Open Access Article
Open Access ArticleiTRAQ-based quantitative proteomic analysis of dark-germinated soybeans in response to salt stress†
Yongqi Yin ,
Fei Qi,
Lu Gao,
Shengqi Rao,
Zhenquan Yang and
Weiming Fang*
,
Fei Qi,
Lu Gao,
Shengqi Rao,
Zhenquan Yang and
Weiming Fang*
College of Food Science and Technology, Yangzhou University, Yangzhou, Jiangsu 210095, People’s Republic of China. E-mail: wmfang@yzu.edu.cn
First published on 16th May 2018
Abstract
Soybean germination under stressful conditions, especially salt stress, has been verified to be an effective way of accumulating gamma-aminobutyric acid (GABA) in dark-germinated soybeans. In this study, a combination of physiological characteristics and isobaric tags for relative and absolute quantitation (iTRAQ) in a proteomic-based approach was used to investigate the protein changes in dark-germinated soybeans under salt stress. A total of 201 differentially abundant proteins (DAPs) were identified and divided into 13 functional groups. Under salt stress, 20 metabolic pathways were significantly enriched in dark-germinated soybeans. GABA content and antioxidase activity were increased while the growth and development of soybeans were inhibited by the salt stress. Promoting the synthesis of ROS-scavenging enzymes, maintaining the protein metabolic balance and re-establishing cellular homeostasis were very important strategies for growth stimulation in response to salt stress. In summary, these results showed comprehensive proteome coverage of dark-germinated soybeans in response to salt treatment, and increased our understanding of the molecular processes involved in plant networks responding to stresses.
1. Introduction
Bean sprouts are among the most traditional, widely consumed vegetables in some Asian countries. In general, abiotic stresses are common problems that significantly reduce plant growth, development and productivity.1,2 In recent years, however, much research has been performed focusing on the induction of functional compound production in bean sprouts under abiotic stresses3 such as light-mediated stress,4–6 ultraviolet-B radiation,7 chemical stress,8 hypoxia stress9 and salt stress.10,11 The amounts of functional compounds like γ-aminobutyric acid (GABA) and phenolic antioxidants in bean sprouts obviously increased under abiotic stresses during germination. GABA is an inhibitory neurotransmitter in the brains and spinal cords of mammals.12 It can prevent certain forms of cancer and reduce the risk of coronary heart disease according to clinical research.13 Salt stress is a common and effective approach to enhance GABA levels in bean sprouts. GABA accumulation in soybean sprouts under salt stress was not only caused by the activation of key synthesizing enzymes in GABA synthetic pathways but also by their gene expression up-regulation during dark germination.14 Previous studies were mainly focused on physiology, biochemistry and specific functional genes in soybeans under salt stress.14,15 However, systematic understanding of the molecular metabolic processes of GABA is still unclear, as are the parallel molecular metabolic networks of dark-germinated soybeans in response to salt stress.Proteomic approaches have been widely used to investigate plant resistance to salt stress. However, the mechanism of plant responses to salinity depends on the severity, duration of the stress, the growth conditions and the developmental stage of the plant, thus novel homeostasis between plant and environment at the proteome level will be established.16,17 Up until now, little is known about the salt stress responses of dark-germinated soybeans at the proteome level.18 Meanwhile, previous studies concerning proteomics were limited to 2-dimensional gel electrophoresis (2-DE) analysis. The defects of a low rate of low abundance protein detection and low reproducibility restricted the full potential of 2-DE in the comprehensive analysis of proteomic changes.19 In dark-germinated soybean cotyledons, 72 differentially abundant proteins (DAPs) were identified through 2-DE analysis, but 62.5% of these DAPs were involved in seed storage proteins.18 New methods have been developed for protein analysis with rapid innovations in proteomics. Recently, the isobaric tags for relative and absolute quantitation (iTRAQ) method was considered as a better alternative that provided an accurate quantitation of DAPs, especially for low abundance proteins.20,21 iTRAQ uses isotope labeling combined with multidimensional liquid chromatography and tandem mass spectrometry, simultaneously identifying and quantitatively comparing protein abundance by analyzing the peak intensities of reporter ions.22 Studies have demonstrated that iTRAQ is an effective method to investigate DAPs in soybeans under different stressful physiological conditions.23–26 To date, few studies have examined the dark-germinated soybean proteome under salt stress using iTRAQ methods.
In the present study, an iTRAQ-based proteomic technique was applied to investigate the proteome changes and identify DAPs in dark-germinated soybeans in response to 4 days of 50 mM NaCl treatment. This study provided a new insight into the physiological and biochemical processes induced by salt stress in soybeans during dark germination, as well as molecular and functional characterization at the protein level.
2. Experimental
2.1. Plant growth and NaCl treatment
Dry soybean seeds (Glycine max L. cultivar Yunhe) were surface-sterilized by soaking in 1% (v/v) sodium hypochlorite for 15 min,27 and were then washed and steeped in distilled water at 30 ± 1 °C for 4 h. The soaked seeds were placed in a sprout machine and germinated in a dark incubator at 30 ± 1 °C. The control sprouts were sprayed with distilled water every 24 h,28 while the salt treated sprouts were sprayed with water containing 50 mM NaCl every 24 h. Sprouts were harvested directly into liquid nitrogen 4 d after sowing until analysis.2.2. Measurement of fresh weight and content of malondialdehyde, H2O2, proline, soluble protein and free amino acids
For the determination of the fresh weight (FW) and dry weight (DW), 30 soybeans were weighed, followed by oven-drying at 80 °C for 48 h, and then the average weight was calculated. The extent of lipid peroxidation in terms of malondialdehyde (MDA) formation was measured based on the method of Madhava and Sresty.29 The H2O2 content was determined using a H2O2 Assay Kit (A064, Nanjing Jiancheng Bioengineering Institute, China). The amounts of proline, soluble protein and free amino acids were measured according to the method of Mostofa et al.302.3. Measurement of antioxidant activities
The dark-germinated soybeans (1.0 g) were homogenized with 5 mL of 50 mM phosphate buffer (pH 7.0) containing 1 mM EDTA and 2% PVP at 4 °C. After centrifugation at 4 °C and 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 20 min, the supernatant was collected and used for the enzyme activity assays. The superoxide dismutase (SOD), catalase (CAT), and peroxidase (POD) activities were determined using an SOD Assay Kit (A001, Nanjing Jiancheng Bioengineering Institute, China), a CAT Assay Kit (A007, Nanjing Jiancheng Bioengineering Institute, China), and a Plant POD Assay Kit (A084-3, Nanjing Jiancheng Bioengineering Institute, China) respectively, according to the manufacturer’s instructions. The glutathione peroxidase (GPX) activity was measured following the method of Drotar et al.31
000g for 20 min, the supernatant was collected and used for the enzyme activity assays. The superoxide dismutase (SOD), catalase (CAT), and peroxidase (POD) activities were determined using an SOD Assay Kit (A001, Nanjing Jiancheng Bioengineering Institute, China), a CAT Assay Kit (A007, Nanjing Jiancheng Bioengineering Institute, China), and a Plant POD Assay Kit (A084-3, Nanjing Jiancheng Bioengineering Institute, China) respectively, according to the manufacturer’s instructions. The glutathione peroxidase (GPX) activity was measured following the method of Drotar et al.31
2.4. Protein extraction, digestion, and iTRAQ labeling
The total protein in 4 d dark germinated soybeans was extracted using a P-PER @ Plant Protein Extraction Kit (89803, Thermo Scientific) according to the manufacturer’s instructions. The protein content was determined according to the Pierce™ Coomassie Protein Assay Kit (Thermo Scientific). For each protein sample, 150 μg of protein was reduced, alkylated, and trypsin digested following the method of Jin et al.,32 and labeled using the iTRAQ Reagent-8Plex Kit according to the manufacturer’s instructions (AB Sciex Inc., MA, USA). Three biological replicates were performed. After labeling, the samples were combined and lyophilized.2.5. MS/MS analysis using an LTQ-Orbitrap XL
The peptide mixture was dissolved in solvent A (2% acetonitrile in water, pH 10.0), and then fractionated using a Thermo UHPLC U3000 pump system (Thermo Fisher Scientific, San Jose, CA) equipped with an ACQUITY UPLC BEH C18 RP column (2.1 × 100 mm, 1.7 μm; Waters, USA). The peptides were eluted at a flow rate of 0.2 mL min−1 with a gradient of 97% buffer A for 12 min and 3–98% solvent B (98% acetonitrile in water, pH 10.0) for 40 min. The system was then maintained in 98% buffer B for 15 min. The absorbance at 214 nm was monitored and fractions were collected every 1 min. A total of 12 fractions were collected and dried in a vacuum concentrator.Each fraction was lyophilized and dissolved in solvent A (2% v/v acetonitrile, 0.1% v/v formic acid), and loaded onto a Thermo Scientific EASY column (2 cm × 100 μm, 5 μm-C18) and separated using a Thermo Scientific EASY column (75 μm × 100 mm, 3 μm-C18) at a flow rate of 300 nL min−1. Peptide separation was performed using the following gradient: 0–100 min from 97% A to 72% A, 100–120 min from 72% A to 45% A, 120–125 min from 45% A to 2% A, which was maintained for 19 min, 144–145 min from 2% A at 97% A, and 145–160 min of 97% A. MS/MS analysis was performed using an LTQ-Orbitrap XL mass spectrometer (Thermo Scientific, Bremen, Germany) according to Jin et al.32 Briefly, a full MS survey scan was performed using a mass range of 100–1800 m/z with a resolution of 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. High-energy collision dissociation fragmentation was used for MS/MS, and the five most intense signals in the survey scan were fragmented. A resolution of 7500 at 200 m/z was used with an isolation width of 2 m/z, 1 microscan, a maximum ion accumulation time of 200 ms, and a normalized collision energy of 40 eV.
000. High-energy collision dissociation fragmentation was used for MS/MS, and the five most intense signals in the survey scan were fragmented. A resolution of 7500 at 200 m/z was used with an isolation width of 2 m/z, 1 microscan, a maximum ion accumulation time of 200 ms, and a normalized collision energy of 40 eV.
2.6. Data analysis and interpretation
The raw MS/MS files were processed using the Proteome Discoverer 1.4 (Thermo Scientific Inc., Bremen, Germany) and the MASCOT search engine version 2.3.02 (Matrix Science) and were compared against the UniProt soybean database (Mar 10th, 2017). The following parameters were used in the search: trypsin as the digestion enzyme; one missed cleavage allowed; 10 ppm for the peptide ion tolerance and 0.02 Da for the fragment ion tolerance; the lowest and highest charges were +2 and +3, respectively; oxidation at methionine was set as the potential variable modification, whereas carbamidomethyl at cysteine was set as the fixed modification. Specifically, an automatic decoy database search was performed in SEQUEST by choosing the decoy checkbox in which a random sequence of the database is generated and tested for the raw spectra, as well as the real database. Only peptides identified at the 99% confidence interval by a SEQUEST probability analysis were counted as identified. In order to be identified as a significant DAP, a protein should be quantified with at least one unique peptide in each experimental replicate, with a p-value smaller than 0.05 and a fold change greater than 1.5 or less than 0.67.33Identified proteins were annotated with their biological functions according to KEGG (http://www.genome.jp/kegg/) and the literature. Information on the DAPs was obtained from the Universal Protein Resource (http://www.uniprot.org/). Pathway enrichment analysis was performed using KOBAS 3.0 (http://kobas.cbi.pku.edu.cn/).
2.7. Statistical analyses
All experiments in this study were repeated at least three times in independent experiments. Average values and standard deviations were computed according to the experimental data. One-way analysis of variance (ANOVA) with Tukey’s test was conducted on the data, and a p-value of 0.05 was considered to be significant.3. Results and discussion
3.1. Growth characteristics of dark-germinated soybeans under salt treatment
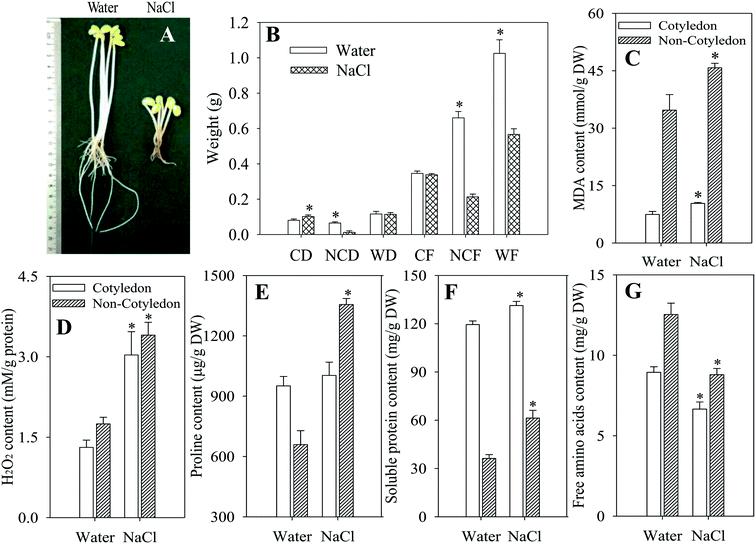
NaCl stress dramatically inhibited the growth and development of soybeans compared with the control (Fig. 1A). It resulted in a significant decrease in the DW and FW of non-cotyledons, as well as in the FW of the whole soybean, in comparison with non-stressed control treatments (p < 0.05). However, the DW of the whole soybean and FW of cotyledons did not differ between those treated with NaCl and the control (Fig. 1B). As indicators of cell damage, MDA and H2O2 content in both organs of dark-germinated soybeans were significantly enhanced by the stress of salt (p < 0.05) (Fig. 1C and D). Meanwhile, compared with the respective control, soluble protein content in sprouts sharply decreased while the free amino acid content significantly decreased after germinating for 4 days under NaCl stress (Fig. 1F and G). The proline level in cotyledons of soybeans was decreased while in non-cotyledons it was increased, relative to that of the control (Fig. 1E).3.2. Effects of salt stress on antioxidant enzyme activities
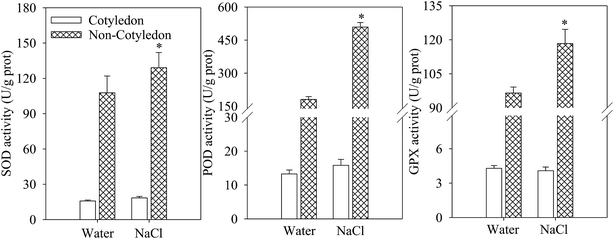
No significant difference was found in the activities of major antioxidant enzymes (SOD, POD, and GPX) in cotyledons of 4 day dark-germinated soybeans between NaCl treatment and the control (Fig. 2). However, in soybean non-cotyledons, the activities of SOD, POD and GPX were significantly increased (Fig. 2A–C).3.3. Effects of salt stress on GABA content
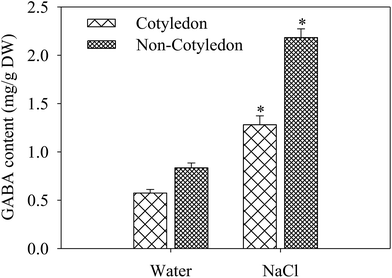
The GABA content in cotyledons and non-cotyledons of 4 day germinated soybeans under salt stress treatment was analyzed. After germinating for 4 days under NaCl stress, the GABA content increased significantly in both organs. It was 1.56- and 1.71-fold of the control in cotyledons and non-cotyledons, respectively (Fig. 3).3.4. iTRAQ analysis and identification of DAPs
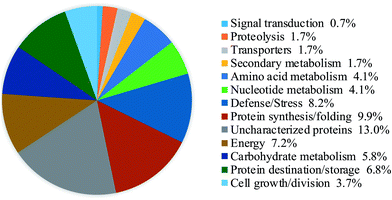
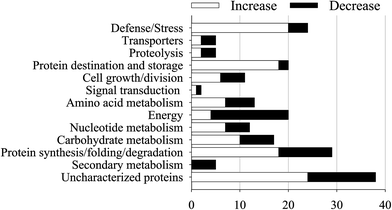
The proteins which had both a fold-change of more than 1.50 or less than 0.67 and a p-value of less than 0.05 were defined as DAPs. A total of 201 DAPs were identified in all 3 replicates (ESI Table S1†). According to the molecular functions listed on the UniProt and KEGG websites, the iTRAQ-quantified DAPs in the salt treatment group were functionally classified into 13 functional classes, i.e., defense/stress (8.2%), protein synthesis/folding (9.9%), energy (7.2%), carbohydrate metabolism (5.8%), protein destination/storage (6.8%), cell growth/division (3.7%), signal transduction (0.7%), nucleotide metabolism (4.1%), transporters (1.7%), secondary metabolism (1.7%), amino acid metabolism (4.1%), etc. (Fig. 4).After dark germinating for 4 days under NaCl stress, 119 DAPs showed a significantly increased abundance while 82 DAPs significantly decreased in soybeans (Fig. 5 and 6). The abundance of most of the defense/stress-related (83.3%), protein destination/storage-related (90.0%), protein synthesis/folding-related (62.1%), and carbohydrate metabolism-related (58.8%) DAPs increased significantly under NaCl treatment, while most of the secondary metabolism-related (100%) and energy-related (80.0%) DAPs decreased, compared with the non-stressed control (Fig. 5 and 6).
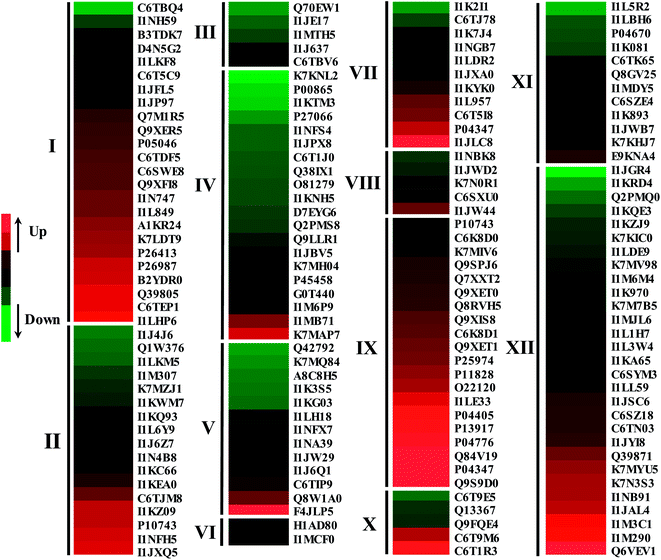 | ||
Fig. 6 Proteins differentially expressed in response to NaCl treatment in 4 day germinated soybeans. The expression change based on the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 transformed expression ratios of proteins was performed using the Gene Cluster 3.0 software. The results were visualized using the JAVA Treeview software. Protein numbers are same as those shown in Table 1. Proteins were involved in defense/stress (I), carbohydrate metabolism (II), secondary metabolism (III), energy (IV), amino acid metabolism (V), signal transduction (VI), cell growth/division (VII), proteolysis (VIII), protein destination/storage (IX), transporters (X), nucleotide metabolism (XI) and protein synthesis/folding (XII). 2 transformed expression ratios of proteins was performed using the Gene Cluster 3.0 software. The results were visualized using the JAVA Treeview software. Protein numbers are same as those shown in Table 1. Proteins were involved in defense/stress (I), carbohydrate metabolism (II), secondary metabolism (III), energy (IV), amino acid metabolism (V), signal transduction (VI), cell growth/division (VII), proteolysis (VIII), protein destination/storage (IX), transporters (X), nucleotide metabolism (XI) and protein synthesis/folding (XII). | ||
In order to analyze the metabolic pathways that responded to salt stress, DAPs were further investigated using the KEGG database. Pathway enrichment analysis demonstrated that 20 KEGG pathways were identified under salt stress with a p-value less than 0.05 as the threshold (Table 1). The top three enrichments of pathways were for ribosome, carbon metabolism and metabolic pathways and 19, 12 and 30 DAPs were found to be enriched in these pathways, respectively.
3.5. Discussion
Under NaCl stress, soybeans have performed a complicated metabolic balance at the cellular, organ, and whole-plant levels. According to our previous study, GABA content increased significantly in soybeans when germinating under 50 mM NaCl treatment, and after 4 d of germination its content was the highest.14 Hence, in the current study, morphological, physiological, and proteome changes in 4 day dark-germinated soybeans under 50 mM NaCl stress were observed. As a salt-sensitive plant, the growth and development of the soybeans were inhibited under NaCl treatment, and this was also reported in previous studies.34 However, the GABA content in germinated soybeans was significantly increased by the NaCl treatment (Fig. 3), which suggested that soybeans can be used as a good material to produce health-supporting functional ingredients. The underlying molecular mechanisms of dark-germinated soybean responses to salt stress need to be discussed further.Salt stress often leads to the plant cells producing reactive oxygen species (ROS), rapidly causing plant toxicity, such as membrane damage, inactive proteins, nucleic acid attack, and so on.35 As indicators of membrane damage and ROS levels, the amounts of MDA and H2O2 in salt-stressed soybeans were dramatically enhanced in the study (Fig. 1C and D). Plants have evolved a set of regulatory mechanisms, along with ROS-scavenging enzymes and antioxidant protection systems, to remove the salinity-triggered oxidative damage.36 Proline accumulation is considered to play a pivotal role in plant responses to salt stress, and it was significantly increased in the non-cotyledon organs of soybeans (Fig. 1E). Meanwhile, the increase in activities of ROS-detoxifying enzymes including SOD, POD and GPX in germinated soybean non-cotyledons was outlined (Fig. 2), and is similar to those of other soybeans37 and salt-responsive species.38,39 Similarly, the proteome results showed that the abundance of two SODs [Cu–Zn] (Q7M1R5, C6SWE8) was increased by salt treatment, implying their significant roles in response to salt stress (ESI Table 1†). NaCl stress significantly enhanced the accumulation of POD (Q9XFI8) while decreasing the POD (C6TBQ4) abundance, suggesting that different POD members may have different functions in salt stress responses in soybeans, which has also been reported in radishes under salt stress.40
In addition, from the present iTRAQ data, some defense-related DAPs were also induced under salt stress conditions, such as stress responsive alpha–beta barrel domain-containing protein (C6T5C9), low-temperature-induced 65 kDa protein (K7LDT9), heat shock 70 kDa protein (P26413) and stress-induced protein SAM22 (P26987). Cellular dehydration can be caused by the decreased water potential of a salt solution. Dehydrins and late embryogenesis abundant (LEA) proteins were among the most ubiquitous dehydration stress-responsive plant proteins.41 In this way, the increase in the levels of two late embryogenesis abundant (LEA) proteins (K7LDT9, I1L849) and two dehydrins (A1KR24, Q39805) under salt stress in the present study might contribute to the enhancement of salt tolerance in dark-germinated soybeans. An enhanced accumulation of dehydrin and LEA proteins was also found in salt-treated plants.41–43 The abundance changes in these DAPs under salt stress implied that the defense system in dark-germinated soybeans was provoked by the NaCl treatment.
The metabolic balance between protein synthesis and degradation played an important role in the plants’ responses to environmental stimuli.44 In translation, the ribosome is the major site where the synthesis of a polypeptide chain takes place, and the ribosomal protein species play a role in translation, ribosomal structure and biogenesis.45 In the present study, the analysis showed that the biggest enrichment of a pathway was in the ribosome and 19 DAPs were found to have been enriched in the pathway (Table 1). The current proteomic analysis showed that ribosomal proteins displayed significant changes in abundance under salt treatment, the species of which mainly belonged to five types with 26 S, 30 S, 40 S, 50 S and 60 S (ESI Fig. S1†). 11 of these ribosomal protein species were increased in abundance. It has been demonstrated that seed storage proteins (SSPs) not only function as storage reserves, but also play additional key roles.46 Under stress conditions, almost all of the SSPs were assigned to β-conglycinin subunits and glycinin was mobilized and its composition and abundance changed.47,48 An obvious trend was observed where most of the DAPs involved in protein destination and storage (18/20, 90%) suffered a marked increase in abundance. In general, SSPs in soybean cotyledons can be mobilized to provide carbon and nitrogen for soybean growth and development during germination, especially when the organic compounds could not be synthesized via photosynthesis in a dark artificial environment. However, our results showed that the dry weight of cotyledons increased dramatically under salt stress, compared with the control (Fig. 1B). This may be reasonably attributed to the germinating soybeans inhibiting use of SSPs for growth under NaCl stress. These results potentially implied that dark germinated soybeans cope with salt stress by promoting protein synthesis and suppressing protein degradation to maintain the protein metabolic balance, which was similar to the results of a previous study.45 Beyond that, two of the four eukaryotic translation initiation factors, associated with protein biosynthesis pathways, were induced by salt treatment.
| Pathway | Pathway ID | Input number | p-value |
|---|---|---|---|
| Ribosome | gmx03010 | 19 | 7.06 × 10−13 |
| Carbon metabolism | gmx01200 | 12 | 1.70 × 10−7 |
| Metabolic pathways | gmx01100 | 30 | 3.51 × 10−6 |
| Carbon fixation in photosynthetic organisms | gmx00710 | 4 | 0.0011 |
| Biosynthesis of secondary metabolites | gmx01110 | 16 | 0.0016 |
| Phagosome | gmx04145 | 4 | 0.0029 |
| RNA transport | gmx03013 | 5 | 0.0032 |
| Biosynthesis of amino acids | gmx01230 | 6 | 0.0034 |
| Beta-alanine metabolism | gmx00410 | 3 | 0.0050 |
| Citrate cycle (TCA cycle) | gmx00020 | 3 | 0.0058 |
| Pentose phosphate pathway | gmx00030 | 3 | 0.0065 |
| Galactose metabolism | gmx00052 | 3 | 0.0067 |
| Photosynthesis | gmx00195 | 3 | 0.0117 |
| Glycolysis/Gluconeogenesis | gmx00010 | 4 | 0.0123 |
| Fructose and mannose metabolism | gmx00051 | 3 | 0.0125 |
| Purine metabolism | gmx00010 | 4 | 0.0153 |
| Sulfur metabolism | gmx00230 | 2 | 0.0203 |
| Pyruvate metabolism | gmx00920 | 3 | 0.0218 |
| Protein processing in the endoplasmic reticulum | gmx00620 | 4 | 0.0425 |
| Alanine, aspartate and glutamate metabolism | gmx04141 | 2 | 0.0465 |
Metabolism represents the basic physiological processes that maintain cell living. In response to NaCl stress, the DAPs were involved in the carbohydrate, nucleotide, amino acid, energy and secondary metabolisms. In the present study, those pathways, such as carbon metabolism, biosynthesis of secondary metabolites, biosynthesis of amino acids, the citrate cycle (TCA cycle) and purine metabolism, were significantly enriched under salt stress (Table 1). In the present study, abundance changes of several enzymes were observed during salt stress, where enzymes were involved in the TCA cycle including dihydrolipoyllysine-residue succinyltransferase, pyruvate dehydrogenase E1 component subunit, acetyl-CoA carboxylase and malate dehydrogenases. However, most of the abundances of those enzymes remaining from TCA cycle-related DAPs decreased under salt stress, which was in agreement with the proteomic findings of salt-stressed radishes.45 Moreover, V-type proton ATPase, ATP synthase subunit alpha and ATP synthase subunit beta were critical to glycolysis and the TCA cycle. NaCl treatment decreased the abundance of all of the proteins identified above, which indicated that glycolysis and the TCA cycle suffered repression to some extent under salt stress. In addition, the abundance of all of these secondary metabolism-related protein species was strikingly decreased by salt stress. It was known that tubulin played a key role in division and growth of plant cells. Our results demonstrated that the abundances of tubulin alpha chain (I1K2I1) and tubulin beta chain (I1K7J4, I1LDR2) were all decreased by salt treatment, while the abundance of tubulin-specific chaperone A (C6T5I8) increased. These indicated that NaCl treatment inhibited plant growth of dark-germinated soybeans, which fully explained the reduced sprout length and the inhibited growth performance. Generally, the differential regulation of metabolism suggested that it might be managed by complex regulatory mechanisms to cope with salt stress in dark germinated soybeans.
4. Conclusions
In the present study, physiological changes were determined in dark germinated soybeans treated with 50 mM NaCl for up to 4 days. Accumulation of GABA in sprouts was enhanced by the salt stress, while the growth and development of soybeans were suppressed. Meanwhile, an iTRAQ-based proteomic technique was applied to compare the abundance of proteins in untreated and salt-stressed soybeans. The results showed that 201 DAPs were identified, and classified into 13 categories. Compared with the water treatment, 20 KEGG pathways were significantly enriched in soybeans under salt stress. Based on the results of the physiological and proteomic analyses, it was suggested that the dark germinated soybeans resisted salt stress through the induction of synthesis of ROS-scavenging enzymes and antioxidants. Promoted protein synthesis and suppressed protein degradation to maintain the protein metabolic balance were required for survival and growth of soybeans under salt stress. Meanwhile, our analysis indicated that carbohydrate, nucleotide and amino acid metabolism, energy supply and cell division were required to re-establish cellular homeostasis under salt stress. Our physiological and proteomic analyses of dark germinated soybeans under salt stress will benefit the understanding of the response process to salt stress of soybeans.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was financially supported by the Natural Science Foundation of China (31501401), the Natural Science Foundation of Jiangsu Province (BK 20150448), the Natural Science Foundation of Colleges and Universities in Jiangsu Province (15KJB550010) and the Natural Science Foundation of Yangzhou City (YZ2016044).References
- R. Pérez-Clemente, V. Vives, S. Zandalinas, M. López-Climent, V. Muñoz and A. Gómez-Cadenas, Biotechnological approaches to study plant responses to stress, BioMed Res. Int., 2013, 2013, 1–10 CrossRef PubMed.
- R. Dolferus, To grow or not to grow: a stressful decision for plants, Plant Sci., 2014, 229, 247–261 CrossRef PubMed.
- M. Świeca, Elicitation with abiotic stresses improves pro-health constituents, antioxidant potential and nutritional quality of lentil sprouts, Saudi J. Biol. Sci., 2015, 22, 409–416 CrossRef PubMed.
- M. Yuan, X. Jia, C. Ding, H. Zeng, L. Du, S. Yuan, Z. Zhang, Q. Wu, C. Hu and J. Liu, Effect of fluorescence light on phenolic compounds and antioxidant activities of soybeans (Glycine max L. Merrill) during germination, Food Sci. Biotechnol., 2015, 24, 1859–1865 CrossRef.
- Q. Bai, R. Yang, L. Zhang and Z. Gu, Salt Stress Induces Accumulation of γ-Aminobutyric Acid in Germinated Foxtail Millet (Setaria italica L.), Cereal Chem., 2013, 90, 145–149 CrossRef.
- R. Gan, M. Wang, W. Lui, K. Wu and H. Corke, Dynamic changes in phytochemical composition and antioxidant capacity in green and black mung bean (Vigna radiata) sprouts, Int. J. Food Sci. Technol., 2016, 51, 2090–2098 CrossRef.
- H. Wang, M. Gui, X. Tian, X. Xin and T. Wang, Effects of UV-B on vitamin C, phenolics, flavonoids and their related enzyme activities in mung bean sprouts (Vigna radiata), Int. J. Food Sci. Technol., 2017, 52, 827–833 CrossRef.
- M. Mendoza-Sánchez, R. G. Guevara-González, E. Castaño-Tostado, E. M. Mercado-Silva, J. A. Acosta-Gallegos, N. E. Rocha-Guzmán and R. Reynoso-Camacho, Effect of chemical stress on germination of cv Dalia bean (Phaseolus vularis L.) as an alternative to increase antioxidant and nutraceutical compounds in sprouts, Food Chem., 2016, 212, 128–137 CrossRef PubMed.
- Y. Guo, R. Yang, H. Chen, Y. Song and Z. Gu, Accumulation of γ-aminobutyric acid in germinated soybean (Glycine max L.) in relation to glutamate decarboxylase and diamine oxidase activity induced by additives under hypoxia, Eur. Food Res. Technol., 2012, 234, 679–687 CrossRef.
- S. Xing, Y. Jun, Z. Hau and L. Liang, Higher accumulation of gamma-aminobutyric acid induced by salt stress through stimulating the activity of diarnine oxidases in Glycine max (L.) Merr. roots, Plant Physiol. Biochem., 2007, 45, 560–566 CrossRef PubMed.
- X. Yan, P. Wang, T. Zhou, Z. Gu and R. Yang, NaCl–CaCl2 treatment enhancing nutritional and functional quality of mung bean sprouts, Emirates Journal of Food and Agriculture, 2017, 29, 123–131 CrossRef.
- Y. Ben-Ari, J. Gaiarsa, R. Tyzio and R. Khazipov, GABA: a pioneer transmitter that excites immature neurons and generates primitive oscillations, Physiol. Rev., 2007, 87, 1215–1284 CrossRef PubMed.
- M. Watanabe, K. Maemura, K. Oki, N. Shiraishi, Y. Shibayama and K. Katsu, Gamma-aminobutyric acid (GABA) and cell proliferation: focus on cancer cells, Histol. Histopathol., 2006, 21, 1135–1141 Search PubMed.
- Y. Yin, R. Yang, Q. Guo and Z. Gu, NaCl stress and supplemental CaCl2 regulating GABA metabolism pathways in germinating soybean, Eur. Food Res. Technol., 2014, 238, 781–788 CrossRef.
- R. Yang, Y. Yin, L. Guo, Y. Han and Z. Gu, Sequence analysis of diamine oxidase gene from fava bean and its expression related to γ-aminobutyric acid accumulation in seeds germinating under hypoxia-NaCl stress, J. Sci. Food Agric., 2014, 94, 1585–1591 CrossRef PubMed.
- Z. Hossain, A. Khatoon and S. Komatsu, Soybean proteomics for unraveling abiotic stress response mechanism, J. Proteome Res., 2013, 12, 4670–4684 CrossRef PubMed.
- W. Ji, R. Cong, S. Li, R. Li, Z. Qin, Y. Li, X. Zhou, S. Chen and J. Li, Comparative proteomic analysis of soybean leaves and roots by iTRAQ provides insights into response mechanisms to short-term salt stress, Front. Plant Sci., 2016, 7, 1–15 Search PubMed.
- Y. Yin, R. Yang and Z. Gu, Organ-specific proteomic analysis of NaCl-stressed germinating soybeans, J. Agric. Food Chem., 2014, 62, 7233–7244 CrossRef PubMed.
- G. Van den Bergh and L. Arckens, Recent advances in 2D electrophoresis: an array of possibilities, Expert Rev. Proteomics, 2005, 2, 243–252 CrossRef PubMed.
- F. C. S. Nogueira, G. Palmisano, V. Schwämmle, F. A. P. Campos, M. R. Larsen, G. B. Domont and P. Roepstorff, Performance of isobaric and isotopic labeling in quantitative plant proteomics, J. Proteome Res., 2012, 11, 3046–3052 CrossRef PubMed.
- S. Wiese, K. A. Reidegeld, H. E. Meyer and B. Warscheid, Protein labeling by iTRAQ: a new tool for quantitative mass spectrometry in proteome research, Proteomics, 2007, 7, 340–350 CrossRef PubMed.
- J. Fan, C. Chen, Q. Yu, R. H. Brlansky, Z. G. Li and F. G. Gmitter, Comparative iTRAQ proteome and transcriptome analyses of sweet orange infected by “Candidatus Liberibacter asiaticus”, Physiol. Plant., 2011, 143, 235–245 Search PubMed.
- W. Ji, R. Cong, S. Li, R. Li, Z. Qin, Y. Li, X. Zhou, S. Chen and J. Li, Comparative proteomic analysis of soybean leaves and roots by iTRAQ provides insights into response mechanisms to short-term salt stress, Front. Plant Sci., 2016, 7, 1–12 Search PubMed.
- W. Ji, J. Koh, S. Li, N. Zhu, C. P. Dufresne, X. Zhao, S. Chen and J. Li, Quantitative proteomics reveals an important role of GsCBRLK in salt stress response of soybean, Plant Soil, 2016, 402, 159–178 CrossRef.
- W. Zeng, Z. Sun, Z. Cai, H. Chen, Z. Lai, S. Yang and X. Tang, Proteomic analysis by iTRAQ-MRM of soybean resistance to Lamprosema Indicate, BMC Genomics, 2017, 18, 444–453 CrossRef PubMed.
- Y. Liu, J. Wu, N. Sun, C. Tu, X. Shi, H. Cheng, S. Liu, S. Li, Y. Wang and Y. Zheng, Intrinsically Disordered Proteins as Important Players during Desiccation Stress of Soybean Radicles, J. Proteome Res., 2017, 16, 2393–2409 CrossRef PubMed.
- Y. Guo, H. Chen, Y. Song and Z. Gu, Effects of soaking and aeration treatment on γ-aminobutyric acid accumulation in germinated soybean (Glycine max L.), Eur. Food Res. Technol., 2011, 232, 787–795 CrossRef.
- H. Zhang, G. Zhang, X. Lü, D. Zhou and X. Han, Salt tolerance during seed germination and early seedling stages of 12 halophytes, Plant Soil, 2015, 388, 229–241 CrossRef.
- K. Madhava Rao and T. Sresty, Antioxidative parameters in the seedlings of pigeonpea (Cajanus cajan(L.) Millspaugh) in response to Zn and Ni stresses, Plant Sci., 2000, 157, 113–128 CrossRef PubMed.
- M. G. Mostofa, M. A. Hossain, M. N. Siddiqui, M. Fujita and L. S. P. Tran, Phenotypical, physiological and biochemical analyses provide insight into selenium-induced phytotoxicity in rice plants, Chemosphere, 2017, 178, 212–223 CrossRef PubMed.
- A. Drotar, P. Phelps and R. Fall, Evidence for glutathione peroxidase activities in cultured plant cells, Plant Sci., 1985, 42, 35–40 CrossRef.
- X. Jin, R. Yang, L. Guo, X. Wang, X. Yan and Z. Gu, iTRAQ analysis of low-phytate mung bean sprouts treated with sodium citrate, sodium acetate and sodium tartrate, Food Chem., 2017, 218, 285–293 CrossRef PubMed.
- H. Ma, L. Song, Y. Shu, S. Wang, J. Niu, Z. Wang, T. Yu, W. Gu and H. Ma, Comparative proteomic analysis of seedling leaves of different salt tolerant soybean genotypes, J. Proteomics, 2012, 75, 1529–1546 CrossRef PubMed.
- W. Kao, T. Tsai, H. Tsai and C. Shih, Response of three Glycine species to salt stress, Environ. Exp. Bot., 2006, 56, 120–125 CrossRef.
- G. Tanou, A. Molassiotis and G. Diamantidis, Induction of reactive oxygen species and necrotic death-like destruction in strawberry leaves by salinity, Environ. Exp. Bot., 2009, 65, 270–281 Search PubMed.
- J. Dat, S. Vandenabeele, E. Vranová, M. Van Montagu, D. Inzé and F. Van Breusegem, Dual action of the active oxygen species during plant stress responses, Cell. Mol. Life Sci., 2000, 57, 779–795 CrossRef PubMed.
- K. Lu, W. Ding, S. Zhu and D. Jiang, Salt-induced difference between Glycine cyrtoloba and G. max in anti-oxidative ability and K+ vs. Na+ selective accumulation, J. Crop Sci., 2016, 4, 129–138 Search PubMed.
- N. L. Ma, W. A. C. Lah, N. A. Kadir, M. Mustaqim, Z. Rahmat, A. Ahmad, S. D. Lam and M. R. Ismail, Susceptibility and tolerance of rice crop to salt threat: Physiological and metabolic inspections, PLoS One, 2018, 13, 1–17 Search PubMed.
- A. Mishra and B. Tanna, Halophytes: potential resources for salt stress tolerance genes and promoters, Front. Plant Sci., 2017, 8, 1–15 Search PubMed.
- X. Sun, Y. Wang, L. Xu, C. Li, W. Zhang, X. Luo, H. Jiang and L. Liu, Unraveling the Root Proteome Changes and Its Relationship to Molecular Mechanism Underlying Salt Stress Response in Radish (Raphanus sativus L.), Front. Plant Sci., 2017, 8, 1–12 Search PubMed.
- C. Ruibal, I. P. Salamó, V. Carballo, A. Castro, M. Bentancor, O. Borsani, L. Szabados and S. Vidal, Differential contribution of individual dehydrin genes from Physcomitrella patens to salt and osmotic stress tolerance, Plant Sci., 2012, 190, 89–102 CrossRef PubMed.
- F. Brini, M. Hanin, V. Lumbreras, S. Irar, M. Pages and K. Masmoudi, Functional characterization of DHN-5, a dehydrin showing a differential phosphorylation pattern in two Tunisian durum wheat (Triticum durum Desf.) varieties with marked differences in salt and drought tolerance, Plant Sci., 2007, 172, 20–28 CrossRef.
- L. Maršálová, P. Vítámvás, R. Hynek, I. T. Prášil and K. Kosová, Proteomic response of Hordeum vulgare cv. Tadmor and Hordeum marinum to salinity stress: similarities and differences between a glycophyte and a halophyte, Front. Plant Sci., 2016, 7, 1–19 Search PubMed.
- I. V. Hinkson and J. E. Elias, The dynamic state of protein turnover: It’s about time, Trends Cell Biol., 2011, 21, 293–303 CrossRef PubMed.
- X. Sun, Y. Wang, L. Xu, C. Li, W. Zhang, X. Luo, H. Jiang and L. Liu, Unraveling the Root Proteome Changes and Its Relationship to Molecular Mechanism Underlying Salt Stress Response in Radish (Raphanus sativus L.), Front. Plant Sci., 2017, 8, 1–15 Search PubMed.
- M. P. Sales, I. R. Gerhardt, M. F. Grossi-de-Sá and J. Xavier-Filho, Do legume storage proteins play a role in defending seeds against bruchids?, Plant Physiol., 2000, 124, 515–522 CrossRef PubMed.
- L. Rajjou, M. Belghazi, R. Huguet, C. Robin, A. Moreau, C. Job and D. Job, Proteomic investigation of the effect of salicylic acid on Arabidopsis seed germination and establishment of early defense mechanisms, Plant Physiol., 2006, 141, 910–923 CrossRef PubMed.
- I. Alam, S. A. Sharmin, K. Kim, Y. Kim, J. J. Lee, J. D. Bahk and B. Lee, Comparative proteomic approach to identify proteins involved in flooding combined with salinity stress in soybean, Plant Soil, 2011, 346, 45–62 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra02996b |
| This journal is © The Royal Society of Chemistry 2018 |