Open Access Article
Open Access ArticleEndonuclease IV recognizes single base mismatch on the eighth base 3′ to the abasic site in DNA strands for ultra-selective and sensitive mutant-type DNA detection†
Jiaju Xu a,
Yanqiao Fu*b and
Yan Xiao
a,
Yanqiao Fu*b and
Yan Xiao *a
*a
aDepartment of Anesthesiology, Tongji Hospital, Huazhong University of Science and Technology, Wuhan, 430030, P. R. China. E-mail: 1571454187@qq.com
bDepartment of Otorhinolaryngology, Taihe Hospital, Hubei University of Medicine, Shiyan, 442000, P. R. China. E-mail: wut712sp@163.com
First published on 30th July 2018
Abstract
Since single nucleotide polymorphism (SNP) is related with many diseases and drug metabolic polymorphous and SNP genotyping is rising rapidly in many biological and medical areas, various methods of discriminating SNPs have been developed, one of which is an enzyme-based method. We uncovered a unique property of endonuclease IV due to which it can discriminate single base mismatches in different positions of DNA strands containing an abasic site, and we also discovered a new property: a mismatch in the +8 position could inhibit the cleavage of endonuclease IV. Then, we coupled +8 mismatch with other mismatches along with the discrimination effect of melting temperature to develop a new ultra-selective and sensitive genotyping system, which showed high discrimination factors. The detection limit was as low as 0.05–0.01%. Our new discovery improves the understanding of endonuclease IV. Also, the method could be applied to clinical real samples; thus, it merits further investigation and improvement for application in clinical utilization for early screening of specific diseases.
Single nucleotide polymorphism (SNP) genotyping is playing a vital role in genome mapping, pharmacogenetic studies, and drug discovery.1 One of the greatest important applications of SNP in biomedical research lies in comparing genome regions between cohorts, for example, matched cohorts with and without a disease, in genome-wide association studies. Statistics show that there exist some similar gene mutations in the diseased population of certain diseases, which are generally single gene mutations in multiple discontinuities. The detection of statistically significant single gene mutations is an important means of early screening of diseases, especially cancers.
However, in some cases, such as tissue biopsy and liquid biopsy, detection may encounter obstacles because of low abundance. For instance, tumor DNA is often covered by a large amount of normal DNA; thus, it is difficult to detect. A considerable amount of research has been done to develop genotyping systems during the last decades, providing us with powerful new insights into mutation detection.
Fluorescent probes are applied in genotyping and have become widely used tools; for example, molecular beacon (MB),2–4 branch-migration-based probe,5,6 cell penetrating peptide (CPP)–DNA fluorescent probe7 and triple-stem probe.8 Enzymatic tools have also been developed such as nicking endonuclease,9 DNase I,10,11 λ exonuclease12,13 and DNA ligases.14 Endonuclease IV (Endo IV) is one of the enzymes used for genotyping.12,15–20 Endo IV recognizes apurinic/apyrimidinic (AP) sites and is eliminated in base excision repair (BER).21,22 It prefers double-stranded DNA (dsDNA) to single-stranded DNA (ssDNA) while cleaving the phosphodiester bond.23,24
The excellent property of Endo IV has attracted many researchers. In 2013, Xiao et al. uncovered a novel property of Endo IV due to which it can discriminate mismatches next to the AP site in DNA strands;25 they found that Endo IV rapidly cleaved dsDNA containing a mismatch 3′ to the AP site (3′ mismatch) or a mismatch 5′ to the AP site (5′ mismatch), whereas it hardly cleaved dsDNA containing both the mismatches, i.e., 3′ and 5′ mismatches. Through simple design, they synthesized a fluorophore- and quencher-labeled DNA probe containing an AP site that had 3′ mismatch to mutant-type (MT) DNA and 3′ and 5′ mismatches to wild-type (WT) DNA for the detection of single base mutation DNA with a detection limit of 0.01%.17 Without redundant components, sophisticated design and complex procedures, the method offers us an excellent biosensing platform with a relatively low limit of detection (LOD), which cannot be reached by most other assays. To simplify the narration, we define the mismatch at the mutant base as functional mismatch and the mismatch that probe-WT and probe-MT duplex both have as intrinsic mismatch; for the biosensing system mentioned above, 3′ mismatch is intrinsic mismatch, and 5′ mismatch is functional mismatch.
Unfortunately, the study has a defect: when a sample shows positive result, it cannot assure that the slow cleavage has resulted from the mismatch at the position next to and 5′ to the AP site, that means, it cannot rule out the possible interference caused by a mismatch at another position in the DNA strand. Notably, for a hotspot of mutation, several adjacent bases are relatively high in mutation rate. A point mutation near the base of target mutation may be found in a sample gained from the patient; thereby, a mismatch other than target mismatch may be led in. Herein, we have discussed the following points under separate conditions: (a) if the unexpected mutant base lies in the intrinsic mismatch base, leaving the hybridization with a functional mismatch (5′ mismatch), a rise in fluorescence can be found, which produces a false positive result; (b) if the unexpected mutant base lies in other bases, three mismatches are formed by the probe and MT DNA, making the results unpredictable. The deficiency reduces the validity of clinical trials, which may be developed using this assay in the future.
For condition (a), the reason for the predicament is that the inhibitory effect occurs if and only if two mismatches occur collectively, which is a double regulation; the reason is as follows: consecutive three mismatches that consist of an AP site and its two adjacent mismatches together result in a locally non-hybrid single-stranded status at the AP site, which can hardly be cleaved by Endo IV, as is mentioned above.23 Thus, the enzymatic activity is inhibited. However, our current understanding of Endo IV limits further expansion of its usage.
To solve this problem, we must find a mechanism such that the inhibitory effect of the enzyme is only regulated by a single mismatch. We speculate that some single-base mismatches at bases other than those next to the AP site may inhibit enzymatic cleavage although single-base mismatch next to the AP site cannot produce inhibitory effects, which has never been studied before.
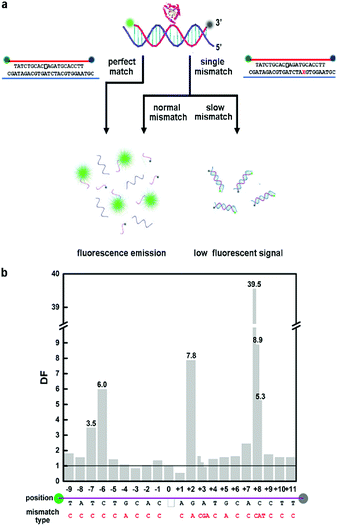
Therefore, we investigated the effect of single mismatch in every base in an AP-site-containing DNA duplex. We synthesized a 21-nt fluorophore- and quencher-labeled probe with an AP site, which is the same as Xiao et al.’s design,25 and tested perfect matched and mismatched DNA strand (see Fig. 1a). Endo IV recognized AP-site-containing DNA strands and cleaved them at diverse rates, and the two resultant single strands detached from the target strand due to thermal instability, thus emitting fluorescent signals. Based on the predicted melting temperatures of the probe and two resultant strands, we set the experimental temperature at 42 °C; at this temperature, the melting temperature (see Table S1†) of the probe-target duplex was not reached due to which they were tightly bound. The discrimination ability of fluorescent probes originated from the thermodynamic difference of single-base mismatches, but the thermodynamic difference caused by different types of base mismatches varied. We term the mismatches that lead to small thermodynamic difference as “stable single-mismatch”, and these yield small discrimination factors.26,27 Stable mismatches are reported to be more difficult to detect; thus, we chose unstable mismatch: for base G, we studied G:A mismatch and for base C, A, and T, we selected C:C, A:C, and T:C mismatches, respectively.
We surveyed mismatches in every position in the DNA strands containing an AP site, and the results showed that mismatches at different positions to the AP site had distinct influence on the cleavage rate of Endo IV. We denoted +x as the xth nucleotide 3′ to the AP site and −x as the xth nucleotide 5′ to the AP site. A −3 A:C mismatch slightly accelerated the reaction similar to +1 A:C mismatch. We observed that −9 T:C, −8 A:C, −5 T:C, −4 G:A, −2 A:C, −1 C:C, +3 A:C, +4 T:C, +5 G:A, +6 C:C, +9 C:C, +10 T:C and +11 T:C mismatches had negligible deaccelerating effect on the cleaving rate. All of the above-mentioned mismatches exhibited no significant difference compared to perfect match; thus, we call them normal mismatches. We observed that −7 T:C, −6 C:C, +2 G:A, +7 A:C, and +8 C:C mismatches clearly decelerated the reaction process; thus, we termed them as slow mismatches. The discrimination factor (DF) can be defined as the cleaving rate ratio of perfect match (PM) target to mismatch (MM) target; the influence on DF led by mismatch position is shown in Fig. 1b.
Remarkably, the cleavage rate of +8 mismatch was extremely slow with DF of 39.5, which was feasible to perform genotyping. A novel method can be roughly developed when we regard +8 C:C mismatched DNA strand as a WT-probe duplex and PM DNA strand as a MT-probe duplex.
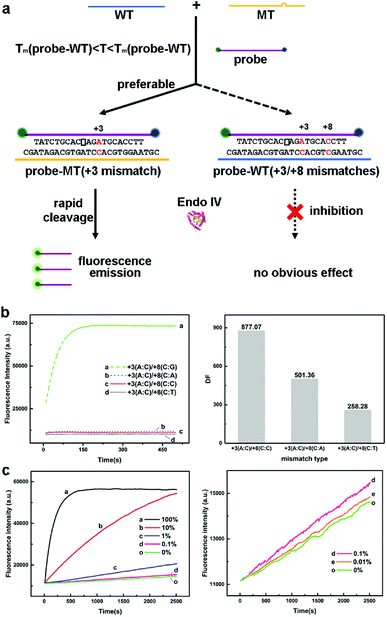
The DF value was still not impressive enough. Then, we designed DNA strands with double mismatches consisting of +8 mismatch and a normal mismatch, and we experimented with their cleaving rates. More than expected, we found that the inhibition effect of +8 mismatch was even enhanced by adding a new normal mismatch; thus, we could expect an even larger discrimination effect. Then, we paired the normal mismatch and corresponding double mismatch (add +8 mismatch) as they had only 1 bp difference and raised the temperature between the melting temperature (Tm) of single mismatch strands and double mismatch strands to compare the cleaving rates (see Table S2†). In this case, the resultant DFs were the co-effect of the property of Endo IV and the differentiation of thermodynamics. It should be noted that the conception of DF of a double mismatch target needs a minor modification, which defines the quotient of the cleaving rate of the corresponding single mismatch and that of double mismatches. The results are shown in Table S3.† An exceptional discrimination was acquired between the pairs +1 A:C mismatch and +1 A:C with +8 C:C mismatch [denoted +1(A:C)/+8(C:C) mismatch hereinafter, DF = 62.30], −2 A:C and −2(A:C)/+8(C:C) (DF = 61.86), −1 C:C and −1(C:C)/+8(C:C) (DF = 127.04), and +3 A:C and +3(A:C)/+8(C:C) (DF = 877.07).
We then investigated other types of base mismatches at the same positions and calculated their DFs (see Table S3†). We concluded that for all types of +3 with +8 mismatches, DFs were remarkable, especially for +3 A:C with all three types of +8 mismatches, and their DFs ranged from 258.28 to 877.07; hence, they had the greatest potential for genotyping among all double mismatches (see Fig. 2a and b). From the results, we could deduce that DFs were indeed mainly determined by the mismatch position of a nucleotide. Interestingly, the hybridization stability of mismatch types determined DFs. It is known that for A:V (V = A, C, G) mismatch, A:G > A:A > A:C and for C:H (H = A, C, T) mismatch, C:T > C:A > C:C.28–30 Our results (Fig. 2b) showed that DF increased with the hybridization stability; thus, we inferred that the recognition ability of Endo IV was partially derived from the thermodynamic property of base pairs. For different normal mismatches with all types of +8 mismatch, DFs were usually C:C > C:A > C:T.
Based on our previous study, to investigate the detection limits of the pair selected, we mixed up each double mismatch with its unique corresponding single mismatch to simulate low abundance mutant-type DNA sample, in which single mismatch represented MT target, and double mismatches represented the WT target (see Fig. 2a and c). The detection of 0.01% MT was available for +3(A:C)/+8(C:C) and +3(A:C) pair. For different directions of mutation, we could detect MT target in abundance as low as 0.05–0.01% in 42 min (see Fig. 2c, S1 and S2†). We also examined the effect after applying it to PCR product treated with exonuclease I and λ exonuclease, and DF was the same as mentioned below (see Fig. S3†). Three hours were required for the whole assay. We also studied the cleavage effect of Endo IV while encountering three mismatched strands containing at least one slow mismatch. The rate of cleavage was even slower than that for single slow mismatch; thus, the mismatch at an untargeted position does not have any impact on the result of the trial while using this method.
We have fully proven the application of the synthesized DNA and then, we demonstrated the applicability of the method to real clinical samples. We extracted genome DNA of a colorectal adenocarcinoma sample, which was detected by BRAF gene exon 15 V600G mutation as MT DNA; then, it was diluted by wild-type genomic DNAs extracted from a normal tissue without that mutation to prepare a series of mixed samples with MT DNA at different abundances. After PCR amplification and two-step enzymatic treatments mentioned below, single-stranded DNA was produced. Then, a customized fluorescent probe was added to the solution, which formed double mismatched duplex with WT DNA and single mismatched duplex with MT DNA (see Fig. 3a). After adding Endo IV, as shown in Fig. 3b, the detection limit was 0.01%. Thus, the method is feasible for a sample gained from biopsy. The data firmly proved the applicability of the method to real clinical samples.
The new properties we discovered enriched our understanding of Endo IV. For AP site binding, Endo IV forms an eight-stranded α-β barrel fold (TIM barrel), and the active site includes three metal ions near the center of the barrel.31 Our current understanding is that only the bases near the center of the barrel, i.e., adjacent to the AP site affect the enzymatic cleavage as nucleotides in the vicinity of the AP site (−2, −1, +1, +2) participate in anchoring the flipped-out abasic nucleotide to the active site of the enzyme.32 However, our newly discovered +8 mismatch that is quite far from the AP site still inhibits the cutting effect of endonuclease IV. The possible reason may be conformational changes. The intrinsic mechanism is worth further study.
The property newly found by us could be applied for constructing DNA sensing platforms with extremely high selectivity when a double mismatched strand is regarded as having single base mismatch in comparison to its corresponding single mismatch strand, which can be used for the study of single nucleotide polymorphisms (SNPs). As shown in Table S3,† for +3(A:C)/+8 and +3(A:C) mismatched pairs, DFs ranged from 258.28 to 877.07 for all types of mismatches, which are remarkable values for genotyping.
It is clear that our design does not exhibit requirements regarding the sequence of the target as we have found a mismatched pair having remarkable DFs for all types of mismatches. The fluorescence-quencher probe can be flexibly designed in accordance to the target strand. It can be an improvement and complement for the method developed by Xiao et al. For some specific sequence, the method may exhibit better performance than the method developed before. For non-targeted mutations, the positive rate reported using this assay is greatly reduced. Thus, our new findings improve the recognition library of Endo IV, covering a wider variety of mutations combined with other Endo-IV-based methods.
Conclusions
Generally, we uncovered the unique property of Endo IV, which recognizes mismatch at the eighth position in AP-site-containing DNA strands. Then, we designed DNA strands with single/double mismatched pairs and linked them to melting temperature differentiation to deduce the background signal for obtaining a better DF value. The application to the real clinical sample was also feasible. Thus, we extended the property of Endo IV, and a new ultra-selective DNA sensing platform was exploited. The method we developed for genotyping of SNPs exhibited extremely high DFs ranging from 258.28 to 877.07 and a detection limit of 0.05–0.01% of target DNA strands from an enormous background of double mismatched strands.Conflicts of interest
The authors declare no conflicts of interest.Notes and references
- K. Yanase, S. Tsukahara, J. Mitsuhashi and Y. Sugimoto, Cancer Lett., 2006, 234, 73–80 CrossRef PubMed.
- D. S. Xiang, K. Zhai, W. J. Xiang and L. Z. Wang, Talanta, 2014, 129, 249–253 CrossRef PubMed.
- S. Hu, W. Tang, Y. Zhao, N. Li and F. Liu, Chem. Sci., 2017, 8, 1021–1026 RSC.
- J. Zheng, R. Yang, M. Shi, C. Wu, X. Fang, Y. Li, J. Li and W. Tan, Chem. Soc. Rev., 2015, 44, 3036–3055 RSC.
- X. Xiao, T. Wu, L. Xu, W. Chen and M. Zhao, Nucleic Acids Res., 2017, 45, e90 CrossRef PubMed.
- X. Zhou, Z. Cui, L. Liu, Z. Sun, M. Lin, Q. Hu, H. Wang and X. Xiao, Analyst, 2018, 143, 2755–2759 RSC.
- L. Chen, S. Fang, X. Xiao, B. Zheng and M. Zhao, Anal. Chem., 2016, 88, 11306–11309 CrossRef PubMed.
- Y. Xiao, K. J. I. Plakos, X. Lou, R. J. White, J. Qian, K. W. Plaxco and H. T. Soh, Angew. Chem., Int. Ed., 2009, 48, 4354–4358 CrossRef PubMed.
- W. Xu, X. Xue, T. Li, H. Zeng and X. Liu, Angew. Chem., Int. Ed., 2009, 48, 6849–6852 CrossRef PubMed.
- T. B. Wu, X. J. Xiao, Z. Zhang and M. P. Zhao, Chem. Sci., 2015, 6, 1206–1211 RSC.
- X. Xiao, T. Wu, F. Gu and M. Zhao, Chem. Sci., 2016, 7, 2051–2057 RSC.
- X. Xiao, C. Zhang, X. Su, C. Song and M. Zhao, Chem. Sci., 2012, 3, 2257–2261 RSC.
- T. Wu, Y. Yang, W. Chen, J. Wang, Z. Yang, S. Wang, X. Xiao, M. Li and M. Zhao, Nucleic Acids Res., 2018, 46, 3119–3129 CrossRef PubMed.
- J. T. Ren, J. H. Wang, J. Wang and E. K. Wang, Biosens. Bioelectron., 2014, 51, 336–342 CrossRef PubMed.
- J. H. Nelson, G. A. Hawkins, K. Edlund, M. Evander, L. Kjellberg, G. Wadell, J. Dillner, T. Gerasimova, A. L. Coker, L. Pirisi, D. Petereit and P. F. Lambert, J. Clin. Microbiol., 2000, 38, 688–695 Search PubMed.
- I. V. Kutyavin, D. Milesi, Y. Belousov, M. Podyminogin, A. Vorobiev, V. Gorn, E. A. Lukhtanov, N. M. Vermeulen and W. Mahoney, Nucleic Acids Res., 2006, 34, e128 CrossRef PubMed.
- X. Xiao, C. Song, C. Zhang, X. Su and M. Zhao, Chem. Commun., 2012, 48, 1964–1966 RSC.
- X. Xiao, Y. Liu and M. Zhao, Chem. Commun., 2013, 49, 2819–2821 RSC.
- J. Xu, L. Li, N. Chen, Y. She, S. Wang, N. Liu and X. Xiao, Chem. Commun., 2017, 53, 9422–9425 RSC.
- X. Xiao, A. Xu, J. Zhai and M. Zhao, Methods, 2013, 64, 255–259 CrossRef PubMed.
- C. D. Mol, D. J. Hosfield and J. A. Tainer, Mutat. Res., DNA Repair, 2000, 460, 211–229 CrossRef.
- O. D. Schärer, Angew. Chem., Int. Ed., 2003, 42, 2946–2974 CrossRef PubMed.
- D. M. Wilson Iii and D. Barsky, Mutat. Res., DNA Repair, 2001, 485, 283–307 CrossRef.
- R. Asano, H. Ishikawa, S. Nakane, N. Nakagawa, S. Kuramitsu and R. Masui, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2011, 67, 149–155 CrossRef PubMed.
- X. Xiao, Y. Liu and M. Zhao, Chem. Commun., 2013, 49, 2819–2821 RSC.
- A. G. Frutos, S. Pal, M. Quesada and J. Lahiri, J. Am. Chem. Soc., 2002, 124, 2396–2397 CrossRef PubMed.
- Z. Mazouz, N. Fourati, C. Zerrouki, A. Ommezine, L. Rebhi, N. Yaakoubi, R. Kalfat and A. Othmane, Biosens. Bioelectron., 2013, 48, 293–298 CrossRef PubMed.
- X. Piao, L. Sun, T. Zhang, Y. Gan and Y. Guan, Acta Biochim. Pol., 2008, 55, 713–720 Search PubMed.
- J. SantaLucia Jr and D. Hicks, Annu. Rev. Biophys. Biomol. Struct., 2004, 33, 415–440 CrossRef PubMed.
- H. Urakawa, S. El Fantroussi, H. Smidt, J. C. Smoot, E. H. Tribou, J. J. Kelly, P. A. Noble and D. A. Stahl, Appl. Environ. Microbiol., 2003, 69, 2848–2856 CrossRef PubMed.
- E. D. Garcin, D. J. Hosfield, S. A. Desai, B. J. Haas, M. Bjoras, R. P. Cunningham and J. A. Tainer, Nat. Struct. Mol. Biol., 2008, 15, 515–522 CrossRef PubMed.
- D. J. Hosfield, Y. Guan, B. J. Haas, R. P. Cunningham and J. A. Tainer, Cell, 1999, 98, 397–408 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra04552f |
| This journal is © The Royal Society of Chemistry 2018 |