Open Access Article
Open Access ArticleCharacterization of single-stranded DNA-binding protein SsbB from Staphylococcus aureus: SsbB cannot stimulate PriA helicase
Kuan-Lin Chena,
Jen-Hao Chenga,
Chih-Yang Linab,
Yen-Hua Huanga and
Cheng-Yang Huang *ac
*ac
aSchool of Biomedical Sciences, Chung Shan Medical University, No. 110, Sec. 1, Chien-Kuo N. Rd., Taichung City, Taiwan. E-mail: cyhuang@csmu.edu.tw; Tel: +886-4-24730022 ext. 11472
bSchool of Medicine, College of Medicine, Chung Shan Medical University, No. 110, Sec. 1, Chien-Kuo N. Rd., Taichung City, Taiwan
cDepartment of Medical Research, Chung Shan Medical University Hospital, No. 110, Sec. 1, Chien-Kuo N. Rd., Taichung City, Taiwan
First published on 7th August 2018
Abstract
Single-stranded DNA-binding proteins (SSBs) are essential to cells as they participate in DNA metabolic processes, such as DNA replication, repair, and recombination. The functions of SSBs have been studied extensively in Escherichia coli. Unlike E. coli, which contains only one type of SSB (EcSSB), some bacteria have more than one paralogous SSB. In Staphylococcus aureus, three SSBs are found, namely, SsbA, SaSsbB, and SsbC. While EcSSB can significantly stimulate EcPriA helicase, SaSsbA does not affect the SaPriA activity. It remains unclear whether SsbBs can participate in the PriA-directed DNA replication restart process. In this study, we characterized the properties of SaSsbBs through structural and functional analyses. Crystal structure of SaSsbB determined at 2.9 Å resolution (PDB entry 5YYU) revealed four OB folds in the N-terminal DNA-binding domain. DNA binding analysis using EMSA showed that SaSsbB binds to ssDNA with greater affinity than SaSsbA does. Gene map analysis demonstrated that SAAV0835 encoding SaSsbB is flanked by unknown genes encoding hypothetical proteins, namely, putative Sipho_Gp157, ERF, and HNHc_6 gene products. Structure-based mutational analysis indicated that the four aromatic residues (Phe37, Phe48, Phe54, and Tyr82) in SaSsbB are at positions that structurally correspond to the important residues of EcSSB for binding to ssDNA and are also critical for SaSsbB to bind ssDNA. Similar to EcSSB and other SSBs such as SaSsbA and SaSsbC, SaSsbB also exhibited high thermostability. However, unlike EcSSB, which can stimulate EcPriA, SaSsbB did not affect the activity of SaPriA. Based on results in this study and previous works, we therefore established that SsbA and SsbB, as well as SsbC, do not stimulate PriA activity.
Introduction
Single-stranded DNA-binding proteins (SSBs) play crucial roles in DNA metabolic processes, such as DNA replication, repair, and recombination in prokaryotes and eukaryotes.1,2 During these reactions, SSB is necessary to maintain the transient unwinding of duplex DNA in a single-stranded state.3 SSB binds to ssDNA with high affinity in a sequence-independent manner. Bacterial SSBs are typically homotetramers, in which four oligonucleotide/oligosaccharide-binding folds (OB fold) form a DNA-binding domain. In additional to ssDNA, SSB also binds to many DNA metabolism proteins that constitute the SSB interactome.4,5 The C-terminal acidic tail (DDDIPF) and the intrinsically disordered linker (IDL) of SSB are necessary to mediate protein–protein interactions.4 The entire C-terminal domain of SSB is disordered even in the presence of ssDNA.6The structure, DNA binding properties, and functions of SSB have been studied extensively in Escherichia coli (EcSSB).7,8 EcSSB has three distinct DNA binding modes that are dependent on protein and salt concentrations in a solution.9 ssDNA unwrapping analysis shows that EcSSB can diffuse along ssDNA in the different binding modes, indicating a highly dynamic complex.10
Several bacteria have two paralogous SSBs, namely, SsbA and SsbB.11 Based on the sequence identity and the DNA binding properties, the third SSB (SsbC) is also identified in Staphylococcus aureus.12 S. aureus, a Gram-positive pathogen, causes serious problems to public health worldwide.13 Some SSB inhibitors as broad-spectrum antibacterial agents targeting S. aureus and other pathogens have been discovered.12,14
SsbA is referred to as a counterpart of EcSSB. SsbA and SsbB are essential for genome maintenance and transformational recombination, respectively.15–18 Significant differences for SsbBs are found in their C-terminal sequences and DNA binding properties. In Bacillus subtilis, SsbB binds to ssDNA with lesser affinity than BsSsbA does.17 However, Streptomyces coelicolor SsbB (ScSsbB) exhibits greater DNA-binding affinity than ScSsbA does.18 Unlike Streptococcus pneumonia SsbB (SpSsbB), BsSsbB and ScSsbB lack the C-terminal acidic tail of SSB for protein–protein interactions.16–18 Thus, SsbBs from different organisms exhibit different protein–DNA and protein–protein interaction specificities.
PriA is a DEXH-type helicase used for replication restart in bacteria.19–22 PriA is a poor helicase and needs some specific loading proteins to reload the replicative DnaB helicase back onto the chromosome. In E. coli, accessory proteins PriB and SSB are known to stimulate PriA helicase activity.23,24 However, SaSsbA, a counterpart of EcSSB, does not trigger SaPriA.25 Instead, SaDnaD is found to enhance the ATPase activity of SaPriA.26 The manner by which SaSsbA and SaSsbB participate in SaPriA-directed primosome assembly and in DNA replication restart remains unclear.
SSB has mainly been studied in Gram-negative E. coli, and, to a lesser extent, in Gram-positive bacteria. Little is known about the fundamental function of SsbB for the assembly of the replication restart primosome. For instance, nothing is known whether or not SsbB can stimulate PriA helicase. Because of lacking experimental evidences, whether SsbB is thermostable and whether SsbB has the typical C-terminal acidic tail of SSB for protein–protein interactions also remain unclear. Whether PriB, an EcSSB-like ssDNA-binding protein lacking the C-terminal domain of SSB,27–29 is a counterpart of SsbB still needs to be further elucidated.
In this study, we have cloned, expressed, purified, and biochemically characterized SaSsbB. We also have crystallized SaSsbB and determined its molecular structure. Unlike EcSSB, SsbB could not enhance PriA activity. Thus, we established that these three EcSSB-like proteins in S. aureus (SsbA, SsbB, and SsbC) do not stimulate PriA activity.
Experimental
Construction of plasmids for SaSsbA, SaSsbB, SaDnaD, and SaPriA expression
SaSsbA,25 SaDnaD,26 and SaPriA30 expression plasmids have been constructed in other studies. SAAV0835, the gene encoding a putative SaSsbB, was amplified through PCR by using the genomic DNA of S. aureus subsp. aureus ED98 as a template. The primers used for the construction of the pET21-SaSsbB plasmid are summarized in Table 1.| Oligonucleotide | Primer |
|---|---|
| a These plasmids were verified by DNA sequencing. Underlined nucleotides indicate the designated site for the restriction site or the mutation site. | |
| SaSsbB-NdeI-N | GGGCATATGTTAAACAGAGTAGTTTTAGTA |
| SaSsbB-XhoI-C | GGGCTCGAGGAACGGGAGGTCTGAAAAATC |
| SaSsbB(F37A)-N | ACATTAGCAGTAAACAGAACAGCCACGAATGCTCAA |
| SaSsbB(F37A)-C | CTCGCCTTGAGCATTCGTGGCTGTTCTGTTTACTGC |
| SaSsbB(F48A)-N | GGCGAGCGTGAAGCAGAGCTTATAAACGTAGTAGTGTTC |
| SaSsbB(F48A)-C | GAACACTACTACGTTTATAAGCTCTGCTTCACGCTCGCC |
| SaSsbB(F54A)-N | TTTATAAACGTAGTAGTGGCCAAAAAACAAGCTGAAAAC |
| SaSsbB(F54A)-C | TTCAGCTTGTTTTTTGGCCACTACTACGTTTATAAAATC |
| SaSsbB(Y82A)-N | CGACTACAAACACGTAACGCCGAAAACAAAGACGGGCAA |
| SaSsbB(Y82A)-C | TTGCCCGTCTTTGTTTTCGGCGTTACGTGTTTGTAGTCG |
Protein expression and purification
Recombinant SaSsbA,25 SaDnaD,26 and SaPriA30 have been purified in other studies. Recombinant SaSsbB was expressed and purified in accordance with a previously described protocol for PriB.27,28 In brief, E. coli BL21(DE3) cells were transformed with the expression vector, and the overexpression of the plasmids was induced by incubating with 1 mM isopropyl thiogalactopyranoside. The protein was purified from a soluble supernatant through Ni2+ affinity chromatography (HiTrap HP; GE Healthcare Bio-Sciences), eluted with Buffer A (20 mM Tris–HCl, 250 mM imidazole, and 0.5 M NaCl, pH 7.9) and dialyzed against a dialysis buffer (20 mM HEPES and 100 mM NaCl, pH 7.0; Buffer B). Protein purity remained at >97% as determined by SDS-PAGE (Mini-PROTEAN Tetra System; Bio-Rad, CA, USA).Preparation of dsDNA substrate
The double-stranded DNA substrate (dsDNA) PS4/PS3-dT30 for ATPase assay was prepared at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 concentration ratio.25,26 PS4/PS3-dT30 was formed in 20 mM HEPES (pH 7.0) and 100 mM NaCl by briefly heating at 95 °C for 5 min and by slowly cooling to room temperature overnight.
1 concentration ratio.25,26 PS4/PS3-dT30 was formed in 20 mM HEPES (pH 7.0) and 100 mM NaCl by briefly heating at 95 °C for 5 min and by slowly cooling to room temperature overnight.
Electrophoretic mobility shift assay (EMSA)
EMSA for SaSsbB was conducted in accordance with a previously described protocol for SSB.31 In brief, various lengths of ssDNA oligonucleotides were radiolabeled with [γ32P] ATP (6000 Ci/mmol; PerkinElmer Life Sciences, Waltham, MA) and T4 polynucleotide kinase (Promega, Madison, WI, USA). The protein (0, 0.01, 0.02, 0.039, 0.078, 0.1563, 0.3125, 0.625, 1.25, and 2.5 μM; tetramer) was incubated for 30 min at 25 °C with 1.7 nM DNA substrates in a total volume of 10 μL in 20 mM Tris–HCl (pH 8.0) and 100 mM NaCl. Aliquots (5 μL) were removed from each of the reaction solutions and added to 2 μL of gel-loading solution (0.25% bromophenol blue and 40% sucrose). The resulting samples were resolved on 8% native polyacrylamide gel at 4 °C in TBE buffer (89 mM Tris borate and 1 mM EDTA) for 1 h at 100 V and visualized through phosphorimaging. A phosphor storage plate was scanned, and data regarding complex and free DNA bands were digitized for quantitative analysis. The ssDNA binding ability of the protein was estimated through linear interpolation from the concentration of the protein that bound 50% of the input DNA.ATPase assay
SaPriA ATPase assay25,26 was performed with 0.4 mM [γ-32P] ATP and 0.12 μM SaPriA in a reaction buffer containing 40 mM Tris (pH 8.0), 10 mM NaCl, 2 mM DTT, 2.5 mM MgCl2, and 0.1 μM PS4/PS3-dT30 DNA substrate. Aliquots (5 μL) were taken and spotted onto a polyethyleneimine cellulose thin-layer chromatography plate, which was subsequently developed in 0.5 M formic acid and 0.25 M LiCl for 30 min. Reaction products were visualized by autoradiography and quantified with a phosphorimager.Site-directed mutagenesis
SaSsbB mutants were generated with a QuikChange Site-Directed Mutagenesis kit in accordance with the manufacturer's protocol (Stratagene, LaJolla, CA, USA) by using the primers (Table 1) and the wild-type plasmid pET21b-SaSsbB as the template. The presence of mutation was verified through DNA sequencing.Bioinformatics
The amino acid sequences of 150 sequenced SSB homologs were aligned using ConSurf,32 and the structures were visualized by using PyMol.Crystallography
Before crystallization was performed, SaSsbB was concentrated to 15 mg mL−1 in Buffer B. Crystals were grown at room temperature through hanging drop vapor diffusion in 30% PEG 4000, 100 mM Tris, and 200 mM sodium acetate at pH 8.5. Data were collected with an ADSC Quantum-315r CCD area detector at SPXF beamline BL13C1 at NSRRC (Taiwan, ROC). Data were integrated and scaled with HKL-2000.33 Four SaSsbB monomers per asymmetric unit were present. The crystal structure of SaSsbB was determined at 2.98 Å resolution with the molecular replacement software Phaser-MR34 by using SaSsbA as a model (PDB entry 5XGT).25 A model was built and refined with PHENIX35 and Coot.36 The final structure was refined to R-factor of 0.2139 and Rfree of 0.2995. The atomic coordinates and related structure factors have been deposited in the PDB with the accession code 5YYU.Results and discussion
Sequence analysis of SaSsbB
SAAV0835, which encodes SaSsbB of 141 aa, was found on the basis of the nucleotide sequence similar to BsSsbB and EcSSB. The amino acid sequence of SaSsbB shared 36% identity to that of SaSsbA. The ConSurf analysis reveals that the C-terminal region of SaSsbB was variable (Fig. 1). Like EcSSB, SaSsbB also had a long flexible region, but its flexible region was composed of few proline and glycine residues. SaSsbB (109–141 aa) had one Gly residue and two Pro residues (Fig. 1), which are significantly less than those of EcSSB (116–178 aa; 15 Gly residues and 10 Pro residues). In addition, SaSsbB did not have a C-terminal acidic peptide tail. The C-terminal acidic tail DDDIPF in EcSSB involved in protein–protein interactions was FSDLPF in SaSsbB.Analysis of ssb (SAAV0835)
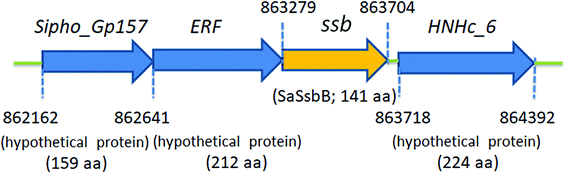
Fig. 2 shows the gene map of S. aureus chromosomal region with the ssb gene SAAV0835, which is flanked by unknown genes encoding hypothetical proteins with similarity to Sipho_Gp157, ERF, and HNHc_6. Unlike E. coli, which contains one type of SSB, S. aureus have three paralogous SSBs (SsbA, SsbB, and SsbC).12 The gene map analyses of ssb show significant differences.12,25 Unlike EcSSB located adjacent to uvrA, SaSsbA is flanked by rpsF and rpsR,25 which encode the ribosomal proteins S6 and S18, respectively. SaSsbC is flanked by the putative SceD, the putative YwpF, and fabZ genes, which code for a transglycosylase, a hypothetical protein, and a β-hydroxyacyl-ACP dehydratase, respectively.12 The gene regulation for SaSsbB is still unknown. Given that SsbB is essential for transformational recombination, these function-undetermined genes (Fig. 2) in S. aureus may be regulated with SaSsbB in a single signaling control and may be also involved in transformational recombination. However, this hypothesized relationship must be further confirmed by a detailed transcription analysis.SaSsbB bound to ssDNA
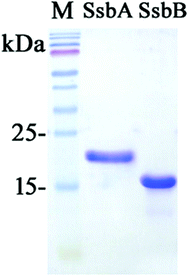
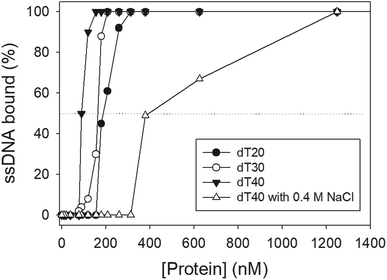
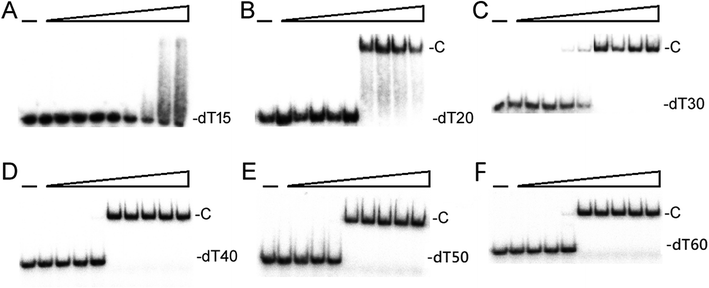
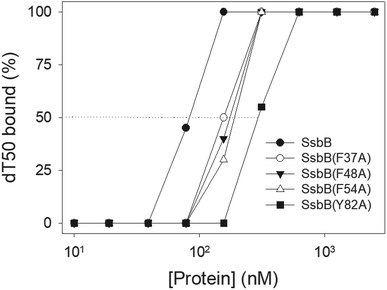
We studied the binding of purified SaSsbB (Fig. 3) to ssDNA (dT15-40) at various protein concentrations by using EMSA. To compare the DNA-binding abilities of SaSsbB, we quantified [Protein]50 through linear interpolation from the protein concentration (Fig. 4 and Table 2). The binding ability of SaSsbB to dT40 in the presence of 0.4 M NaCl was also analyzed (Fig. 4). [SaSsbB]50 of dT40 binding was 90 ± 4 nM, which was about fourfold lower than that in the presence of 0.4 M NaCl (382 ± 16 nM). Thus, the binding ability of SaSsbB to ssDNA is salt-dependent. Under the condition, only one band shift was found for dT20-60 (Fig. 5).| DNA | [Protein]50 (nM) |
|---|---|
| a [Protein]50 was calculated from the titration curves of EMSA by determining the concentration of the protein needed to achieve the midpoint value for input DNA binding. Errors are standard deviations determined by three independent titration experiments. | |
| dT15 | >2000 |
| dT20 | 190 ± 8 |
| dT30 | 164 ± 7 |
| dT40 | 90 ± 4 |
| dT40 (with 0.4 M NaCl) | 382 ± 16 |
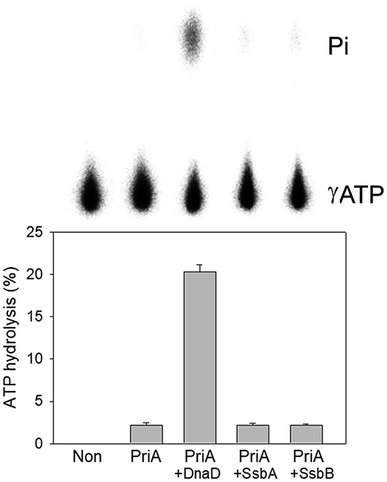
Stimulation of the ATPase activity of SaPriA by SaSsbB
To date, it remains unclear whether SsbB can stimulate the activity of the primosomal protein PriA. To investigate the possible effect of SaSsbB, we performed an ATPase assay for SaPriA. SaDnaD,26 which stimulates the SaPriA activity, was used as a positive control. In contrast to SaDnaD,26 we found that the ATPase activity of SaPriA in the presence of SaSsbB was not changed (Fig. 6). Given that the C-terminal domain of SsbB did not contain the acidic tail (Fig. 1), SsbB may not bind to PriA. Thus, no stimulation occurred (Fig. 6).Crystal structure of SaSsbB
In this study, we have shown that unlike EcSSB, SaSsbB did not contain the C-terminal acidic peptide and could not stimulate SaPriA helicase. To deeply understand the structure–function relationship of SaSsbB, we crystallized SaSsbB through hanging drop vapor diffusion and determined its structure at a resolution of 2.98 Å (Table 3). The secondary structural element of SaSsbB is similar to that of SaSsbA (Fig. 7A). The amino acids 107–141 in SaSsbB ternary structure were not observed. Consistent with the result from gel filtration analysis (data not shown), four monomers of SaSsbB per asymmetric unit were present (Fig. 7B). The SaSsbB monomer has an OB-fold domain similar to EcSSB, and the core of the OB-fold domain possesses a β-barrel capped with an α-helix. Unlike ScSSB, which contains an additional strand (β6),18 SaSsbB does not contain β6. Additional β6 strands clamp two neighboring subunits together in a tetrameric SSB.18 Thus, SsbBs from different organisms may exhibit different protein–DNA and protein–protein interaction specificities.| a Values in parentheses are for the highest resolution shell.b Rsym = Σ|I − ‘I’|/ΣI, where I is the observed intensity, ‘I’ is the statistically weighted average intensity of multiple observations of symmetry-related reflections. | |
|---|---|
| Data collection | |
| Crystal | SaSsbB |
| Wavelength (Å) | 0.975 |
| Resolution (Å) | 30–2.98 |
| Space group | P212121 |
| Cell dimension (Å) | a = 63.99, α = 90 |
| b = 84.74, β = 90 | |
| c = 84.86, γ = 90 | |
| Completeness (%) | 99.8 (99.8)a |
| 〈I/σI〉 | 13 (2.5) |
| Rsym or Rmerge (%)b | 0.125 (0.541) |
| Redundancy | 3.8 (4.0) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|
| Refinement | |
| Resolution (Å) | 30–2.98 |
| No. reflections | 9334 |
| Rwork/Rfree | 0.2139/0.2995 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|
| No. atoms | |
| Protein | 399 |
| Water | 16 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|
| R.m.s deviation | |
| Bond lengths (Å) | 0.011 |
| Bond angles (°) | 1.385 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|
| Ramachandran plot | |
| In preferred regions | 359 (93.25%) |
| In allowed regions | 20 (5.19%) |
| Outliers | 6 (1.56%) |
| PDB entry | 5YYU |
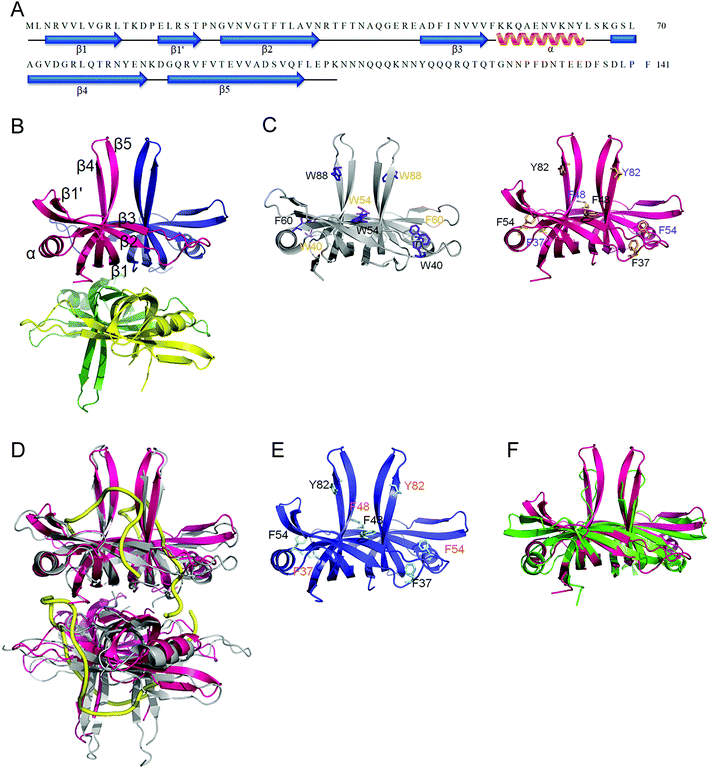 | ||
| Fig. 7 Crystal structure of SaSsbB. (A) The secondary structural element of SaSsbB. The secondary structural element of SaSsbB is shown above its sequence. (B) Crystal structure of SaSsbB. Four monomers of SaSsbB per asymmetric unit were present. The entire C-terminal domain was disordered. (C) ssDNA-binding mode of SaSsbB. In the EcSSB–ssDNA complex (PDB entry 1EYG), Trp40, Trp54, Phe60, and Trp88 participated in ssDNA binding via stacking interactions. The corresponding residues in SaSsbB, namely, Phe37, Phe48, Phe54, and Tyr82, might play roles in ssDNA binding similar to those of EcSSB. For clarity, only a dimer of EcSSB and SaSsbB is shown. (D) Superposition of SaSsbB and EcSSB. The N-terminal domains of SaSsbB and EcSSB (gray) are similar. (E) Crystal structure of SaSsbA. The residues proposed for binding DNA in SaSsbB are also identical to those in SaSsbA. (F) Superposition of SaSsbB and KpPriB. The N-terminal domain of SaSsbB and KpPriB (PDB entry 4APV; green) are similar, in which the only significant difference is in the lengths of the β4 and β5 sheets. | ||
Trp40, Trp54, Phe60, and Trp88 in EcSSB participate in ssDNA binding via stacking interactions (Fig. 7C). Correspondingly, Phe37, Phe48, Phe54, and Tyr82 in SaSsbB might play roles in ssDNA binding (Fig. 7D). These residues proposed for binding DNA in SaSsbB were also identical to those in SaSsbA (Fig. 7E). SaSsbB structurally resembles PriB,27,28 but significant differences in the lengths of β4- and β5-sheets were found (Fig. 7F).
Mutational analysis
According to crystal structure of SaSsbB, we speculated that Phe37, Phe48, Phe54, and Tyr82 in SaSsbB allow nucleic acids to wrap around the whole SaSsbB. We constructed and analyzed alanine substitution mutants (i.e., F37A, F48A, F54A, and Y82A) through EMSA (Fig. 8). Table 4 summarizes [Protein]50 of the binding of these SaSsbB variants to dT50. These SaSsbB mutants have [Protein]50 higher than that of the wild-type SaSsbB. The mutational effect on the ssDNA binding activity of SaSsbB followed the order Y82A > F54A > F48A > F37A. Structure-based mutational analysis indicated that SaSsbB might bind to ssDNA in a manner similar to that of EcSSB (Table 4).| dT50 | [Protein]50 (nM) |
|---|---|
| a [Protein]50 was calculated from the titration curves of EMSA by determining the concentration of the protein (tetramers) needed to achieve the midpoint value for input DNA binding. Errors are standard deviations determined by three independent titration experiments. | |
| SaSsbB | 83 ± 7 |
| SaSsbB(F37A) | 155 ± 12 |
| SaSsbB(F48A) | 176 ± 10 |
| SaSsbB(F54A) | 191 ± 16 |
| SaSsbB(Y82A) | 296 ± 18 |
Thermostability
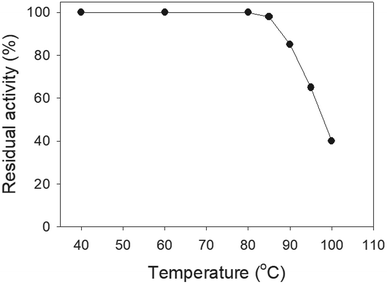
SSB proteins have high thermostability.37 SsbA and SsbC are highly thermostable.12 It is still unknown whether SsbB has high thermostability. We performed indirect thermostability experiments (Fig. 9). The activity of SaSsbB incubated at 100 °C, 95 °C, 90 °C, and 85 °C for 30 min decreased by 60%, 35%, 15%, and 2%, respectively. Given that the activity of EcSSB decreased by 50% after 30 min incubation at 95 °C,37 we determined that the thermostability of these SSBs followed the order SaSsbA = SaSsbB > SaSsbC > EcSSB (Table 5). Thus, SaSsbB also exhibited high thermostability.| Temperature | The decreased activity (%) | |||
|---|---|---|---|---|
| SaSsbA | SaSsbB | SaSsbC | EcSSB | |
| a Protein (1 μM) was incubated at temperatures ranging from 40 °C to 100 °C for 30 min. The resultant protein solution was incubated at 25 °C for 30 min with dT30. The phosphor storage plate was scanned, and the data for complex and free DNA bands were digitized for quantitative analysis. Results of SaSsbA,12 SaSsbC,12 and EcSSB37 are adapted from previous works for comparison. | ||||
| 85 °C | 2 | 2 | 2 | |
| 90 °C | 15 | 15 | 20 | |
| 95 °C | 35 | 35 | 40 | 50 |
| 100 °C | 60 | 60 | 70 | |
SsbB is not a counterpart of PriB
PriB is a dimeric ssDNA-binding protein with two OB folds,27–29 only found in some Gram-negative bacteria.21,22 Our crystal structure reveals that the N-terminal DNA-binding domain of SaSsbB structurally resembles PriB, although they significantly differ in the lengths of β4- and β5-sheets (Fig. 7F). Like SaSsbB, PriB also lacks the acidic tail. Because E. coli has only one SSB, it may raise a question whether PriB is the second SSB in E. coli and plays a functional role that is similar to SsbB in S. aureus. Sequence comparisons and operon organization analyses also show that PriB evolves from the duplication of the SSB gene.38 However, PriA activity can be significantly stimulated by PriB but not by SsbB (Fig. 6). Thus, SaSsbB and EcPriB have different functions, and PriB is not a counterpart of SsbB. Considering that the mechanisms of action of primosomes involved in DNA replication restart differ between E. coli21,22 and Gram-positive bacteria,39 we should elucidate the process by which PriA can cooperate with various loading factors to reactivate the same stalled forks.Recently, we have identified and characterized the third SSB (SsbC) in S. aureus.12 The structure and ssDNA-binding mode of SaSsbA,25 SaSsbB (this study), and SaSsbC12 are similar. Further studies are still needed to understand why SSB in S. aureus is necessary to evolve three similar but different SSBs.
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
We thank the experimental facility and the technical services provided by the Synchrotron Radiation Protein Crystallography Facility of the National Core Facility Program for Biotechnology, Ministry of Science and Technology and the National Synchrotron Radiation Research Center, a national user facility supported by the Ministry of Science and Technology, Taiwan, ROC. This research was supported by a grant from the Ministry of Science and Technology, Taiwan (MOST 107-2320-B-040-014 to C. Y. Huang).References
- D. J. Richard, E. Bolderson and K. K. Khanna, Crit. Rev. Biochem. Mol. Biol., 2009, 44, 98–116 CrossRef PubMed.
- E. Antony and T. M. Lohman, Semin. Cell Dev. Biol., 2018 DOI:10.1016/j.semcdb.2018.03.017.
- R. R. Meyer and P. S. Laine, Microbiol. Rev., 1990, 54, 342–380 Search PubMed.
- P. R. Bianco, Prog. Biophys. Mol. Biol., 2017, 127, 111–118 CrossRef PubMed.
- A. Costes, F. Lecointe, S. McGovern, S. Quevillon-Cheruel and P. Polard, PLoS Genet., 2010, 6, e1001238 CrossRef PubMed.
- S. N. Savvides, S. Raghunathan, K. Futterer, A. G. Kozlov, T. M. Lohman and G. Waksman, Protein Sci., 2004, 13, 1942–1947 CrossRef PubMed.
- T. H. Dickey, S. E. Altschuler and D. S. Wuttke, Structure, 2013, 21, 1074–1084 CrossRef PubMed.
- S. Raghunathan, A. G. Kozlov, T. M. Lohman and G. Waksman, Nat. Struct. Biol., 2000, 7, 648–652 CrossRef PubMed.
- T. M. Lohman and M. E. Ferrari, Annu. Rev. Biochem., 1994, 63, 527–570 CrossRef PubMed.
- S. Suksombat, R. Khafizov, A. G. Kozlov, T. M. Lohman and Y. R. Chemla, eLife, 2015, 4, e08193 CrossRef PubMed.
- C. Lindner, R. Nijland, M. van Hartskamp, S. Bron, L. W. Hamoen and O. P. Kuipers, J. Bacteriol., 2004, 186, 1097–1105 CrossRef PubMed.
- Y. H. Huang and C. Y. Huang, Oncotarget, 2018, 9, 20239–20254 Search PubMed.
- N. Koyama, J. Inokoshi and H. Tomoda, Molecules, 2012, 18, 204–224 CrossRef PubMed.
- J. G. Glanzer, J. L. Endres, B. M. Byrne, S. Liu, K. W. Bayles and G. G. Oakley, J. Antimicrob. Chemother., 2016, 71, 3432–3440 CrossRef PubMed.
- S. Ayora, B. Carrasco, P. P. Cardenas, C. E. Cesar, C. Canas, T. Yadav, C. Marchisone and J. C. Alonso, FEMS Microbiol. Rev., 2011, 35, 1055–1081 CrossRef PubMed.
- L. Attaiech, A. Olivier, I. Mortier-Barriere, A. L. Soulet, C. Granadel, B. Martin, P. Polard and J. P. Claverys, PLoS Genet., 2011, 7, e1002156 CrossRef PubMed.
- T. Yadav, B. Carrasco, A. R. Myers, N. P. George, J. L. Keck and J. C. Alonso, Nucleic Acids Res., 2012, 40, 5546–5559 CrossRef PubMed.
- T. Paradzik, N. Ivic, Z. Filic, B. A. Manjasetty, P. Herron, M. Luic and D. Vujaklija, Nucleic Acids Res., 20, 41, 3659–3672 CrossRef PubMed.
- G. C. Allen Jr, N. E. Dixon and A. Kornberg, Cell, 1993, 74, 713–722 CrossRef.
- C. A. Ouzounis and B. J. Blencowe, Nucleic Acids Res., 1991, 19, 6953 CrossRef PubMed.
- Y. H. Huang and C. Y. Huang, BioMed Res. Int., 2014, 2014, 195162 Search PubMed.
- T. A. Windgassen, S. R. Wessel, B. Bhattacharyya and J. L. Keck, Nucleic Acids Res., 2018, 46, 504–519 CrossRef PubMed.
- C. J. Cadman, M. Lopper, P. B. Moon, J. L. Keck and P. McGlynn, J. Biol. Chem., 2005, 280, 39693–39700 CrossRef PubMed.
- C. J. Cadman and P. McGlynn, Nucleic Acids Res., 2004, 32, 6378–6387 CrossRef PubMed.
- Y. H. Huang, H. H. Guan, C. J. Chen and C. Y. Huang, PLoS One, 2017, 12, e0182060 CrossRef PubMed.
- Y. H. Huang, Y. Lien, C. C. Huang and C. Y. Huang, PLoS One, 2016, 11, e0157593 CrossRef PubMed.
- Y. H. Huang, Y. H. Lo, W. Huang and C. Y. Huang, Genes Cells, 2012, 17, 837–849 CrossRef PubMed.
- C. Y. Huang, C. H. Hsu, Y. J. Sun, H. N. Wu and C. D. Hsiao, Nucleic Acids Res., 2006, 34, 3878–3886 CrossRef PubMed.
- J. H. Liu, T. W. Chang, C. Y. Huang, S. U. Chen, H. N. Wu, M. C. Chang and C. D. Hsiao, J. Biol. Chem., 2004, 279, 50465–50471 CrossRef PubMed.
- Y. H. Huang, C. C. Huang, C. C. Chen, K. J. Yang and C. Y. Huang, Protein J., 2015, 34, 169–172 CrossRef PubMed.
- C. Y. Huang, Determination of the binding site-size of the protein-DNA complex by use of the electrophoretic mobility shift assay, in Stoichiometry and Research: The Importance of Quantity in Biomedicine, ed. Innocenti A, InTech Press, Rijeka, Croatia, 2012, pp. 235–242 Search PubMed.
- M. Landau, I. Mayrose, Y. Rosenberg, F. Glaser, E. Martz, T. Pupko and N. Ben-Tal, Nucleic Acids Res., 2005, 33, W299–W302 CrossRef PubMed.
- Z. Otwinowski and W. Minor, Methods Enzymol., 1997, 276, 307–326 Search PubMed.
- A. J. McCoy, R. W. Grosse-Kunstleve, P. D. Adams, M. D. Winn, L. C. Storoni and R. J. Read, J. Appl. Crystallogr., 2007, 40, 658–674 CrossRef PubMed.
- J. J. Headd, N. Echols, P. V. Afonine, R. W. Grosse-Kunstleve, V. B. Chen, N. W. Moriarty, D. C. Richardson, J. S. Richardson and P. D. Adams, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2012, 68, 381–390 CrossRef PubMed.
- P. Emsley and K. Cowtan, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2004, 60, 2126–2132 CrossRef PubMed.
- M. Nowak, M. Olszewski, M. Spibida and J. Kur, BMC Microbiol., 2014, 14, 91 CrossRef PubMed.
- V. A. Ponomarev, K. S. Makarova, L. Aravind and E. V. Koonin, J. Mol. Microbiol. Biotechnol., 2003, 5, 225–229 CrossRef PubMed.
- M. Velten, S. McGovern, S. Marsin, S. D. Ehrlich, P. Noirot and P. Polard, Mol. Cell, 2003, 11, 1009–1020 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2018 |