Open Access Article
Open Access ArticleSynthesis and evaluation of new sterol derivatives as potential antitumor agents†
Xiang Chen a,
Yong Jun Ganb,
Yu Yu*a and
Yuan Zhang*c
a,
Yong Jun Ganb,
Yu Yu*a and
Yuan Zhang*c
aResearch Laboratory of Pharmaceutical Chemistry School of Pharmacy, Chongqing Medical University, Chongqing, People's Republic of China 400016. E-mail: 86487165@qq.com; Tel: +86-023-68485983
bExperimental Teaching Center, Chongqing Medical University, Chongqing, People's Republic of China 401331. E-mail: cqjy0708@126.com
cDepartment of Pharmacy, The First Affiliated Hospital of Chongqing Medical University, Chongqing, People's Republic of China 40016. E-mail: 36360496@qq.com
First published on 24th July 2018
Abstract
The current optimization of tetrazanbigen (TNBG) on the C-ring provided a series of new sterol derivatives 2a–2n. All new synthesized compounds were screened for their anti-proliferation activities against five human cancer cell lines (HepG2, QGY-7701, SMMC-7721, A-431 and NCI-H23 cell lines) in vitro. Among them, 2a, 2b, 2c, 2m and 2n exhibited high anti-proliferation activities on SMMC-7721, and their IC50 values approach that of the positive control drug cisplatin. Compound 2a not only showed strong anti-proliferation activities against QGY-7701 and HepG2 cell lines, with IC50 values (IC50: 6.81 ± 0.24 μM, 7.69 ± 0.87 μM) better than that of cisplatin (IC50: 8.75 μM, 18.89 ± 2.01 μM), but also exhibited good aqueous solubility (0.15–15 mg mL−1 at pH 7.4 and 2.0). On the most sensitive QGY-7701 cell line, Oil red O staining and western blot analysis were performed. The results suggested that 2a can inhibit the growth of cancer cells possibly by interfering with the lipid metabolism balance of tumor cells, resulting in lipid accumulation and cell apoptosis (lipotoxicity). Moreover, after being treated with 2a, lipid accumulation of QGY-7701 cell was increased in a time and dose dependent manner. Based on these promising results, 2a was selected for drug formulation and further pre-clinical development.
Introduction
As we all know, malignant tumors have become the main cause of death from disease in the world and now threaten all of us.1,2 Although there are many drugs and methods that can be available for cancer treatment, the successful treatment of malignant tumours continues to be a challenge on account of the serious side effects of drugs and the drug resistance of malignant tumors. Therefore, new anti-tumor drugs with lower side effects and better efficacy have become an urgent requirement at present.3Malignant cells are characterized by uncontrolled growth and proliferation. For rapid growth and proliferation, proliferating cells must take up enough of the right nutrients and energy to duplicate all macromolecular components and assemble the components into daughter cells.4,5 Unlike the energy source of normal cells, the main energy source of cancer cells relies on a non-oxidation energy source, such as anaerobic glycolysis that was observed in most cancer cells.6–8 Besides anaerobic glycolysis, a large number of research results showed that in order to meet the requirements of tumor cells growth, the biosynthesis of lipids in tumor cells is far more than that of normal cells.9,10 Highly proliferating tumor cells need enough lipids for membrane production and energy supplement.11,12 Lipid is the main resource of energy and nutrients to keep the rapid growth and proliferation of cancer cells.6–8,13 But, there are evidences suggested that an excessive accumulation of lipids in cancer cells may also cause deleterious effects.14,15 As a matter of fact, excess lipid accumulation in cancer cells may not only affect fundamental cellular processes, but also induce cancer cells apoptosis (collectively termed lipotoxicity).16,17 All the results indicated that the critical role of lipids in the growth and proliferation of cancer cells represents a potential candidate target that could be exploited for cancer therapy. Therefore, treating cancer through interfering with the lipid metabolism balance has been supposed to a good strategy by more and more researchers.15,18–20
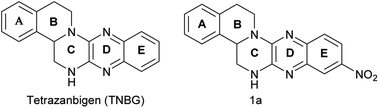
Subsequently, tetrazanbigen (TNBG, Fig. 1) was found to the effect which could disturb the lipid metabolism balance of tumor cells by inducing lipid accumulation.21 TNBG, a novel sterol derivative with isoquinoline moieties, exhibits lower side effects and strong antitumor efficacy against common solid tumors, including the liver cancer, the bladder cancer and human epidermoid carcinoma.22 in vivo experiment, once-daily administration of TNBG (1.5 mg kg−1 day−1) produces significant tumor growth inhibition in tumour volume and tumour weight. Percentage tumor growth inhibition (TGI%) was 87%. It was interesting to note that the results from pathological analysis in nude mice demonstrated that TNBG could induce lipid accumulation specifically in cancer tissue rather than in other normal organs (such as: heart, kidney, lung, blood vessels, brain, etc.).23,24 TNBG has been undergoing preclinical development as potential antitumor agent. However, a problem has limits its clinical development for a long time: the very poor water solubility; further study of drug formulation cannot be conducted. But these findings indicated that TNBG may serve as structural templates for designing and developing novel antitumor drug. Hence, transforming the structure of TNBG so as to significantly increase the potential druggability while maintaining or increasing the anti-tumor activity is the goal of this work.
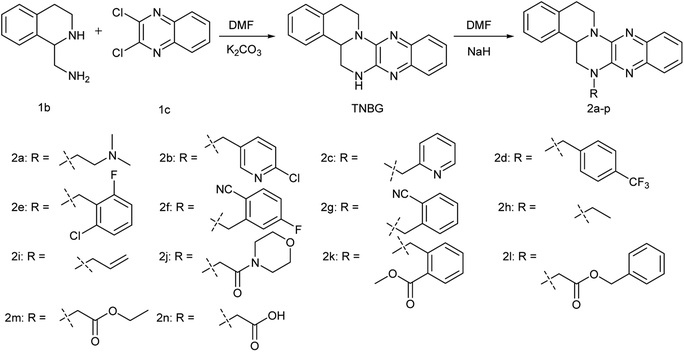
In our previous work, a series of TNBG derivatives were synthesized and their antiproliferative activities against four tumour cell lines were evaluated by 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide (MTT) assay.25 Unfortunately, these compounds have not shown sufficiently water-soluble for further clinical development. But, these work provided the following information about the structure–activity relationship (SAR) of TNBG: in the TNBG molecular structure, the isoquinoline moiety is essential for the anti-tumor effect. In fact, some steroid derivatives with isoquinoline moieties exhibited the ability to induce cellular lipid accumulation in vitro.26 In addition, when the substituent(s) group on E-ring of TNBG was a mono electron-withdrawing group (mono-EWGs), such as: nitro, cyano and so on, the antiproliferative activity showed an enhanced trend. It indicated that the introduction of mono-EWGs on E-ring of TNBG may have a positive effect (such as compound 1a, the structure of compound 1a was shown in Fig. 1). But, when the substituent(s) group on the E-ring or the A-ring of TNBG were mono-donating groups (mono-EDGs) or double electron- withdrawing groups (double-EWGs), the antiproliferative activity of TNBG derivatives showed the weakening trend.22–24 And thus, regulating electron cloud density on benzopyrazine ring to a proper degree was an important factor which is expected to maintain or increase the anti-tumor activity. Based on above results, the structural optimization of TNBG was mainly focused on C-ring in the current work. And then, R substituent(s) which are the polar or nonpolar or ionizable groups were introduced to the nitrogen atom of the C-ring. Accordingly, a series of TNBG derivatives with different R groups on the C-ring are reported herein (Fig. 2).
Results and discussion
Synthetic procedures
As was shown in Fig. 2, (1,2,3,4-tetrahydroisoquinolin-1-yl)methanamine (1b) was allowed to react with 2,3-dichloroquinoxaline (1c) in DMF in the presence of excess K2CO3 at 110 °C to produce TNBG with a yield 60%. This chemical reaction was facilitated by solid alkali. Therefore, a number of classical solid alkalis were initially screened, including potassium carbonate, sodium hydroxide, sodium carbonate and sodium bicarbonate, with the highest yield observed for potassium carbonate. Subsequently, some solvents, such as toluene and xylene were examined, but none of them surpassed N,N-dimethylformamide (DMF). The most probably reason of this situation is that DMF was the excellent solvent for the reactants. Finally, the effects of temperature were tested, revealing that decreasing the reaction temperature to 80 °C or increasing it to 145 °C diminished the yield.All target compounds were synthesized by TNBG reacting separately with different halogenated R groups in DMF in the presence of excess NaH at room temperature with the yield at 30–70%. All target compounds were chromatographed on silica gel. Elution with EtOAc/petroleum ether gave faint yellow solid. The TNBG are not novel and their NMR and MS spectra are in agreement with previous reports in the literature.21 The structures of compounds 2a–n were confirmed by analytical and spectral data (1H, 13CNMR and ESI-HRMS).
Determination of aqueous solubility: pH = 7.4 and 2.0
We measured the aqueous solubility (S) of all new synthesized compounds by using a shake-flask + UV-Vis spectrophotometry method at pH = 7.4 and 2.0 (the results are presented in Table 1). The aqueous solubility of 2a, 2b, 2c and 2n were better than that of TNBG at pH = 7.4 and 2.0. Among them, 2a had the best aqueous solubility (S: 15.68 mg mL−1) at pH 2.0 which represents a 150 fold increase over that of TNBG (S: 102.55 μg mL−1 at pH 2.0). At pH 7.4, the aqueous solubility of 2a was 152.55 μg mL−1 and the aqueous solubility of TNBG cannot be determined by using the shake-flask + UV-Vis spectrophotometry method at pH = 7.4. The most probably reason is that the R substituent of the C-ring has the presence of the amine group which can serve as H-bond acceptors improving aqueous solubility. Compared with TNBG, the others of the new compounds showed equivalent aqueous solubility.| Comp. | Sa (μg mL−1) | IC50b (μM) | ||||
|---|---|---|---|---|---|---|
| pH 7.4/2.0 | HepG2 | QGY-7701 | SMMC-7721 | A-431 | NCI-H23 | |
| a The aqueous solubility (S) of all new synthesized compounds were measured at pH 7.4/2.0 at 25 °C ± 1 °C by the shake-flask + UV-Vis spectrophotometry method. The phosphate buffer solution with pH = 2.0 and pH = 7.4 was chosen as solvent.b IC50 of the compounds in each cell lines were measured by CCk8 assay after 72 h of treatment. Experiments were repeated at least three times.c The inhibitory percentage of these cell lines at 20 μM was less than 45%.d “—” stands for not determined or cannot be determine. | ||||||
| 2a | 152.95/15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 680 680 |
7.69 ± 0.87 | 6.81 ± 0.24 | 10.19 ± 0.91 | — | — |
| 2b | 10.25/710.12 | 8.79 ± 0.91 | 7.58 ± 0.93 | 10.78 ± 0.96 | 18.64 ± 2.42 | 17.37 ± 2.15 |
| 2c | 21.30/750.60 | 8.64 ± 1.31 | 7.29 ± 0.77 | 9.34 ± 0.91 | >20c | >20c |
| 2d | —/— | 18.0 ± 1.04 | — | — | >20c | >20c |
| 2e | —/— | 18.22 ± 0.84 | — | >20c | >20c | >20c |
| 2f | —/— | — | — | — | >20c | >20c |
| 2g | —/— | >20c | — | >20c | — | >20c |
| 2h | —/535.96 | 11.28 ± 1.12 | — | — | >20c | 17.88 ± 0.59 |
| 2i | —/105.85 | 12.63 ± 0.95 | — | >20c | >20c | >20c |
| 2j | 9.21/— | 12.91 ± 0.75 | 11.31 ± 1.42 | 13.90 ± 0.99 | >20c | >20c |
| 2k | —/— | 14.37 ± 1.21 | 11.52 ± 0.63 | 18.18 ± 1.5 | >20c | >20c |
| 2l | —/— | 13.72 ± 0.62 | — | — | >20c | >20c |
| 2m | 12.65/— | 9.66 ± 0.91 | 9.0 ± 0.9 | 11.30 ± 0.76 | >20c | >20c |
| 2n | 806.32/150.21 | 8.04 ± 1.41 | 7.42 ± 0.63 | 9.15 ± 0.80 | 18.03 ± 0.61 | 16.32 ± 0.57 |
| TNBG | —/102.55 | 7.9 ± 0.28 | 7.16 ± 0.13 | 9.79 ± 1.1 | >20c | >20c |
| Cisplatin | —/— | 18.89 ± 2.01 | 8.75 ± 0.26 | 9.32 ± 0.68 | — | — |
CCK-8 cell proliferation assay
All new compounds were tested against five tumour cells (HepG2, QGY-7701, SMMC-7721, A-431 and NCI-H23 cell lines) for antiproliferative activity by the standard Cell Counting Kit-8 (CCK-8) assay. As shown in Table 1, among them, 2a, 2b, 2c, 2m and 2n exhibited highly anti-proliferation activities on SMMC-7721, and their IC50 values approach that of the positive control drugs cisplatin. And compound 2a showed strong anti-proliferation activities against QGY-7701, the IC50 values (IC50: 6.81 μM) better than that of cisplatin (IC50: 8.75 ± 0.26 μM). It was interesting to note that the A-431 cell and NCI-H23 cell showed much less sensitivity to all new synthesized compounds compared with QGY-7701 cell line. For example, the inhibitory percentage of 2c, 2j, 2k and 2m on A-431 and NCI-H23 cell lines was less than 45% at 20 μM, and the IC50 values of 2c, 2j, 2k, 2m on HepG2, QGY-7701 and SMMC-7721 cell lines were in the range between 7 to 12 μM. In addition, the sensitivity of HepG2 cell line to TNBG derivatives and the positive control drugs (cisplatin) was considerably different. The HepG2 cells were more sensitive to TNBG derivatives than to cisplatin, when comparing the IC50 of TNBG derivatives to that of cisplatin. The IC50 (7.69 ± 0.87 μM) of 2a on HepG2 cells was far less than that of cisplatin (18.89 ± 2.01 μM).Careful examination the differences in activity caused by different substituents, some new information about the structure–activity relationship (SAR) of TNBG derivatives were found. Firstly, the structural frameworks of 2b–c and 2d–g are the same (R substituent with aromatic rings), but compounds 2b and 2c were more active than the compounds 2d–g; this may be due to the nitrogen atom of the pyridine ring can serve as H-bond acceptors, and may form hydrogen bonds between the pyridine groups of 2b–2c and the target of drug action. Furthermore, the alkaline characteristics of pyridine may help compounds enriched around tumor cells. Although R substituent(s) of 2a did not have the aromatic rings, it could be viewed as a specific form of 2b and 2c; compared with 2b and 2c, its steric hindrance was rather low and its alkaline was much stronger. However, compared with TNBG, the activities fall significantly when nonpolar groups are introduced into the nitrogen atom of the C-ring such as compounds 2d–g and 2k. Apart from the amine group, carboxyl also exhibited highly anti-proliferation activities on HepG2, QGY-7701 and SMMC-7721 cell lines; the IC50 values of 2n approach that of TNBG.
Then, we estimated the IC50 of 2a on SMMC-7721, QGY-7701 and the normal liver cell line (LO2) under the same conditions.
As shown in Table 2, the IC50 values on LO2 cells were 2 times higher than those on SMMC-7721, QGY-7701 cell lines. This finding indicated that LO2 cell showed much less sensitivity to 2a compared with SMMC-7721, QGY-7701 cell lines. And then, based on these promising results, compound 2a was selected for further study.
Compound 2a could induce lipid accumulation
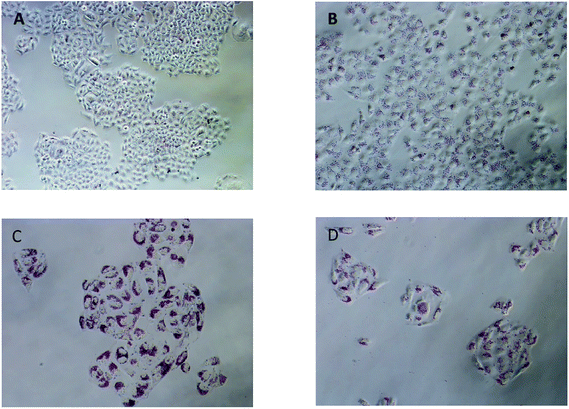
Lipid metabolism homeostasis is essential to maintain fundamental cellular processes. Lipid metabolism disorder may lead to multiple deleterious effects, such as lipid droplet (an excessive accumulation of the lipids); and then, an excessive accumulation of these lipids in cancer cells can also induce cancer cells apoptosis (lipotoxicity). In our previous study, Yuan et al. pointed out that “the TNBG antitumor mechanism is probably to influence tumor lipid metabolism, resulting in accumulation of LDs in tumor cells”.24 Zheng et al. further pointed out that there is a positive correlation between lipid accumulation induced by TNBG and tumor cell apoptosis in tumor cells.23During the in vivo antitumor activities of 2a was tested by the CCK-8 assay, a large number of red granular substances were found in the tumor cells. Oil Red O is a lipid specific dye which could be used to quickly and easily quantify the extent of lipid accumulation.27 Therefore, the Oil Red O was employed. After treatment with 2a for 24 h, a large number of red granular substance were found in QGY-7701 cells; Oil red O staining confirmed it that was accumulated lipid droplets (Fig. 3B). The lipid droplets were not found in untreated group (Fig. 3A). After treatment with 2a for 48 h and 72 h, respectively, the lipid droplet volumes grow larger and increasingly obvious (Fig. 3C and D); eventually, the accumulated lipid droplets caused cell membrane rupture, cell apoptosis, and cell number reduction.
For the sake of further understanding the correlation between the lipid accumulation and the antiproliferative activity of 2a, the quantitative analysis of lipid accumulational factors were performed. QGY-7701 cells were exposed to different concentrations (0, 1, 2, 4, 8 μg mL−1) of compound 2a for 24, 48, and 72 hour, respectively. In different concentration and time, the accumulated lipid was stained with Oil red O, and then isopropyl alcohol was added to the stained culture dish and determining the amount of extracted dye at 510 nm. The time- and dose-dependent curves of lipid droplets accumulation in QGY-7701 cell lines were obtained (Fig. 4).
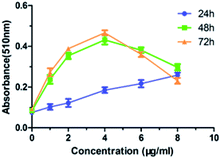 | ||
| Fig. 4 The time- and dose-dependent curves of lipid droplets accumulation in QGY-7701 cells. (n = 3). | ||
As shown in Fig. 4, in the range between 0 to 8 μg mL−1, the lipid droplets increased with increasing concentration of 2a in the 24 h treatment group. In the other two treatment groups (48 h and 72 h), the absorbance curves of lipid accumulation were analogical. And in the range between 0 and 4 μg mL−1, the lipid droplets increased with increasing concentrations of compound 2a; however, in the range between 4 and 8 μg mL−1, the absorbance curves of lipid accumulation decreased with increasing concentrations of compound 2a. And it was probably due to cell apoptosis and cell number reduction caused by the powerful antiproliferative activity of compound 2a. At the same concentration conditions, the absorbance curves of lipid accumulation increased with time (Fig. 4). All these results showed that the amount of accumulated lipid droplets induced by compound 2a was proportional to time and dose below certain concentration.
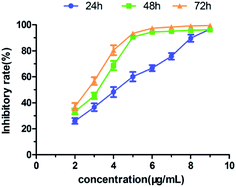
At the meantime, QGY-7701 cells were exposed to different concentrations (2, 3, 4, 5, 6, 7, 8, 9 μg mL−1) of compound 2a for 24, 48, and 72 hour, respectively. In different concentration and time, the relative inhibition rates of tumor cell growth were evaluated by the CCK-8 assay. As shown in the Fig. 5, it was found that the relative inhibition rates of QGY-7701 cell growth increased with increasing time and dose. All these results further indicated that the anti-proliferation activity of 2a increased in a time- and dose- dependent manner below certain concentration.
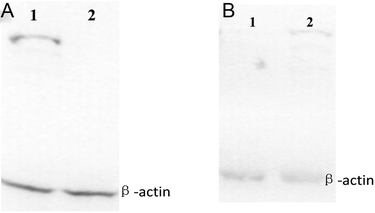
Western blot analysis
Sterol regulatory element binding protein-1 (SREBP-1), a transcription factor, plays a vital role in lipid homeostasis and lipoprotein metabolism. Upregulation of SREBP-1 can promote intracellular lipid synthesis.28 Microsomal triglyceride transfer protein (MTTP) is a lipid-binding protein, which is involved in the transport and metabolism of intracellular lipid and mainly exists in the endoplasmic reticulum lumen of cell. Absolute or relative absence of MTTP in cell may result in decrease of the intracellular lipid transport activity. The lipid accumulates when the pathway of lipid excreted from cells by the form of lipoprotein was suppressed.29,30 Western blot analysis was used to detect the changes in SREBP-1 and MTTP in QGY-7701 cell. As shown in the Fig. 6, western blot analysis demonstrated that SREBP-1 was in up-regulation and MTTP was in relative absence in the 2a-treated QGY-7701 cell line. These results indicated that 2a can result in the lipid droplet accumulation of the cancer cell by inducing upregulation of SREBP-1 and down regulation of MTTP. Excess lipid droplet accumulations in cancer cells may interfere with the lipid metabolism balance of tumor cells and result in further lipid accumulation and cells apoptosis (lipotoxicity).Experimental
Reagents, cell lines and culture and equipment
All reagents were purchased from Sigma Chemical Co. (St. Louis, MO, USA) and the solvents were purchased from Chengdu Chron Chemicals Co,. Ltd (Chengdu, China). (1,2,3,4-Tetrahydroisoquinolin-1-yl)methanamine (1b, purity > 95%) and 2,3-dichloroquinoxaline (1c, purity > 95%) were synthesized by Research Laboratory of Pharmaceutical Chemistry School of Pharmacy, Chongqing Medical University, China. Unless otherwise noted, all reagents were used as received without further purification. The HRMS analysis was performed on Hybrid Quadrupole-TOF LC/MS/MS Mass Spectrometer. The NMR spectrum was recorded by Innovative Drug Research Centre, School of Pharmaceutical Sciences Chongqing University, China. All the five human cancer cell lines were obtained from Sichuan Kelun Pharmaceutical Research Institute (Sichuan, China).General procedure for the synthesis of compounds 2a–n
To a suspension of 0.3 g of NaH (60% oil dispersion, 0.008 mol) in dimethylformamide (DMF 10 mL) was added TNBG (0.58 g 0.002 mol). The mixture was cooled to 0 °C, and then halogenated R groups (0.008 mol) was slowly added dropwise between 0 to 5 °C. After the reaction mixture was stirred for 3 h at 0 °C, and ethanol (20 mL) was carefully added dropwise to the reaction mixture. Then, the reaction mixture was poured into 100 mL ice-water; the mixture was extracted with methylene dichloride (50 mL × 3). And the organic phase was washed with H2O (20 mL × 3), brine (20 mL × 3), and then dried with anhydrous sodium sulfate and filtered. The filtrate was removed under vacuum to give amber oil. This was chromatographed on silica gel. Elution with 0–15% EtOAc/petroleum ether gave a faint yellow solid.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–), 5.38–5.16 (m, 3H, CH2
CH–), 5.38–5.16 (m, 3H, CH2![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) , c-CH), 4.84 (d, J = 8.35 Hz, 1H, c-CH2), 4.46 (s, 2H, CH2), 3.85 (dd, J = 8.45 Hz, 2.82 Hz, 1H, c-CH2), 3.44 (t, J = 10.25 Hz, 1H, c-ArCH2), 3.18–3.04 (m, 2H, c-BnCH2), 2.97–2.86 (m, 1H, c-ArCH2); 13C-NMR (600 MHz, DMSO-d6): δ 143.4, 142.9, 137.2, 136.8, 135.4, 133.8, 133.2, 129.4, 127.4, 126.7, 125.9, 125.2, 125.0, 124.6, 124.2, 117.8, 52.6, 50.9, 50.0, 28.4; HRMS (ESI) m/z calcd for C21H21N4 (M + H)+ 329.1761, found 329.1769.
, c-CH), 4.84 (d, J = 8.35 Hz, 1H, c-CH2), 4.46 (s, 2H, CH2), 3.85 (dd, J = 8.45 Hz, 2.82 Hz, 1H, c-CH2), 3.44 (t, J = 10.25 Hz, 1H, c-ArCH2), 3.18–3.04 (m, 2H, c-BnCH2), 2.97–2.86 (m, 1H, c-ArCH2); 13C-NMR (600 MHz, DMSO-d6): δ 143.4, 142.9, 137.2, 136.8, 135.4, 133.8, 133.2, 129.4, 127.4, 126.7, 125.9, 125.2, 125.0, 124.6, 124.2, 117.8, 52.6, 50.9, 50.0, 28.4; HRMS (ESI) m/z calcd for C21H21N4 (M + H)+ 329.1761, found 329.1769.General procedure: determination of aqueous solubility: pH = 1.0 and pH = 7.4
The phosphate buffer solution with pH = 2.0 and pH = 7.4 was chosen as solvent. According to the “Chinese Pharmacopoeia (2015)”, the phosphate buffer solution with pH = 2.0 was prepared using H3PO4 and Na2HPO4, and the phosphate buffer solution with pH = 7.4 was prepared using NaOH and KH2PO4. A (0.2 mg mL−1) stock solution of each compound in pH 7.4 and pH 2.0 phosphate buffered saline was prepared, respectively; and the stock solutions were diluted to a theoretical concentration of 2, 4, 6, 8 and 10 μg mL−1 to generate a standard curve. The spectra were recorded in the region 200–400 nm using a Shimadzu UV-1800 with a 10 mm quartz cell. Proper absorbance wavelengths for calculations were selected for each compound manually based on absorbance maxima.5 mL of phosphate buffer solution was accurately measured and saturated with an excess of the test compound, and then equilibrated at 25 °C on a water-bathing constant temperature vibrator for 24 h. The mixed solutions were filtered. According to experiment needs, the filtrate may be diluted to the appropriate concentration and tested by UV–Vis spectro-photometry at the proper absorbance wavelengths. The solubility of compounds was calculated by Microsoft Excel. For each compound, three measurements were performed.
Cell proliferation assay
The anti-tumor growth effects of all new synthesized compounds in vitro were studied via measuring the viability of five tumour cells (HepG2, QGY-7701, SMMC-7721, A-431 and NCI-H23 cell lines) using the Cell Counting Kit-8 (CCK8) according to the manufacturer's instruction. In short, tumour cells were seeded in 96-well plates (1 × 104 of cells per well in a final volume of 200 μL); followed by exposure to different concentrations (0, 2, 3, 4, 5, 6, 7, 8 μg mL−1) of compounds for 24, 48 and 72 h, respectively. After washed with PBS, (10 μL) CCK-8 was added into each well, and the cells were incubated for an additional 1 h at 37 °C. The OD values were determined at 450 nm using a microplate reader. The relative inhibition rates of cancer cell growth and IC50 values were calculated by GraphPad Prism 5. Experiments were repeated at least three times.Oil red O staining and quantification of lipids
QGY-7701 cells were seeded in 96-well plates (1 × 104 of cells per well in a final volume of 200 μL); the cells were divided into (0, 1, 2, 4, 8 μg mL−1) 5 treatment groups. Each treatment group was treated with the corresponding concentration of compound 2a for 24 hours, 48 hours, and 72 hours, respectively. And then, the cell culture solution was decanted. The cells were carefully washed with phosphate-buffered saline (PBS). Cultures were fixed for at least 1 h with 10% formalin at room temperature and rinsed with bidistilled water. The fixed cells were stained for 1 h by complete immersion in the working solution of Oil red O. After the Oil Red O staining was performed, the morphological changes in the formation and accumulation of lipid droplets were observed by imaging with Olympus inverted microscope. In order to determine the extent of lipid accumulation, 200 μL of isopropyl alcohol was added per well, the extracted dye was immediately removed by gentle pipetting and its absorbance was determined at 510 nm.Preparation of the working solution of Oil red O: 1.05 g oil red O was accurately weighed and dissolved in isopropanol (300 mL). Then, the solution was left overnight at room temperature and filtered. 225 mL double distilled water was added and left at 4 °C for the night and then filtered. The filtrate was stored for use.
Western blot assay
QGY-7701 cells were seeded in 6-well plates (1 × 104 of cells per well in a final volume of 200 μL), and followed by exposure to 3 μg mL−1 2a for 72 h. And then, the cell culture solution was decanted. The cells were carefully washed with ice-cold phosphate-buffered saline (PBS) and lysed with lysis buffer. And then according to the instruction of Subcellular Structure Protein Extraction Kit, the membrane protein was extracted. The membrane proteins were separated by 12% SDS-PAGE and transferred to polyvinylidene fluoride (PVDF) membrane. After the membranes were incubated with 5% nonfat milk at 4 °C for the night, the corresponding primary antibodies (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 3% BSA PBST dilutions) were added and incubated at 4 °C for the night and then rinsed in PBST (15 min × 1, 5 min × 4). The corresponding secondary antibody (1
200 3% BSA PBST dilutions) were added and incubated at 4 °C for the night and then rinsed in PBST (15 min × 1, 5 min × 4). The corresponding secondary antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2500 3% BSA dilutions) were added and shook at 25 °C for 1 h and then rinsed in PBST (15 min × 1, 5 min × 4). The protein expression was detected with the Super Signal West Pico Substrate (Pierce, Rockford, IL, USA).
2500 3% BSA dilutions) were added and shook at 25 °C for 1 h and then rinsed in PBST (15 min × 1, 5 min × 4). The protein expression was detected with the Super Signal West Pico Substrate (Pierce, Rockford, IL, USA).
Conclusions
In summary, our earlier work directed us to design and synthesize 14 new sterol derivatives. All new synthesized compounds were screened for their anti-proliferation activities against five human cancer cell lines (HepG2, QGY-7701, SMMC-7721, A-431 and NCI-H23 cell lines) in vitro. Among them, 2a, 2b, 2c, 2m and 2n exhibited highly anti-proliferation activities on SMMC-7721, and their IC50 values approach that of the positive control drugs cisplatin. And compound 2a not only showed strong anti-proliferation activities against QGY-7701 and HepG2 cell lines, the IC50 values (IC50: 6.81 ± 0.24 μM, 7.69 ± 0.87 μM) better than that of cisplatin (IC50: 8.75 μM, 18.89 ± 2.01 μM), but also exhibited good aqueous solubility (0.15–15 mg mL−1 at pH 7.4 and 2.0). In addition, the sensitivity of five human cancer cell lines to TNBG derivatives was considerably different. The A-431 cell and NCI-H23 cell showed much less sensitivity to all new synthesized compounds compared with HepG2, QGY-7701 and SMMC-7721 cell lines. The most sensitivity to TNBG derivatives was QGY-7701 cell line.Some new information about the structure–activity relationship (SAR) of TNBG derivatives were found that the R substituent on the 6-positions of the ring C was modifiable and could maintain or increase the anti-tumor activity; when the R substituent is an amino group, the highest antitumor activity and the appropriate water-solubility are observed. The most probably reason of this situation is that the R substituent of the C-ring have the presence of the amine group which can serve as H-bond acceptors improving aqueous solubility and anti-proliferation activities.
Further studies of 2a on the most sensitive QGY-7701 cell line suggested that 2a can inhibit the growth of the cancer cell possibly by interfering with the lipid metabolism balance of tumor cells and resulting in lipid accumulation and cells apoptosis (lipotoxicity). And after being treated with 2a, lipid accumulation of QGY-7701 cell was increased in a time and dose dependent manner. So far, 2a exhibited the requisite biological properties necessary for a useful drug. Based on these promising results, compound 2a was selected for drug formulation and further pre-clinical development.
Conflicts of interest
The authors declare no conflict of interest associated with this manuscript.Acknowledgements
This research work was financially supported through grants from National Science Foundation of China (Grants 81172097, 30171070, 30371632, 30772595 and 81201693).Notes and references
- K. Devinder, S. Kuldeep, V. Vikas, Y. Kavita, B. Avinash and K. Vinod, Design, regioselective synthesis and cytotoxic evaluation of 2-aminoimida zoleequinoline hybrids against cancer and primary endothelial cells, Eur. J. Med. Chem., 2014, 87, 150–158 CrossRef PubMed.
- G. Z. Yang, L. W. Fu, Y. Ai, Y. J. Liang, J. C. Liu, H. W. He, Y. Chen and C. Tang, Synthesis and in vitro antiproliferative evaluation of pyrimido[5,4-c]quinoline-4-(3H)-one derivatives, Eur. J. Med. Chem., 2012, 47, 206–213 CrossRef PubMed.
- L. B. O. Freitas, T. F. Borgati, R. P. D. Freitas, A. L. T. G. Ruiz, G. M. Marchetti, J. E. D. Carvalho, E. F. F. D. Cunha, T. C. Ramalho and R. B. Alves, Synthesis and antiproliferative activity of 8-hydroxyquinoline derivatives containing a 1,2,3-triazole moiety, Eur. J. Med. Chem., 2014, 84, 595–604 CrossRef PubMed.
- R. J. DeBerardinis, J. J. Lum, G. Hatzivassiliou and C. B. Thompson, The biology of cancer: metabolic reprogramming fuels cell growth and prolifera-tion, Cell Metab., 2008, 7, 11–20 CrossRef PubMed.
- R. J. DeBerardinis, N. Sayed, D. Ditsworth and C. B. Thompson, Brick by brick: metabolism and tumor cell growth, Curr. Opin. Genet. Dev., 2008, 18, 54–61 CrossRef PubMed.
- C. S. Yeh, J. Y. Wang, T. L. Cheng, C. H. Juana, C. H. Wua and S. R. Lina, Fatty acid metabolism pathway play an important role in carcinogenesis of human colorectal cancers by microarray-bioinformatics analysis, Cancer Lett., 2006, 233, 297–308 CrossRef PubMed.
- E. Furuta, H. Okuda, A. Kobayashi and K. Watabe, Metabolic genes in cancer: their roles in tumor progression and clinical implications, Biochim. Biophys. Acta, 2010, 1805, 141–152 Search PubMed.
- M. G. Vander, L. C. Cantl and C. B. Thompson, Understanding the Warburg effect: the metabolic requirements of cell proliferation, Science, 2009, 324, 1029–1033 CrossRef PubMed.
- J. A. Menendez, Fine-tuning the lipogenic/lipolytic balance to optimize the metabolic requirements of cancer cell growth: molecular mechanisms and therapeutic perspectives, Biochim. Biophys. Acta, 2010, 1801, 381–391 CrossRef PubMed.
- D. K. Nambiar, G. Deep, R. P. Singh, C. Agarwal and R. Agarwal, Silibinin inhibits aberrant lipid metabolism, proliferation and emergence of androgen-independence in prostate cancer cells via primarily targeting the sterol response element binding protein 1, Oncotarget, 2014, 5, 10017–10033 CrossRef PubMed.
- M. T. Accioly, P. Pacheco, C. M. Maya-Monteiro, N. Carrossini, B. K. Robbs, S. S. Oliveira, C. Kaufmann, J. A. Morgado-Diaz, P. T. Bozza and J. P. B. Viola, Lipid bodies are reservoirs of cyclooxygenase-2 and sites of prostaglandin-E2 synthesis in colon cancer cells, Cancer Res., 2008, 68, 1732–1740 CrossRef PubMed.
- S. Kanno, K. Kurauchi, A. Tomizawa, S. Yomogida and M. Ishikawa, Albumin modulates docosahexaenoic acid-induced cytotoxicity in human hepatocellular carcinoma cell lines, Toxicol. Lett., 2011, 200, 154–161 CrossRef PubMed.
- T. Mashima, H. Seimiya1 and T. Tsuruo, De novo fatty-acid synthesis and related pathways as molecular targets for cancer therapy, Br. J. Cancer, 2009, 100, 1369–1372 CrossRef PubMed.
- N. A. Herpen and V. B. Schrauwen-Hinderling, Lipid accumulation in nonadipose tissue and lipotoxicity, Physiol. Behav., 2008, 94, 231–241 CrossRef PubMed.
- K. J. Williams, J. P. Argus, Y. Zhu, M. Q. Wilks, B. N. Marbois, A. G. York, Y. Kidani, A. L. Pourzia, D. Akhavan, D. N. Lisiero, E. Komisopoulou, A. H. Henkin, H. Soto, B. T. Chamberlain, L. Vergnes, M. E. Jung, J. Z. Torres, L. M. Liau, H. R. Christofk, R. M. Prins, P. S. Mischel, K. Reue, T. G. Graeber and S. J. Bensinger, An essential requirement for the SCAP/SREBP signaling axis to protect cancer cells from lipotoxicity, Cancer Res., 2013, 73, 2850–2862 CrossRef PubMed.
- N. Zaidi, L. Lupien, N. B. Kuemmerle, W. B. Kinlaw, J. V. Swinnen and K. Smans, Lipogenesis and lipolysis: the pathways exploited by the cancer cells to acquire fatty acids, Prog. Lipid Res., 2013, 52, 585–589 CrossRef PubMed.
- A. Reinartz, J. Ehling, A. Leue, C. Liedtke, U. Schneider, J. Kopitz, T. Weiss, C. Hellerbrand, R. Weiskirchen, R. Knüchel and N. Gassler, Lipid-induced up-regulation of human acyl-CoA synthetase 5 promotes hepatocellular apoptosis, Biochim. Biophys. Acta, 2010, 1801, 1025–1035 CrossRef PubMed.
- S. Miaoxian, H. Y. Chung and Y. L. Li, Deoxyelephantopin from Elephantopus scaber L. induces cell-cycle arrest and apoptosis in the human nasopharyngeal cancer CNE cell, Biochem. Biophys. Res. Commun., 2011, 411, 342–347 CrossRef PubMed.
- W. L. Lee and L. F. Shyur, Deoxyelephantopin impedes mammary adenocarcinoma cell motility by inhibiting calpain-mediated adhesion dynamics and inducing reactive oxygen species and aggresome formation, Free Radical Biol. Med., 2012, 52, 1423–1436 CrossRef PubMed.
- Y. Salma, E. Lafont, N. Therville, S. Carpentier, M. J. Bonnafé, T. Levade, Y. Génisson and N. Andrieu-Abadie, The natural marine anhydrophytosphingosine, Jaspine B, induces apoptosis in melanoma cells by interfering with ceramide metabolism, Biochem. Pharmacol., 2009, 78, 477–485 CrossRef PubMed.
- Y. Yu, Preparation and used of a kind of azagonane compound, Chinese Patent, CN1124251A, 2001.
- L. L. Li, A survey on the mechanism of a new antitumor drug TNBG with novel configuration. Doctor thesis, Chongqing Medical University, Chinese Chongqing, 2005.05.
- X. H. Zheng, W. Li, Z. P. Lan, X. L. Yang, L. J. Li, Y. H. Yuan, Z. Xia, X. G. Chen, X. Y. Zhang and Y. Yu, Antitumour effects of tetrazanbigen against human hepatocellular carcinoma QGY-7701 through inducing lipid accumulation in vitro and in vivo, J. Pharm. Pharmacol., 2015, 67, 1593–1602 CrossRef PubMed.
- Y. H. Yuan, W. Li, L. J. Li, X. L. Yang, R. Gu, H. B. Liu, K. H. Huang and Y. Yu, Effects of Tetrazanbigen on the Protein Expression in Human Hepatocellular Carcinoma Cell Line QGY-7701, J. Huazhong Univ. Sci. Technol., Med. Sci., 2009, 29(3), 304–308 CrossRef PubMed.
- M. M. Zhu, The structure–activity relationship and interaction of the new anti-tumor TNBG and its derivatives, Master thesis, Chongqing Medical University, Chinese Chongqing, 2013.05.
- Q. Zhou, Z. K. Melkoumian, A. Lucktong, M. Moniwa, J. R. Davie and J. S. Strobl, Rapid induction of histone hyperacetylation and cellular differentiation in human breast tumor cell lines following degradation of histone deacetylase-1, J. Biol. Chem., 2000, 275, 35256–35263 CrossRef PubMed.
- J. L. Ramirez-Zacarias, F. Castro-Mufiozledo and W. Kuri-Harcuch, Quantitation of adipose conversion and triglycerides by staining intracytoplasmic lipids with Oil red O, Histochemistry, 1992, 97, 493–497 Search PubMed.
- M. S. Brown and J. L. Goldstein, The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor, Cell, 1997, 89(3), 331–340 CrossRef PubMed.
- A. Atzel and J. R. Wetterau, Mechanism of Microsomal Triglyceride Transfer Protein catalyzed lipid transport, Biochemistry, 1993, 32(39), 10444–10450 CrossRef PubMed.
- S. Bernard, S. Touzet and I. Personne, Association between microsomal triglyceride transfer protein gene polymorphism and the biological features of liver steatosis in patients with type II diabetes, Diabetologia, 2000, 43(8), 995–999 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra04152k |
| This journal is © The Royal Society of Chemistry 2018 |