence of the target influenza A (H1N1) virus, the influenza A (H1N1) virus-aptamer binding and the release of G-DNA induce the restoration of fluorescence signals, turning the biosensor to the ‘ON’ state. The proposed PC was designed in such a way that the reflected wavelength from the structure overlaps the fluorescence wavelength of the quantum dot (Qdot) for highly sensitive detection.15–17 Therefore, the output fluorescence signals with specific wavelengths can be enhanced by the emission light guide in 3D PC and enhanced output signals are detected by a low-end charge-coupled device (CCD).
Results and discussion
Highly sensitive detection using emission light guide
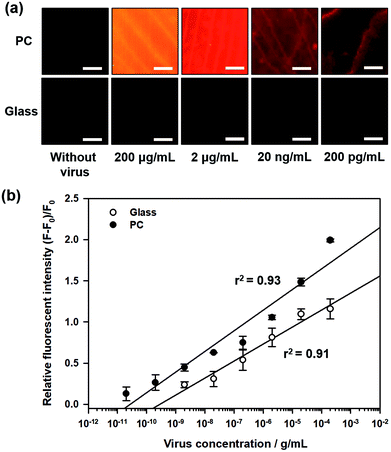
To enhance the fluorescence signals and improve the sensitivity of the proposed sensor, the reflected peak wavelength in 3D PC was engineered to be similar to the emission peak wavelength of the fluorophore. This was realized by changing the pore size of 3D PC, which was directly associated with the size of nanoparticles. The resulting reflected wavelength in the proposed sensor could be predicted by Bragg's equation, which has been thoroughly discussed in our previous studies (more details in ESI†). Assuming that the PEG-DA material did not swell, the peak wavelength value in the wet state was 673 nm.As described above, the emission fluorescent signals from Qdot were enhanced by the light guide in 3D PC, whose reflected wavelength needed to be matched with the emission wavelength of Qdot. In other words, to maximize the effect of the emission light guide, fluorescence probes with emission peak wavelength of around 673 nm would be appropriate. However, because of ineffective quenching by using Qdot with long wavelength emission (e.g., emission maximum > 650 nm) as FRET donors (due to large core sizes), low quantum yields, and significantly high absorption coefficient in aqueous solution, we tried detecting influenza A (H1N1) virus using Qdot 605-aptamer conjugates for further improvement.18,19 Among the different species of Qdot-streptavidin conjugates, which were purchased from Invitrogen Inc. (more details in ESI†), Qdot 605 was considered the best alternate fluorescence probe.
Fig. 2a shows the fluorescence images during influenza A (H1N1) virus detection on 3D PC and 2D glass surfaces, with different concentrations of influenza A (H1N1) virus from 200 pg mL−1 to 200 μg mL−1. For quantitative analysis, the relative fluorescence intensity was calculated as follows:
| η = (F − F0)/F0 | (1) |
| Reference | Method | Assay time | LOD |
|---|---|---|---|
| a 30 PFU mL−1 = ∼3 × 103 particles per mL = ∼1.5 pg mL−1.b 1 × 102 TCID50/100 μL = ∼1 × 104 particles per mL = ∼50 pg mL−1.c 0.032 HAU/20 μL = ∼6.4 × 106 particles per mL = ∼8 ng mL−1.23 | |||
| Su et al.,20 2012 | Surface plasmon resonance | 20 min | 1.5a pg mL−1 |
| Feng et al.,21 2015 | PCR-based SELEX | 80 min | 50b pg mL−1 |
| Tseng et al.,22 2016 | Fluorescence | 9 steps, total 30 min | 8c ng mL−1 |
| This work | Fluorescence & light guiding | 40 min | 138 pg mL−1 |
Selectivity of PC-based biosensor
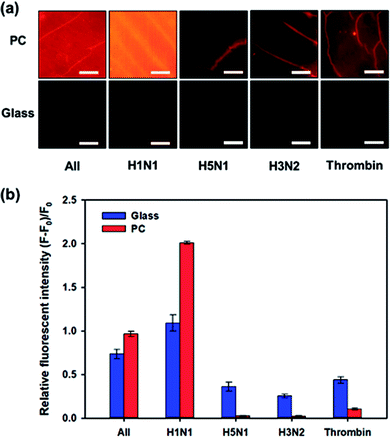
One of the important factors of the biosensor is selectivity to target molecules. The specificity of the influenza A (H1N1) virus-binding aptamer enables our proposed biosensor to have high selectivity for influenza A (H1N1) virus detection. We compared the variation in fluorescence intensity in case of different types of viruses or biomolecules: H1N1, H5N1, H3N2, thrombin from human plasma (Sigma-Aldrich, Korea) and their mixture with the same concentration of 200 μg mL−1. Fig. 3a shows a comparison of the fluorescent images after these samples were added to the biosensor individually and incubated for 2 h. Quantitative analysis was performed by comparing the relative fluorescence intensities ((F − F0)/F0), as shown in Fig. 3b. When nontargeted viruses or biomolecules (H5N1, H3N2 or thrombin) were introduced to the biosensor, only small changes in fluorescence intensity were observed. On the contrary, in the presence of influenza A (H1N1) virus, the intensity of the fluorescence signal from Qdot 605 increased significantly and the relative fluorescence intensity reached to about 1.99. When a mixture of H1N1, H5N1, H3N2, and thrombin was introduced, the relative fluorescence intensity decreased compared to pure H1N1 but showed high values compared to those of other untargeted biomolecules. Interestingly, the selectivity performance using 3D PC was better than the selectivity using 2D glass, that is, high signal-to-noise ratio could be achieved through the light guide of 3D PC. This result can be explained using a simple ‘signal plus noise’ model, in which the measured data F is composed of a signal component Fs and a noise component ηi, where i stands for the substrate used for detection, i.e., p for 3D PC and g for 2D glass. Hence, the relative fluorescence intensity (F − F0)/F0 can be derived to Fs/F0 + ηi/F0 − 1. When only nontargeted viruses or biomolecules existed in samples, the signal Fs was the same as the original signal F0, and the noise ηi came from the nontargeted molecules. The term Fs/F0 becomes nearly 1; hence, the relative fluorescence intensity could be simplified to ηi/F0. In the case of the proposed PC-based biosensor, F0 was higher than that in the 2D glass, as shown in Fig. 3a. Hence, the relative fluorescence intensity (F − F0)/F0 in H5N1, H3N2 or thrombin was much smaller on 3D PC than on the 2D glass surface, indicating higher signal-to-noise ratio. These results indicated that our proposed PC-based biosensor had higher selectivity for influenza A (H1N1) virus than other analyses.Optimization of assay time
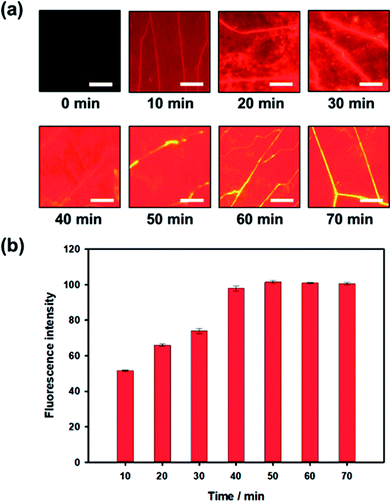
During the influenza A (H1N1) virus-aptamer binding process, the dark quencher-labeled G-DNA was released from the aptamer. To optimize the analysis time, variations in fluorescence signals from Qdot 605 were recorded for 70 min during the assay process. When influenza A (H1N1) virus with concentration 200 μL mL−1 was introduced, the fluorescence intensity started to increase after 10 minutes. Subsequently, the fluorescence intensity increased gradually and then reached a saturation state after nearly 40 min, as shown in Fig. 4. Based on these results, the influenza A (H1N1) virus could be quantified within 40 min using our proposed detection method.Quantitative analysis in low-cost portable home-made setup
The detection method proposed in this study could precisely detect influenza A (H1N1) virus using professional instruments, which were expensive and complicated, as well as a simple home-made setup, which was of low-cost and portable, as shown in Fig. 5a. The fluorescence images at different concentrations of influenza A (H1N1) virus were captured with a smartphone camera on the proposed home-made setup, as shown in Fig. 5b. Then, the fluorescence images were analyzed using ImagePro Plus software. Using the home-made setup and a smartphone camera, we obtained LOD of 70 ng mL−1 for the PC and 3.68 μg mL−1 for the 2D glass surface, as shown in Fig. 5c.Virus detection in a real sample
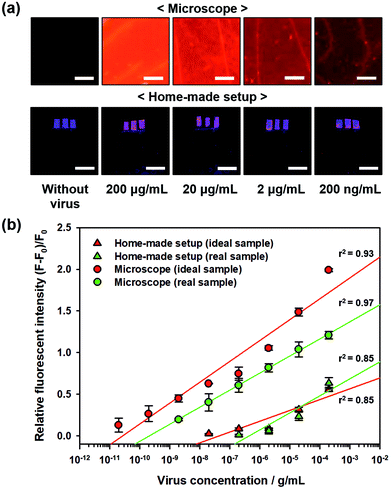
For practical applications of biosensors, real samples, i.e., blood or serum, contain abundant proteins and biomolecules that make detecting environment more complicated than the ideal sample. Hence, such samples limit the performance of the biosensors in terms of sensitivity. To quantitatively evaluate the sensitivity of the proposed PC-based biosensor in real samples, sterile-filtered human serum was used with different H1N1 virus concentrations, and the fluorescent images were captured using a microscope as well as home-made setup with a smartphone, as shown in Fig. 6a. The measured relative fluorescence intensity during virus detection in real samples (human serum) compared to those of ideal samples (pure PBS solution) is shown in Fig. 6b. In addition, we determined the LOD of the biosensor under these detection conditions. The LODs of the proposed PC-based biosensor with a microscope and home-made setup were 560 pg mL−1 and 600 ng mL−1, respectively. Due to non-specific binding of other protein molecules, the relative fluorescence intensity decreased slightly, but the LODs were still higher than the LOD on the 2D glass surface using the ideal sample (897 pg mL−1 under microscopic observation and 3.68 μg mL−1 in the home-made setup) owing to the high signal-to-noise ratio of the proposed PC-based biosensor.Experimental
Fabrication process of 3D photonic crystal
The fabrication process of 3D nanoporous PC has been described in our previous studies, as shown in Fig. S1†:24,25 First, a drop of colloidal silica nanoparticle dispersion (diameter: 300 nm, Polysciences, Inc., Warrington, PA) was spread on a slide glass and then, the droplet was covered with cover glass (25 × 25 mm). During evaporation of the solution, the silica nanoparticles were self-assembled.26,27 After getting fully dried, the colloidal crystals were immersed in photocurable poly(ethylene glycol)-diacrylate (PEG-DA) solution for 5 min, allowing sufficient penetration into the interstices between nanoparticles. PEG-DA solution was prepared by mixing 99 wt% PEG-DA solution (average MW 250, Sigma-Aldrich, Korea) with 1 wt% water-soluble photoinitiator 2-hydroxy-2-methylpropiophenone (Sigma-Aldrich, Korea). Then, a polyethylene terephthalate (PET) film modified with urethane groups was covered on it, and the sample was exposed to UV light (250–400 nm) for a few tens of seconds at 90 mW cm−2. Then, the polymerized PEG-DA layer with PET film was peeled off from the slide glass. The resultant inverse opal structures were obtained by wet etching of the silica nanoparticles in buffer oxide etchant (BOE). The inverse opal structures with PEG-DA hydrogels were used in this study because hydrogels provide homogeneous water environment and have higher capacity for biomolecule immobilization of proteins than the traditional 2D glass surface. Moreover, the environment was conducive for covalent attachment of proteins and had greater probability of interacting with the target ligand.28,29Preparation of quenched Qdot-aptamer conjugates
First, the Qdot-aptamer conjugates were prepared by conjugating streptavidin-functionalized Qdot 605 (Qdot® ITK™ streptavidin conjugate, Invitrogen, Carlsbad, CA, USA) to 3′ biotin-modified aptamer (IDT Inc., Korea) at a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20. The DNA sequence of influenza A (H1N1) virus-binding aptamer was 5′-/5AmMC12/GGG TTT GGG TTG GGT TGG GTT TTT GGG TTT GGG TTG GGT TGG GAA AAA/3Bio/-3′. Before conjugation, the biotinylated aptamer was heated to 70 °C for 10 min to unfold the DNA strain and cooled to 25 °C for 10 min to block the DNA in its unfolded structure. The Qdot-aptamer conjugates were synthesized in phosphate-buffered saline (PBS: 10 mM phosphate buffer, 2.7 mM KCl, and 137 mM NaCl, pH 7.4) in tubes for 4.5 h at 25 °C in the dark. Then, Iowa black FQ conjugated G-DNA (Mbiotech, Korea) was added to Qdot-aptamer conjugates with G-DNA-to-aptamer molar ratio of 20
20. The DNA sequence of influenza A (H1N1) virus-binding aptamer was 5′-/5AmMC12/GGG TTT GGG TTG GGT TGG GTT TTT GGG TTT GGG TTG GGT TGG GAA AAA/3Bio/-3′. Before conjugation, the biotinylated aptamer was heated to 70 °C for 10 min to unfold the DNA strain and cooled to 25 °C for 10 min to block the DNA in its unfolded structure. The Qdot-aptamer conjugates were synthesized in phosphate-buffered saline (PBS: 10 mM phosphate buffer, 2.7 mM KCl, and 137 mM NaCl, pH 7.4) in tubes for 4.5 h at 25 °C in the dark. Then, Iowa black FQ conjugated G-DNA (Mbiotech, Korea) was added to Qdot-aptamer conjugates with G-DNA-to-aptamer molar ratio of 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The sequence of G-DNA conjugated with double dark quencher was 5′-/5IABkFQ//ZEN/ACC CAA CCC AAA CCC-3′. To conjugate G-DNA with aptamer, the PEG-DA surface immersed in G-DNA solution was first baked to 70 °C for 5 min and then cooled to 25 °C for 3 h in the dark. The final concentrations of aptamer, Qdot and G-DNA in the as-prepared conjugates were 1 μM, 0.05 μM and 20 μM, respectively. Finally, unconjugated G-DNA was removed by centrifugal filtration at 13
1. The sequence of G-DNA conjugated with double dark quencher was 5′-/5IABkFQ//ZEN/ACC CAA CCC AAA CCC-3′. To conjugate G-DNA with aptamer, the PEG-DA surface immersed in G-DNA solution was first baked to 70 °C for 5 min and then cooled to 25 °C for 3 h in the dark. The final concentrations of aptamer, Qdot and G-DNA in the as-prepared conjugates were 1 μM, 0.05 μM and 20 μM, respectively. Finally, unconjugated G-DNA was removed by centrifugal filtration at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 30 min and then refilled with PBS solution.
000 rpm for 30 min and then refilled with PBS solution.
Due to FRET between Qdot 605 and dark quencher Iowa black FQ that were within a certain distance of each other, the donor Qdot 605 transferred energy to the acceptor dark quencher and the fluorescence emission signals from Qdot 605 reduced compared to the original fluorescence intensity. The Iowa black FQ had a broad absorbance spectrum ranging from 420 to 620 nm, which overlapped the emission wavelength of Qdot 605.30
Influenza A (H1N1) virus detection in 3D photonic crystal
To perform the experiment for detecting the target virus in the ideal sample and the real (or non-ideal) sample, the influenza A (H1N1) virus (MyBiosource, Inc., USA) in PBS solution and sterile-filtered human serum (Sigma Aldrich, Korea) were introduced to the as-prepared quenched Qdot-aptamer conjugates and incubated in tubes for 2 h at 37 °C in the dark. The final concentrations of influenza A (H1N1) virus solution in the bioassays were 20 pg mL−1 to 200 μg mL−1. As control groups, other species of virus (H5N1 and H3N2 of the influenza A virus) were also purchased from MyBiosource Inc. The fluorescence images during virus detection were captured by an inverted fluorescence microscope (IX71, Olympus Optical Co., Ltd., Tokyo, Japan) with a color CCD camera (CoolSNAP, Photometrics, Tucson, AZ, USA) and the fluorescence intensity of Qdot 605 in the images was evaluated using ImagePro Plus software (MediaCybernetics, Bethesda, MD, USA).Fabrication process of low-cost portable home-made setup
In the proposed biosensor, the output fluorescence signals could be significantly enhanced owing to the emission light guide in 3D PC. Hence, we propose a low-cost portable home-made chamber for quantitative analysis with a smartphone camera, as shown in Fig. 5a. The proposed home-made chamber was made with acrylic plates of 5 mm thickness, cut by a laser cutting machine. The size of the home-made chamber was 20 × 10 × 10 cm. UV-light emitting diodes (UV-LEDs) with wavelength of 395 nm, which was within the excitation wavelength range of Qdot 605, were used as light source in the chamber. The total cost of the home-made setup was only 20 US dollars, and the components such as acrylic plates or UV LEDs can be easily purchased.Conclusions
The proposed biosensor allows end-users to detect influenza A (H1N1) virus without complex assay processes and long analysis time. The processes for end-users include collecting target samples from either flu-containing aerosols (typically coughing or sneezing from infected patients) or throat and nasal swabs, and mixing the target biomolecules with the as-prepared Qdot-aptamer conjugates, and incubating them for 40 min. The proposed biosensor showed high sensitivity (LOD of 138 pg mL−1) owing to enhanced output signal through the emission light guide for specific wavelength in the 3D PC structure. Moreover, high selectivity against other species of influenza A virus (H5N1, H3N2) or biomolecules (thrombin from human plasma) was confirmed by quantitative analysis based on 3D PCthat showed improved signal-to-noise ratio over 2D surfaces. Furthermore, the target influenza A (H1N1) virus could be easily visualized through a low-cost portable home-made setup (total cost of only 20 US dollars) and a smartphone camera, which showed an LOD of 70 ng mL−1. This portable detection setup allows end-users to measure fluorescence intensity ubiquitously without expensive and complicated instruments. Therefore, the acquired information can be easily transmitted and analyzed through integration of IT technologies such as smartphones and online databases for real-time analysis and in-time diagnosis, and it can be applied versatilely in medical, pharmaceutical, and military environments.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIP) (NRF-2016R1D1A1B03936000, 2017R1A4A1015564, 2017S1A5B6054769).Notes and references
- M. I. Nelson and E. C. Holmes, Nat. Rev. Genet., 2007, 8, 196–205 CrossRef PubMed.
- J. Ginsberg, M. H. Mohebbi, R. S. Patel, L. Brammer, M. S. Smolinski and L. Brilliant, Nature, 2009, 457, 1012–1014 CrossRef PubMed.
- F. Carrat, E. Vergu, N. M. Ferguson, M. Lemaitre, S. Cauchemez, S. Leach and A.-J. Valleron, Am. J. Epidemiol., 2008, 167, 775–785 CrossRef PubMed.
- C. Cao, R. Dhumpa, D. D. Bang, Z. Ghavifekr, J. Høgberg and A. Wolff, Analyst, 2010, 135, 337–342 RSC.
- J. C. Pedersen, in Avian influenza virus, Springer, 2008, pp. 53–66 Search PubMed.
- P. R. Woolcock and C. J. Cardona, Avian Dis., 2005, 49, 477–481 CrossRef PubMed.
- D. S. Song, Y. J. Lee, O. M. Jeong, Y. J. Kim, C. H. Park, J. E. Yoo, W. J. Jeon, J. H. Kwon, G. W. Ha and B. K. Kang, J. Vet. Sci., 2009, 10, 323–329 CrossRef PubMed.
- R. A. Fouchier, T. M. Bestebroer, S. Herfst, L. Van Der Kemp, G. F. Rimmelzwaan and A. D. Osterhaus, J. Clin. Microbiol., 2000, 38, 4096–4101 Search PubMed.
- R. Dhumpa, Rapid detection of Avian Influenza Virus – Towards point of care diagnosis, Technical University of Denmark (DTU), 2011 Search PubMed.
- Y. Li, M. Hong, B. Qiu, Z. Lin, Y. Chen, Z. Cai and G. Chen, Biosens. Bioelectron., 2014, 54, 358–364 CrossRef PubMed.
- I. Shiratori, J. Akitomi, D. A. Boltz, K. Horii, M. Furuichi and I. Waga, Biochem. Biophys. Res. Commun., 2014, 443, 37–41 CrossRef PubMed.
- S. John, Phys. Rev. Lett., 1987, 58, 2486 CrossRef PubMed.
- E. Yablonovitch, Phys. Rev. Lett., 1987, 58, 2059 CrossRef PubMed.
- C. Wang, C. Y. Lim, E. Choi, Y. Park and J. Park, Sens. Actuators, B, 2016, 223, 372–378 CrossRef.
- P. Lodahl, A. F. Van Driel, I. S. Nikolaev and A. Irman, Nature, 2004, 430, 654 CrossRef PubMed.
- A. R. Clapp, I. L. Medintz, J. M. Mauro, B. R. Fisher, M. G. Bawendi and H. Mattoussi, J. Am. Chem. Soc., 2004, 126, 301–310 CrossRef PubMed.
- D. Geißler, L. J. Charbonnière, R. F. Ziessel, N. G. Butlin, H. G. Löhmannsröben and N. Hildebrandt, Angew. Chem., Int. Ed., 2010, 49, 1396–1401 CrossRef PubMed.
- R. M. Pope and E. S. Fry, Appl. Opt., 1997, 36, 8710–8723 CrossRef PubMed.
- H. Buiteveld, J. Hakvoort and M. Donze, Optical properties of pure water, 1994 Search PubMed.
- L.-C. Su, C.-M. Chang, Y.-L. Tseng, Y.-F. Chang, Y.-C. Li, Y.-S. Chang and C. Chou, Anal. Chem., 2012, 84, 3914–3920 CrossRef PubMed.
- X. Feng, K. Liu, Y. Ning, L. Chen and L. Deng, J. Nanomed. Nanotechnol., 2015, 6, 1 Search PubMed.
- Y.-T. Tseng, C.-H. Wang, C.-P. Chang and G.-B. Lee, Biosens. Bioelectron., 2016, 82, 105–111 CrossRef PubMed.
- F. Vollmer, S. Arnold and D. Keng, Proc. Natl. Acad. Sci., 2008, 105, 20701–20704 CrossRef PubMed.
- J. H. Kim, J. H. Moon, S.-Y. Lee and J. Park, Appl. Phys. Lett., 2010, 97, 103701 CrossRef.
- E. Choi, Y. Choi, Y. H. P. Nejad, K. Shin and J. Park, Sens. Actuators, B, 2013, 180, 107–113 CrossRef.
- Y. Xia, B. Gates, Y. Yin and Y. Lu, Adv. Mater., 2000, 12, 693–713 CrossRef.
- P. A. Hiltner and I. M. Krieger, J. Phys. Chem., 1969, 73, 2386–2389 CrossRef.
- Y. Zhao, X. Zhao, B. Tang, W. Xu and Z. Gu, Langmuir, 2010, 26, 6111–6114 CrossRef PubMed.
- M. F. Ali, R. Kirby, A. P. Goodey, M. D. Rodriguez, A. D. Ellington, D. P. Neikirk and J. T. McDevitt, Anal. Chem., 2003, 75, 4732–4739 CrossRef PubMed.
- E. Petryayeva, W. R. Algar and I. L. Medintz, Appl. Spectrosc., 2013, 67, 215–252 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra02596g |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2018 |