Open Access Article
Open Access ArticlePolydopamine nanoparticles kill cancer cells
Celia Nieto,
Milena A. Vega,
Gema Marcelo * and
Eva M. Martín del Valle*
* and
Eva M. Martín del Valle*
Department of Chemical Engineering, Universidad de Salamanca, Plaza de los Caídos s/n 37008, Salamanca, Spain. E-mail: gemamarcelo@usal.es; emvalle@usal.es
First published on 24th October 2018
Abstract
Polydopamine (PD) is a synthetic melanin analogue of growing importance in the field of biomedicine, especially with respect to cancer research, due, in part, to its biocompatibility. But little is known about the cytotoxic effects of PD on cancer cell lines. PD is a UV-vis absorbing material whose absorbance overlaps with that of formazan salts, which are used to assess cell viability in MTT assays. In this study, a protocol has been established to eliminate the contributing absorbance of PD at 550 nm, and has been applied to characterize the cytotoxicity of PD nanoparticles in both healthy and breast cancer cell lines. Once the protocol is applied, it was found that PD is per se an antineoplastic system, meaning it selectively kills cancer cells, especially those of breast cancer, but it has no toxic effect on healthy cells. The mechanism of action could be related to the production of ROS and the alteration of iron homeostasis in lysosomes. To the best of our knowledge there are only a few examples of nanoparticle systems devoid of drugs that selectively kill cancer cells.
Introduction
Polydopamine (PD) is a synthetic melanin analogue with properties similar to those occurring in natural systems.1,2 In cancer research, polydopamine plays a central role in devising new materials for diagnosis and/or therapy due to its: (i) biocompatibility;3 (ii) physical properties, such as thermal deactivation3,4 and chemical versatility,5–7 which permits further functionalization strategies as well as the chelation of diverse metallic cations;8–10 and (iii) its coating ability regardless of the substrate material.6,7The biocompatibility of PD has been shown by previous studies using MTT assays on different cell lines.3,11–13 Additionally, its long-term toxicity has been examined by intravenous injection in rats, where the injected animals remain alive for one month with no abnormal effects observed.3 However, little is known about the cytotoxic effects of PD in cancer cells.
Radical oxygen species (ROS) are produced endogenously in multiple and normal cellular metabolic processes. Lysosomes are acid cell compartments that act as centers for cell degradation, among other functions. Therefore, they are the main intracellular stores for Fe3+, Fe2+, Zn2+ and Cu2+ cations.14,15 Many of these metals participate in the production of ROS via Fenton chemistry.16 Lysosomes contain certain levels of ROS, which are essential for normal cell function.17 However, an aberrant accumulation of metals and/or ROS is linked to immediate cell damage and degenerative diseases.16,18
Recently, new strategies for targeting cancer treatment have emerged that are linked to the use of nanosystems/drugs capable of altering the lysosomal homeostasis of certain metallic cations, essentially iron.19 This in turn leads to ROS production and subsequent cancer cell death.20–22 The mechanisms of these systems are thought to act differently, where nanosystems (metal–organic frameworks) can deliver Fe2+, degrading lysosomes through the production of ROS. Additionally, salinomycin molecules sequester Fe2+ in the lysosome leading to its depletion, which is linked to ROS production and causes cancer cell death. This latter mechanism is known as ferroptosis, and a similar mechanism occurs with the use of ultra-small (ethylene glycol)-coated silica nanoparticles.
The MTT assay is a standard test frequently used in cytotoxicity studies.23 It is based on the fact that only metabolic active cells, i.e. viable cells, are able to reduce and convert the yellow water-soluble tetrazolium dye (3-[4,5-dimethyl-2-thiazolyl]-2,5-diphenyl-2H-tetrazolium bromide) into purple-colored formazan crystals ([E,Z]-5-[4,5-dimethylthiazol-2-yl]-1,3-diphenyl-formazan) due to the action of mitochondrial dehydrogenases. The formazan product is spectrophotometrically analyzed at 550 nm and its absorbance can be related to the number of viable cells. However, PD is an UV-vis absorbing material, and its broad absorption band overlaps with the absorbance band of the formazan salts. If not taken into account, this overlap would lead to the overestimation of cell viability and the real toxic effect of PD would not be correctly assessed.
Here we show that the absorbance of PD overlaps with the absorbance of formazan salts and the consequences of underestimating the true absorbance in cell viability assays. A general protocol eliminating the contribution of the absorbance of PD from the MTT absorbance measurement has been designed. Also, the toxic effect of PD on both a healthy cell line and a cancer cell line has been assessed following this proposed protocol.
Melanins have an active role in the regulation of the free iron concentration in neurons of the substantia nigra.24 Similar to natural melanins, PD can sequester many metallic cations in the lysosomes of rapidly proliferating cancer cells which often contain more metals than normal cells. Likewise, PD may also alter metal homeostasis which could adversely affect cell survival.
Results and discussion
Synthesis and characterization of PD nanoparticles
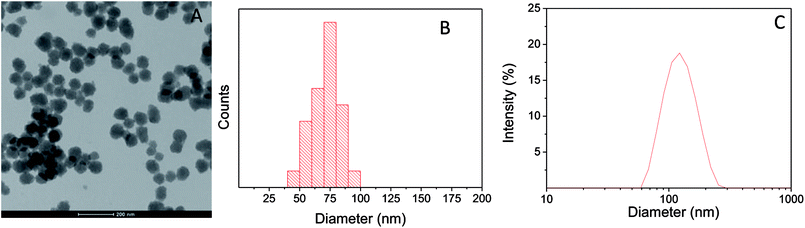
PD nanoparticles were synthesized by oxidative polymerization of dopamine in a basic aqueous medium containing ethanol, water and ammonium hydroxide.3 A TEM image and a histogram exhibiting the range of sizes are shown in Fig. 1A and B. The average size is 71 ± 11 nm. PD particles remained stable in water with an average hydrodynamic diameter of 116 ± 20 nm (PdI = 0.04), as determined by DLS (from the intensity-averaged size distribution shown in Fig. 1C).UV-vis spectroscopy characterization of PD nanoparticles and the MTT reagent
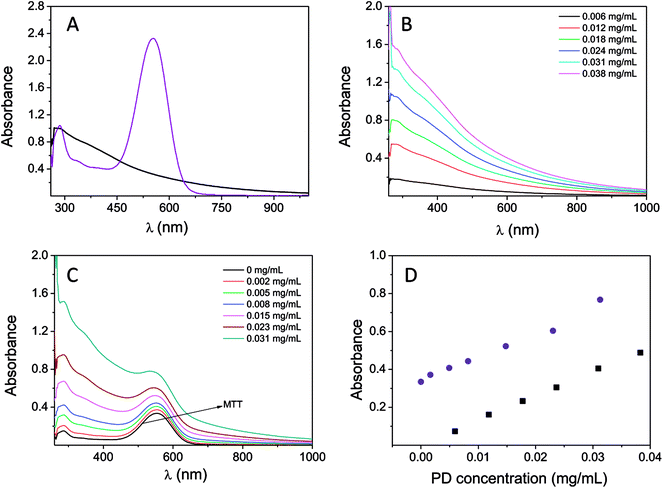
The interference caused by the absorption of PD together with the absorption of the MTT reagent was studied by UV-vis spectroscopy. The PD particle dispersion in water (concentrations ranging from 0.005 to 0.04 mg mL−1) and a solution containing formazan in DMSO (0.025 mM) were used for this analysis. The absorption spectra (normalized at 280 nm) of PD (0.012 mg mL−1) and the MTT reagent are shown in Fig. 2A. PD has a broad and featureless absorption band ranging from 200 to 1000 nm, with the maximum absorption at 280 nm. In addition, absorption at 550 nm was also shown to be significant. The variation observed in the absorption spectrum with respect to concentration is shown in Fig. 2B. The extinction coefficient at 550 nm was 12.79 mL mg−1 cm−1, which could not be expressed in molar units because the chemical structure of PD still remains under discussion.25,26 Additionally, the UV-vis spectrum of the MTT reagent is characterized by an absorption band located at 554 nm, which is approximately the wavelength of absorption used in the MTT assay. Furthermore, formazan does not show absorption above 700 nm. Therefore, PD absorbance overlaps with the absorbance of the MTT reagent. The effect of the concentration of PD on the MTT absorption spectrum is shown in Fig. 2C. The addition of PD (final concentrations in the range of 0.001–0.031 mg mL−1) to the MTT solution leads to an increase in the MTT absorption band, Fig. 1C. Thus, the final spectrum is the result of the merging absorption bands of PD and the MTT located at 554 nm spectra. The change in absorbance at 550 nm using a mixture containing the MTT reagent (at a fixed concentration, 0.025 mM) and PD at different concentrations is shown in Fig. 2D. The absorbance at 550 nm increases from 0.34 (in the absence of PD nanoparticles) to 0.77 when PD is at a concentration of 0.031 mg mL−1. The variation observed is linear, and the slope is close to the PD extinction coefficient 12.91 mL mg−1 cm−1. As a result, the degree to which cellular viability would be overestimated is based on the concentration of PD. That is to say, a two-fold increase in viability would be obtained when the concentration of PD is 0.04 mg mL−1.Moreover, the absorbance of the mixture MTT + PD is the result of combining the absorbances of both PD nanoparticles and the MTT reagent at a particular PD concentration. Consequently, the contribution of PD can be eliminated from the absorbance determined by the MTT assay if either the PD absorbance or concentration is known.
PD particles present a great capacity for adhering to different types of materials. PD is comprised of catechol groups, which are versatile adhesives able to bind to either organic or inorganic substrates using a mechanism that is similar to the adhesion of marine mussels to surfaces.6,27 This characteristic makes evaluating toxicity using UV-vis absorption spectroscopy difficult. It has been observed that PD remains adhered to the bottom of the cell culture plate when carrying out the MTT assay, which makes it practically impossible to eliminate PD from the cell plate when the medium is discarded. Moreover, part of the PD adhered to the plate may be re-dispersed when DMSO is added to dissolve the formazan crystals. As a result, the absorbance obtained at 550 nm would not provide a correct estimate of the quantity of viable cells, but instead would be an overestimate.
Therefore, the contribution of PD to the absorbance determined by the MTT assay could be easily removed if the concentration of the PD remaining in the culture plate is known. Since absorbance could be determined from the concentration by using a calibration curve that relates PD concentration to its absorbance at 550 nm.
Protocol for estimating cell viability using an MTT assay
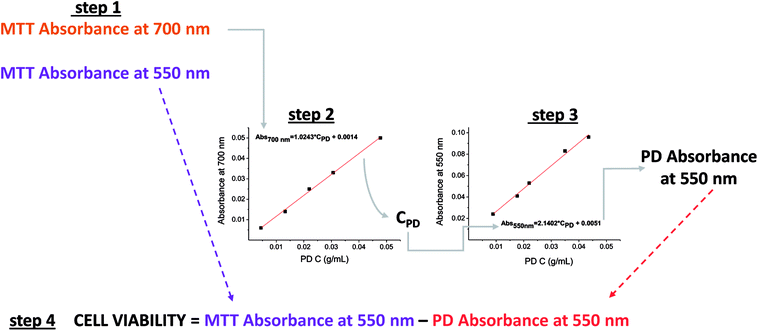
First, it is necessary to determine the absorbance of PD at 550 nm in order to eliminate its contribution from the absorption measured by the MTT assay. This allows the absorbance of formazan at 550 nm to be obtained, which gives an indication of the quantity of viable cells.Fortunately, many of the microplate readers measure wavelengths of absorption within the range of 340 to 800 nm. The protocol proposed in this study requires that calibration curves be established for absorbance versus PD concentration (mg of PD/mL of DMSO) at both 550 and 700 nm using the microplate reader. In addition, the absorbance readings at both 550 nm and 700 nm in the MTT assay are also required.
A wavelength of 700 nm was selected because only PD absorbs and not formazan, as shown in Fig. 2A. Thus, the absorbance read at this wavelength in the MTT assay will determine the remaining concentration of PD in the plate by allowing the absorbance to be interpolated from the corresponding calibration curve of the absorbance of PD (700 nm) vs. the concentration of PD. Once the concentration of PD is known, its contributing absorbance at 550 nm can be estimated by interpolating the PD concentration in the calibration curve PD absorbance (550 nm) vs. PD concentration.
Finally, the PD absorbance value must be subtracted from the absorbance determined at 550 nm in the MTT assay before cell viability can be quantified. The proposed protocol is illustrated in Scheme 1.
Cytotoxicity study of PD nanoparticles
The toxicity of the PD nanoparticles in a healthy cell line (NIH3T3) and a breast cancer (BT474) cell line was evaluated by performing a MTT assay. The procedure previously described for eliminating the contributing absorbance of PD from the MTT absorbance measured at 550 nm was applied to estimate cell viability.The results obtained on the cytotoxic effect of PD (concentration range 0.015–0.070 mg mL−1) in NIH3T3 cells during a period of 72 h are shown in Fig. 3.
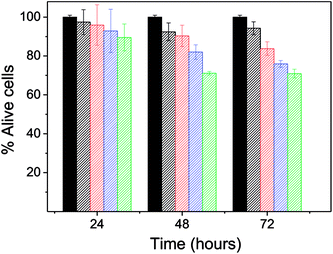 | ||
| Fig. 3 The viability of the cell line NIH3T3 cultured in different concentrations of PD. Green: 0.02 mg mL−1, blue: 0.050 mg mL−1, red: 0.072 mg mL−1 and black: 0.093 mg mL−1. | ||
After 24 h, the percentage of live cells was greater than 90% for all of the concentrations assayed, which supports the idea that PD is a biocompatible system. After 48 hours, cell viability was reduced to less than 80% but only for the concentration 0.093 mg mL−1. After 72 hours of exposure, cell viability was slightly less than 80% for concentrations 0.072 and 0.093 mg mL−1. Therefore, PD appears not to be cytotoxic at concentrations below 0.05 mg mL−1, findings that agree with those of prior studies where PD did not present a toxic effect in vitro or in vivo in rats.3,11–13
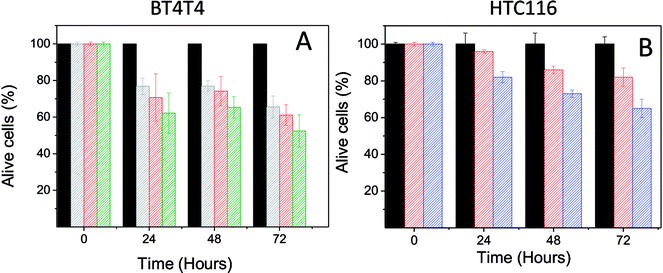
The effect of PD on the BT474 cancer cell line was assessed during a period of 72 hours with a PD concentration range of 0.022–0.044 mg mL−1. These results are shown in Fig. 4A. PD did not exhibit toxic effects on normal cells with the same concentration range. In contrast, PD particles had a significant adverse effect on BT474 cell viability. At 24 hours, cell proliferation was reduced to 77 and 62% for the lower and upper limits of the concentration range, respectively. After 48 hours, the percentage of live cells was between 77–64%, and finally at 72 hours, viability was in the range of 65–52%.
In conclusion, it can be said that PD selectively kills BT474 cancer cells in a concentration range that does not have any adverse effect on NIH3T3 healthy cells.
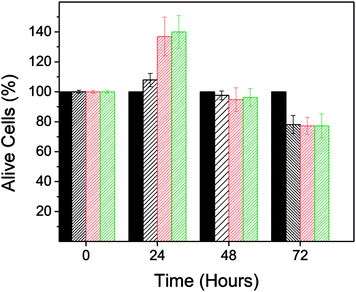
The adverse effect of PD on the viability of the BT474 cell line would not have been detected if the absorbance of PD had been eliminated directly from the absorbance recorded in the MTT assay. Fig. 5 shows the resulting cell viability when the developed protocol was not applied. In this case cell viability ranged between 108–140% after a 24 hours treatment.
Cell internalization
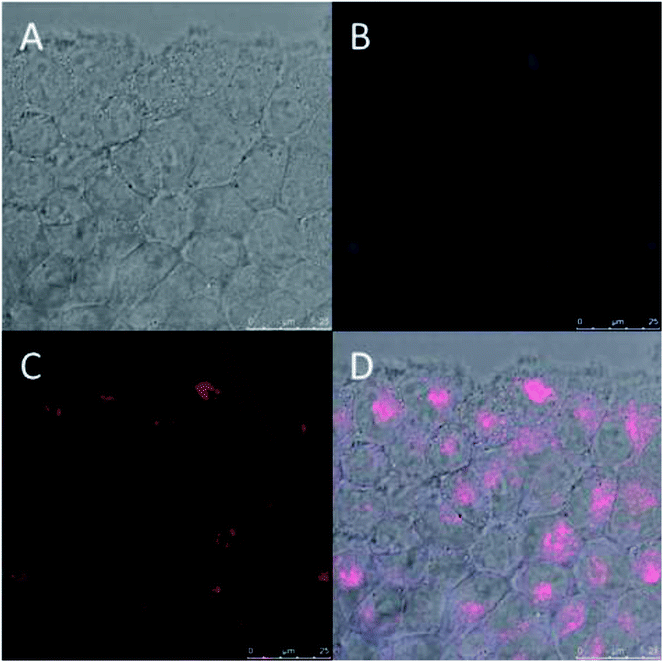
Fluorescent PD particles9,28 were prepared and their internalization in BT474 cancer cells was monitored. BT474 cells were cocultured with fluorescent PD particles. Fig. 6 shows that after 4 hours, the PD fluorescence (blue) co-localized with the fluorescence of the lysosome chemical marker (red), which indicates that PD targets lysosomes via an endocytic mechanism, a result that agrees with prior work reporting the same mechanism for the systematic uptake of PD.11,29PD chelation of metallic cations (Mn+)
The PD chemical structure is still under discussion.25,26,30–32 Despite this, it is accepted the presence of certain functional groups in the PD structure such as alcohol (phenolic), quinone and semiquinone groups, amine group (indole ring), amine groups from uncycled dopamine units and acid groups from a small fraction of pyrrolecarboxylic acid moieties derived from the oxidative degradation of the indole units. Fe2+, Fe3+ and Cu2+, among others, are metallic cations that can be found in lysosomes. Recently, we showed by infrared spectroscopy that these functional groups are responsible for the rich PD complexation chemistry towards many metallic cations and their loading capacity was determined.9 Here, we again show a brief summary of the interaction of PD with Fe2+ and Fe3+ at pH 4.5 (which is the pH of lysosomes).9 PD particles can load iron in both oxidative states through the complexation with catechol groups. The adsorption of Fe3+, expressed as Q (mg of Mn+/g of PD particles), was notable with Q = 370 mg g−1, as compared to Fe2+ with Q = 210 mg g−1, which was smaller but also significant. In conclusion, these results may support the following hypothesis: PD could sequester these metal cations in lysosomes and lead to the modification of its homeostasis.Cytotoxicity mechanism
Lysosomes are the main intracellular stores for many metals and have an important role in the regulation of iron transport through the cell.15 Additionally, iron is involved in the generation of ROS via Fenton reactions in lysosomes, in an abnormal concentration are toxic to the cell. In the former characterizations, we showed that PD particles target lysosomes via an endocytic mechanism where their metal complexation chemistry may alter the homeostasis of iron.Ferroptosis is a cell death mechanism that is related to an iron dependent ROS production that leads to oxidative damage of the membrane lipids of lysosome.16,20,21 ROS are produced dependently upon modifying iron levels in lysosome. However, the precise role of iron in this mechanism is unclear. This action mechanism has been reported for salinomycin molecule and ultra-small (ethylene glycol)-coated silica nanoparticles among others.20,21 Some evidences of this mechanism can be found upon co-treatment with either glutathione (GT) or deferoxamine (DFO). GT is selected because is a potent antioxidant involved in cellular protection against ROS33 and DFO is a lysosomal iron chelating agent used to treat iron overload.34 After GT/DFO co-treatment, the ferroptosis mechanism is blocked and the toxic effect of the drug is suppressed.
Now, we are going to focus our study on providing some insights that could support the idea that both iron level alteration and ROS production are closely related to the mechanism that could explain the toxicity of PD particles in breast cancer cells. To do so, we selected the PD concentration of 0.033 mg mL−1 to study the toxic effect of PD particles in combination with either DFO or glutathione after a 24 hours treatment.
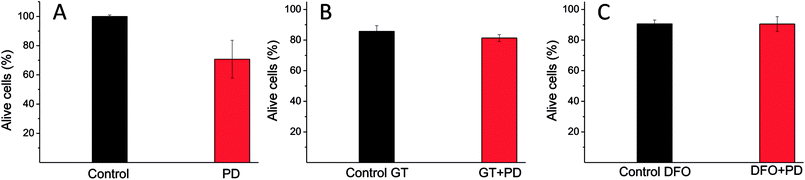
First, cell proliferation was reduced to 70% (related to the control) when they are treated only with PD particles, Fig. 7A. After that, BT4T4 cells were co-cultured with PD particles (0.033 mg mL−1) and GT (50 μM). After 24 hours, cell viability was very similar to the cell viability observed for the control (cells treated only with GT), as shown in Fig. 7B. This fact could indicate that GT protects cancer cells from the effect exerted by PD particles and suggests that the mechanism of toxicity for PD could be related to ROS production.
To evaluate if PD could alter the lysosomal iron homeostasis, we co-treated breast cancer cells with PD (0.033 mg mL−1) and DFO (0.7 μM). The co-treatment with DFO and PD seemed to block the toxic effect of PD since cell viability was very similar to the cell viability observed for the control (cells treated only with DFO), Fig. 7C.
In sum, the toxicity of PD particles in the cancer cell line, BT4T4, could be related to ROS production, which may be associated with the alteration of iron homeostasis in lysosomes as it occurs in the ferroptosis mechanism. As characterized above, PD particles would be able to sequester iron in both oxidative states at lysosomal pH. Nevertheless, additional work is required to know in full the mechanism of action of PD particles.
Finally, we sought to show that the toxic effect of PD is not limited only to the BT474 cancer line. Thus, we examined the toxic effect of PD in a human colorectal carcinoma (HTC16) cell line. The result of this analysis (at 15 and 50 mg mL−1) on HTC16 cells during a 72 hours period is shown in Fig. 4B. After 24 hours, viability was reduced to 82% with a concentration of 50 mg mL−1. After 48 hours of treatment, viability significantly decreased to 86 and 73% with the PD concentrations of 15 and 50 mg mL−1, respectively. Finally, after 72 hours, the viability slightly decreased to values of 82 and 65% for the PD concentrations of 15 and 50 mg mL−1, respectively.
Conclusions
A protocol has been established for evaluating the antiproliferative effect of PD systems on cell lines using a standard MTT assay. This procedure allows the contributing absorbance of PD at 550 nm to be eliminated from the MTT assay used to assess cell viability. This method is easy to implement, since it only requires that calibration curves for absorbance versus PD concentration be established at both 550 and 700 nm, as well as the absorbance readings at these same wavelengths for the MTT assay.In this work, this protocol was applied to assess the cytotoxicity of PD particles in both healthy and breast cancer cell (BT474) lines by means of the MTT assay. It was found that PD kills cancer cells within the concentration range of 0.02–0.04 mg mL−1. By contrast, PD particles were not toxic to healthy fibroblasts with this same concentration range.
It has been shown that PD particles target cellular lysosomes by an endocytic mechanism and exert an action that leads to the selective killing of cancer cells.
Furthermore, PD nanoparticles present rich metal complexation chemistry at lysosomal pH and can load Fe3+ and Fe2+ with Q values of 330 and 210 mg g−1, respectively.
The toxic effect of PD is inhibited in the presence of both DFO (iron chelating agent) and glutathione (radical scavenging agent). The results of the experiments performed using these two compounds may support the idea that the mechanism that could explain the selective toxicity of PD particles against cancer cells might be related to ROS production and an alteration of iron homeostasis in lysosomes.
These results are relevant since, to the best of our knowledge, there are only a few existing examples of nanosystems that are devoid of drugs that selectively kill cancer cells.20
Experimental
Materials and methods
TEM images were taken with a Technai Spirit Twin equipment. Dynamic light scattering (DLS) measurements were performed by using Malvern Zetasizer Nano ZS series equipment. Particles dispersed in deionized water (pH = 6) at less than 0.01 weight% were characterized. Hydrodynamic size was given as the number average size. The correlation functions were analyzed by cumulants method. The equipment used for the iron analysis was an ICP-OES Plasma Emission Spectrometer model ULTIMA 2 of Jobin Yvon. For the equipment calibration, standardized solutions of the different metals (Panreac) of 1000 mg L−1 were used, grouping all the standard solutions in a multi-elemental pattern. The range calibration used was 10–100 ppm. Absorbance in the MTT assay was measured in a microplate reader EZ Reader 2000 of Biochrom. Absorbance spectra were obtained with UV 1800 Shimadzu UV-vis spectrophotometer. 1 cm quartz cuvettes were used for such characterization.Synthesis of polydopamine [PD] nanoparticles
An ammonia aqueous solution (3.3 mL, NH4OH, 28–30%) was mixed with ethanol (40 mL) and deionized water (90 mL) under magnetic stirring at 25 °C for 30 min to synthesize polydopamine (PD) nanoparticles with an average diameter of approximately 100 nm. Dopamine hydrochloride (0.5 g) was dissolved in deionized water (10 mL) and then added to the above solution. The mixture was left to react for 24 h. PD nanoparticles were isolated by centrifugation and purified by centrifugation–redispersion in deionized water (40 mL), a procedure that was repeated at least four times. At the end of this procedure, the nanoparticles were resuspended in PBS at a final concentration of approximately 2.4 mg mL−1.Cell culture
BT-474, HTC 116 and NIH/3T3 cell lines were all obtained from the American Type Culture Collection (lot numbers HTB-20, CCL-247 and CRL-1658, respectively). All of them were cultured as recommended and cells' authenticity was regularly test by short tandem repeat (STR). All of the cell lines used were cultured in a growth medium consisting of high-glucose DMEM supplemented with a 10% of fetal bovine serum (FBS) and a 1% of penicillin–streptomycin at 37 °C in a 5% CO2 humidified environment.MTT assay
Healthy (NIH3T3) and cancer (BT474) cell lines were cultured and seeded in 24-well plates (8000 cells per mL). After 24 hours, different aliquots of a colloidal dispersion of PD nanoparticles (in PBS, pH = 7.4) were added to the cells. The concentration of PD in the culture medium was in the range of 0.01–0.1 mg mL−1.Cell viability was determined every 24 hours, during a 72 hours period, by adding a tetrazolium bromide solution (0.55 mg mL−1) and by dissolving the produced formazan salts in DMSO (500 μL per well). Absorbance was determined at 550 and 700 nm. The protocol that will be described later on in this work was applied to eliminate the contribution of the absorbance of PD at 550 nm in the MTT assay. Each result value is the average of three independent experiments.
Conflicts of interest
The authors declare no conflict of interests.Acknowledgements
This work was supported by a grant from the Regional Government of Castile and Leon (project reference number SA004U16). G. M. and C. N. also thank the Regional Government of Castile and Leon and European Social Fund for being awarded a Post-doctoral and Pre-doctoral fellowship, respectively.References
- P. Meredith and T. Sarna, Pigm. Cell Res., 2006, 19, 572–594 CrossRef CAS PubMed
.
- M. d'Ischia, A. Napolitano, V. Ball, C.-T. Chen and M. J. Buehler, Acc. Chem. Res., 2014, 47, 3541–3550 CrossRef PubMed
.
- Y. Liu, K. Ai, J. Liu, M. Deng, Y. He and L. Lu, Adv. Mater., 2013, 25, 1353–1359 CrossRef CAS PubMed
.
- P. Meredith and J. Riesz, Photochem. Photobiol., 2004, 79, 211–216 CrossRef CAS PubMed
.
- H. Lee, J. Rho and P. B. Messersmith, Adv. Mater., 2009, 21, 431–434 CrossRef CAS PubMed
.
- H. Lee, S. M. Dellatore, W. M. Miller and P. B. Messersmith, Science, 2007, 318, 426–430 CrossRef CAS PubMed
.
- Y. Song, G. Ye, F. Wu, Z. Wang, S. Liu, M. Kopeć, Z. Wang, J. Chen, J. Wang and K. Matyjaszewski, Chem. Mater., 2016, 28, 5013–5021 CrossRef CAS
.
- R. Ge, M. Lin, X. Li, S. Liu, W. Wang, S. Li, X. Zhang, Y. Liu, L. Liu, F. Shi, H. Sun, H. Zhang and B. Yang, ACS Appl. Mater. Interfaces, 2017, 9, 19706–19716 CrossRef CAS PubMed
.
- M. A. Vega, C. Nieto, G. Marcelo and E. M. Martín del Valle, Colloids Surf., B, 2018, 167, 284–290 CrossRef CAS PubMed
.
- Z.-H. Miao, H. Wang, H. Yang, Z.-L. Li, L. Zhen and C.-Y. Xu, ACS Appl. Mater. Interfaces, 2015, 7, 16946–16952 CrossRef CAS PubMed
.
- Y. Huang, Y. Li, Z. Hu, X. Yue, M. T. Proetto, Y. Jones and N. C. Gianneschi, ACS Cent. Sci., 2017, 3, 564–569 CrossRef CAS PubMed
.
- A. Postma, Y. Yan, Y. Wang, A. N. Zelikin, E. Tjipto and F. Caruso, Chem. Mater., 2009, 21, 3042–3044 CrossRef CAS
.
- K.-Y. Ju, Y. Lee, S. Lee, S. B. Park and J.-K. Lee, Biomacromolecules, 2011, 12, 625–632 CrossRef CAS PubMed
.
- H. Xu and D. Ren, Annu. Rev. Physiol., 2015, 77, 57–80 CrossRef CAS PubMed
.
- T. Kurz, J. W. Eaton and U. T. Brunk, Int. J. Biochem. Cell Biol., 2011, 43, 1686–1697 CrossRef CAS PubMed
.
- S. J. Dixon and B. R. Stockwell, Nat. Chem. Biol., 2014, 10, 9–17 CrossRef CAS PubMed
.
- J. D. Lambeth and A. S. Neish, Annu. Rev. Pathol.: Mech. Dis., 2014, 9, 119–145 CrossRef CAS PubMed
.
- D. B. Kell, BMC Med. Genomics, 2009, 2, 2 CrossRef PubMed
.
- A. Tarangelo and S. J. Dixon, Nat. Nanotechnol., 2016, 11, 921 CrossRef CAS PubMed
.
- S. E. Kim, L. Zhang, K. Ma, M. Riegman, F. Chen, I. Ingold, M. Conrad, M. Z. Turker, M. Gao, X. Jiang, S. Monette, M. Pauliah, M. Gonen, P. Zanzonico, T. Quinn, U. Wiesner, M. S. Bradbury and M. Overholtzer, Nat. Nanotechnol., 2016, 11, 977 CrossRef CAS PubMed
.
- T. T. Mai, A. Hamaï, A. Hienzsch, T. Cañeque, S. Müller, J. Wicinski, O. Cabaud, C. Leroy, A. David, V. Acevedo, A. Ryo, C. Ginestier, D. Birnbaum, E. Charafe-Jauffret, P. Codogno, M. Mehrpour and R. Rodriguez, Nat. Chem., 2017, 9, 1025 CrossRef CAS PubMed
.
- D. Wang, J. Zhou, R. Chen, R. Shi, C. Wang, J. Lu, G. Zhao, G. Xia, S. Zhou, Z. Liu, H. Wang, Z. Guo and Q. Chen, Chem. Mater., 2017, 29, 3477–3489 CrossRef CAS
.
- M. V. Berridge, P. M. Herst and A. S. Tan, in Biotechnology Annual Review, Elsevier, 2005, vol. 11, pp. 127–152 Search PubMed
.
- L. Zecca, M. Gallorini, V. Schünemann, A. X. Trautwein, M. Gerlach, P. Riederer, P. Vezzoni and D. Tampellini, J. Neurochem., 2001, 76, 1766–1773 CrossRef CAS PubMed
.
- N. F. Della Vecchia, R. Avolio, M. Alfè, M. E. Errico, A. Napolitano and M. d'Ischia, Adv. Funct. Mater., 2013, 23, 1331–1340 CrossRef CAS
.
- J. Liebscher, R. Mrówczyński, H. A. Scheidt, C. Filip, N. D. Hădade, R. Turcu, A. Bende and S. Beck, Langmuir, 2013, 29, 10539–10548 CrossRef CAS PubMed
.
- J. H. Waite, Int. J. Adhes. Adhes., 1987, 7, 9–14 CrossRef CAS
.
- X. Chen, Y. Yan, M. Müllner, M. P. van Koeverden, K. F. Noi, W. Zhu and F. Caruso, Langmuir, 2014, 30, 2921–2925 CrossRef CAS PubMed
.
- L. Ding, X. Zhu, Y. Wang, B. Shi, X. Ling, H. Chen, W. Nan, A. Barrett, Z. Guo, W. Tao, J. Wu and X. Shi, Nano Lett., 2017, 17, 6790–6801 CrossRef CAS PubMed
.
- N. F. Della Vecchia, A. Luchini, A. Napolitano, G. D'Errico, G. Vitiello, N. Szekely, M. d'Ischia and L. Paduano, Langmuir, 2014, 30, 9811–9818 CrossRef CAS PubMed
.
- D. R. Dreyer, D. J. Miller, B. D. Freeman, D. R. Paul and C. W. Bielawski, Langmuir, 2012, 28, 6428–6435 CrossRef CAS PubMed
.
- S. Hong, Y. S. Na, S. Choi, I. T. Song, W. Y. Kim and H. Lee, Adv. Funct. Mater., 2012, 22, 4711–4717 CrossRef CAS
.
- G. K. Balendiran, R. Dabur and D. Fraser, Cell Biochem. Funct., 2004, 22, 343–352 CrossRef CAS PubMed
.
- P. V. Bernhardt, Dalton Trans., 2007, 3214–3220, 10.1039/B708133B
.
| This journal is © The Royal Society of Chemistry 2018 |