Open Access Article
Open Access ArticleTherapeutic efficacy of polydatin for nonalcoholic fatty liver disease via regulating inflammatory response in obese mice
Juan-Fen Mo a,
Jia-Yuan Wua,
Li Zhenga,
Ya-Wei Yub,
Tian-Xin Zhangc,
Li Guoa and
Yi Bao*a
a,
Jia-Yuan Wua,
Li Zhenga,
Ya-Wei Yub,
Tian-Xin Zhangc,
Li Guoa and
Yi Bao*a
aThe Key Laboratory, The Second Affiliated Hospital of Jiaxing University, 1518 Huancheng North Road, Jiaxing, Zhejiang 314000, China. E-mail: baoyi@jxey.com; Fax: +86-573-82082936; Tel: +86-573-82073185
bDepartment of Pathology, The Second Affiliated Hospital of Jiaxing University, Jiaxing, Zhejiang 314000, China
cClinical Laboratory, The Second Affiliated Hospital of Jiaxing University, Jiaxing, Zhejiang 314000, China
First published on 5th September 2018
Abstract
Polydatin (PD), a natural precursor of resveratrol, has been used to treat several diseases, such as cardiovascular diseases, hepatic diseases and various cancers. In this study, we aimed to investigate the protective effects and underlying mechanisms of PD on non-alcoholic fatty liver disease (NAFLD) using a high fat induced obese mice model. The studied subjects were randomly divided into a lean group, a high fat diet (HFD) group, and a high fat diet with PD (HFD + PD) group. The results showed that PD reduced the body weights in HFD mice. PD also downregulated the serum levels of triglyceride (TG), low density lipoprotein (LDL), aspartate aminotransferase (AST) and alanine aminotransferase (ALT), and upregulated high density lipoprotein (HDL). Moreover, PD significantly alleviated hepatocyte steatosis and reduced Gr-1+ cells in the liver tissues of HFD mice. The mRNA levels of pro-inflammatory factors, such as monocyte chemoattractant protein-1 (MCP-1), tumor necrosis factor alpha (TNF-α), interleukin-6 (IL-6), S100A8 and S100A9 were significantly decreased in the liver tissues of HFD mice with PD treatment, and the downregulation of MCP-1 and S100A9 protein expressions was also observed. In conclusion, PD had beneficial roles in suppressing lipid accumulation in hepatocytes and anti-inflammatory responses in the liver tissue of obese associated NAFLD.
1 Introduction
Polydatin (PD) is a natural component isolated from the root and rhizome of Polygonum cuspidatum, a plant that grows widely in Asia and North America.1,2 PD is the most abundant precursor form of resveratrol in nature, but has a greater bioavailability than resveratrol.3 Previous data have demonstrated that PD has numerous biological properties for mediating oxidative stress, anti-platelet aggregation, cardio-protective activities, and anti-inflammatory functions.4,5 A recent study reported that PD could alleviate insulin resistance and chronic liver damage in a diet-induced obese rat model.6,7 Thus, PD may have protective effects against non-alcoholic fatty liver disease (NAFLD).Currently, NAFLD is the most epidemic liver disease globally and is associated with liver injury and inflammation, which gradually aggravate to become non-alcoholic steatohepatitis (NASH), fibrosis and cirrhosis.8,9 Lipid accumulation in the hepatocytes is the first stage of NAFLD and can be induced by obesity.10 Recently, it has become gradually clear that a wide range of pro-inflammatory proteins and cytokines can be secreted by adipose tissues and cause a chronic inflammation state in obesity.11,12 These risk factors play important roles in the development of NAFLD.13 There is still no satisfactory treatment for NAFLD and novel therapeutic methods need to be further explored.
In the current study, using a high fat diet induced obese mice model, we investigated therapeutic efficacies and underlying mechanisms of PD in the treatment of NAFLD. The results showed that PD could alleviate hepatocyte steatosis and reduce pro-inflammatory factors in NAFLD associated with obesity.
2 Materials and methods
2.1 Animal experiments
30 male C57/BL6 mice (5 weeks of age) were purchased from Shanghai Sippr-BK Laboratory Animal Co., Ltd. (Shanghai, China; animal quality certification number: SCXK 2013-0016). Mice were fed in standard laboratory conditions (23 ± 1 °C, 40–60% relative humidity, and a 12 h light–dark cycle) in a barrier facility with laminar flow cabinets. Mice were randomly divided into three experimental groups: a lean group (n = 10), fed a standard chow diet for 14 weeks; a high fat diet group (HFD, n = 10), fed a high fat diet, high carbohydrate diet with 45% calories from fat (Medicience, China) for 14 weeks; a group of mice (HFD + PD, n = 10), received the same diet as the HFD group and PD was added at week 10 (100 mg kg−1 d−1) for 4 weeks. PD (with a purity ≥ 98%, HPLC; molecular weight: 390.39) was purchased from Beijing Solarbio Science & Technology Co., Ltd. (Solarbio, China).After 14 weeks, all mice were fasted for 12 h and only given deionized water. Subsequently, the mice were sacrificed and whole blood, serum and liver tissues were collected. The whole blood was analyzed by a flow cytometry method within 4 h collection. The serum was separated and stored at 4 °C. A portion of liver tissues was fixed with 10% formalin for 24 h. Other liver tissues were immediately frozen in liquid nitrogen and stored at −80 °C.
All animal experiments were performed in accordance with the Administration of Affairs Concerning Experimental Animals (State Scientific and Technological Commission, P. R. China, 1988), and all animal procedures were approved by the Committee of the Second Affiliated Hospital of Jiaxing University, Jiaxing, China.
2.2 Serum biochemical profiles analysis
Commercial kits for measuring total triglyceride (TG), aspartate aminotransferase (AST) and alanine aminotransferase (ALT) were purchased from Abbott Laboratories (Abbott, USA). Commercial kits for measuring low density lipoprotein-cholesterol (LDL) and high density lipoprotein-cholesterol (HDL) were purchased from Biosino Biotechnology Co., Ltd. (Biosino, China). These parameters were obtained using an automatic biochemical analyzer (Abbott, USA).2.3 Histology staining
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 diluted anti-Gr-1 antibody (R&D, USA) at 4 °C for overnight. Slides were treated with anti-rabbit IgG (Servicebio, China) for 30 min and then incubated with simple stain DAB solution (Servicebio, China). Finally, slides were counterstained with hematoxylin, dehydrated, and mounted.
200 diluted anti-Gr-1 antibody (R&D, USA) at 4 °C for overnight. Slides were treated with anti-rabbit IgG (Servicebio, China) for 30 min and then incubated with simple stain DAB solution (Servicebio, China). Finally, slides were counterstained with hematoxylin, dehydrated, and mounted.The slides were observed and photographed by a light microscope (Zeiss, Germany).
2.4 Flow cytometry
Anti-CD11b-fluorescein isothiocyanate and anti-Gr-1-phycoerythrin were purchased from BD Biosciences (BD, USA). For extracellular staining of immune markers, 50 μL of whole blood were stained with 10 μL of anti-Gr-1 and anti-CD11b, and incubated on ice for 20 minutes. After centrifugation, the cells were made into a suspension and then quantified by FACS Canto II (BD Biosciences, USA). A total of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events were recorded and analyzed by the FACS Canto II diva software (BD Biosciences, USA).
000 events were recorded and analyzed by the FACS Canto II diva software (BD Biosciences, USA).
2.5 RNA isolation and reverse transcription quantitative polymerase chain reaction (qPCR)
Total RNA was extracted from liver tissues using TRZOL reagent (Invitrogen, USA). RNA was precipitated with isopropanol and dissolved in diethyl pyrocarbonate-treated distilled water. First strand cDNA was generated by reverse transcriptase master mix (TaKaRa, Japan). Quantitative PCR reactions comprised a final volume of 20 μL, containing 1 μL of cDNA, 10 nM of forward and reverse primers, and PCR master mixture (TaKaRa, Japan). qPCR was performed in 96-well plates using an ABI Stepone Plus machine (Applied Biosystems, USA). Average gene Ct values were normalised against the housekeeping gene GAPDH. Fold differences were determined using the comparative Ct method. The primer sequences were shown in Table 1.| Gene | Sequences for primers | |
|---|---|---|
| GAPDH | F:AACAGGGTGGTGGACCTCAT | R:GGGATAGGGCCTCTCTTGCT |
| MCP-1 | F:CACAACCACCTCAAGCAC | R:AAGGGAATACCATAACATCA |
| TNF-α | F:AGGGAGAGTGGTCAGGTTGC | R:GTGAGGAAGGCTGTGCATTG |
| IL-6 | F:CCGGAGAGGAGACTTCACAG | R:TCCACGATTTCCCAGAGAAC |
| S100A8 | F:CTTCAAGACATCGTTTGAAAGG | R:ATTCTTGTAGAGGGCATGGT |
| S100A9 | F:TTAGCCTTGAAGAGCAAGAAGATGG | R:AGCTCAGCTGATTGTCCTGGT |
2.6 Western blot analysis
Liver tissues were lysed in RIPA buffer (Sigma, USA) using a homogenizer. The protein concentration was measured by Pierce™ BCA Protein Assay Kit (Thermo Scientific, USA). Samples (20 μg per well) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to poly vinylidene fluoride membranes. After blocking with 3% bovine serum albumin (Sigma, USA) for 1 h at room temperature, membranes were incubated with primary antibodies include monocyte chemoattractant protein-1 (MCP-1) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 dilution, Abcam, USA) and S100A9 (1
400 dilution, Abcam, USA) and S100A9 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution, Abcam, USA) overnight at 4 °C and then subjected to secondary antibody conjugated with horseradish peroxidase (1
1000 dilution, Abcam, USA) overnight at 4 °C and then subjected to secondary antibody conjugated with horseradish peroxidase (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution, Jackson Laboratory, USA) for 2 h at room temperature. β-actin was used as a control (EarthOX, USA). The relative expression of the protein bands was detected by densitometry with ChemiDoc XRS software (Bio-Rad, USA). The quantified immunoblots from three independent experiments were analyzed by Image J software (NIH, USA).
1000 dilution, Jackson Laboratory, USA) for 2 h at room temperature. β-actin was used as a control (EarthOX, USA). The relative expression of the protein bands was detected by densitometry with ChemiDoc XRS software (Bio-Rad, USA). The quantified immunoblots from three independent experiments were analyzed by Image J software (NIH, USA).
2.7 Statistical analysis
All experimental data were expressed as the mean ± standard deviation (S.D.). Statistical analyses were performed using GraphPad Prism 6 (GraphPad Software, USA). Statistical significance of expression changes were calculated by unpaired t test for two groups. A P value < 0.05 was considered as statistically significant.3 Results
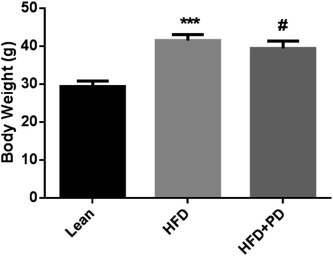
3.1 Effects of PD on body weights and serum biochemical indices
There were no significant differences in the initial body weights among the three groups. After 14 weeks, the body weights of the HFD group were significantly higher compared with the lean group (P < 0.001), but in the HFD group with PD treatment for 4 weeks, a reduced the body weights were observed in the HFD + PD group compared with HFD group (P < 0.05) (Fig. 1). In addition, the serum levels of TG, LDL, ALT and AST were significantly higher in HFD group compared with that in the lean group at week 14 (Table 2). Compared with the HFD group, the TG level was significantly decreased (P < 0.01), and the HDL level was significantly increased (P < 0.05) in the HFD + PD group (Table 2). Moreover, the levels of LDL and ALT in HFD + PD group had a decreased tendency compared with the HFD group, but there was no statistical significance (P > 0.05).| Biochemical indicators | Lean | HFD | HFD + PD |
|---|---|---|---|
| a Data were presented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, lean group vs. HFD group, #P < 0.05, ##P < 0.01, HFD group vs. HFD + PD group. | |||
| TG (mmol L−1) | 0.88 ± 0.045 | 1.36 ± 0.039*** | 1.07 ± 0.074## |
| HDL (mmol L−1) | 1.81 ± 0.029 | 1.91 ± 0.067 | 2.19 ± 0.088# |
| LDL (mmol L−1) | 0.37 ± 0.007 | 0.94 ± 0.039*** | 0.79 ± 0.058 |
| ALT (IU L−1) | 26.00 ± 1.155 | 67.00 ± 10.270** | 44.40 ± 7.132 |
| AST (IU L−1) | 138.60 ± 1.364 | 149.00 ± 4.183* | 152.20 ± 20.270 |
3.2 PD attenuates hepatic steatosis in HFD mice
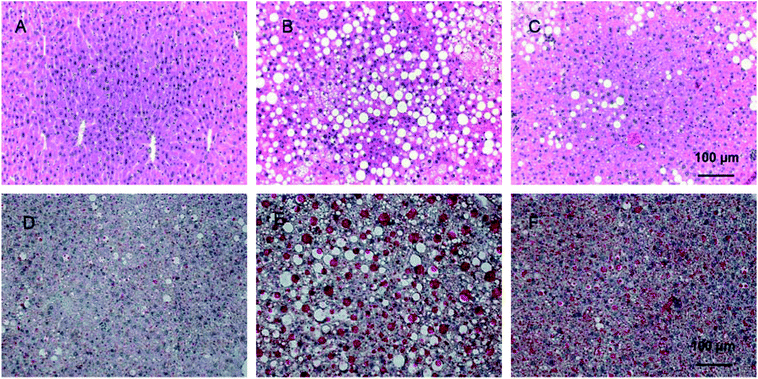
H & E staining was performed to evaluate the protective effects of PD on liver histology in HFD mice. As shown in Fig. 2A, in the lean group, hepatic lobules were clear with uniformed hepatocytes which have clear and neatly lined up nulcei. In the HFD group (Fig. 2B), a manifesting appearance of hepatic steatosis was present in the liver tissue with diffused vacuolar lipid droplets and more ballooning changes in hepatocytes compared with the lean group. In the HFD + PD group, significantly reduced accumulation of hepatic lipid droplets was observed, and the size was smaller compared with the HFD group (Fig. 2C). In Oil Red O staining, hepatic lipid droplets were significantly larger and the number increased in HFD group compared with lean group (Fig. 2D). After PD treatment for 4 weeks in the HFD mice, the hepatic lipid depositions were significantly reduced in size and numbers compared with the HFD mice without PD treatment (Fig. 2E).3.3 PD reduces the percentage of myeloid-derived suppressor cells (MDSCs) in HFD mice
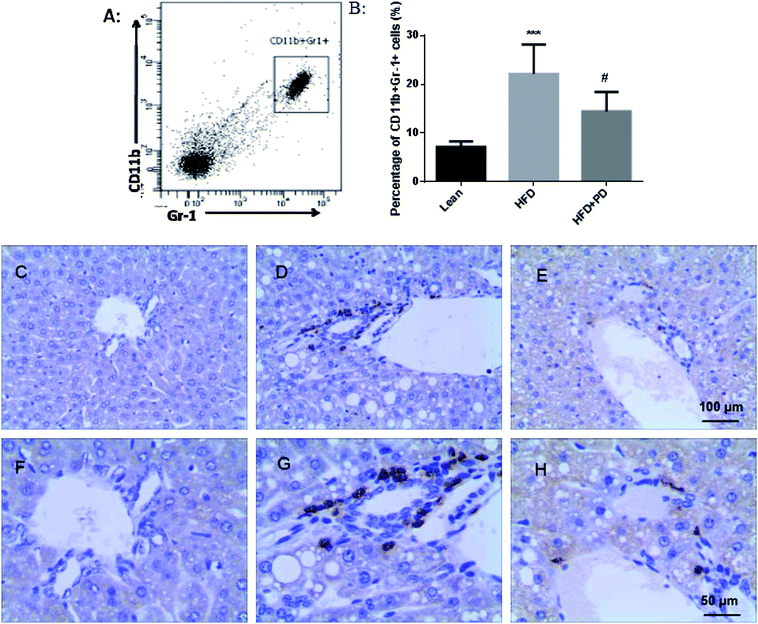
In the present study, CD11b+ Gr-1+ MDSCs were selected as the study object in Fig. 3A. We observed that the percentage of CD11b+ Gr-1+ cells was significantly increased in the peripheral blood of HFD mice compared to the lean group (P < 0.001) (Fig. 3B). After treatment with PD, the percentage of MDSCs was significantly decreased compared with the HFD group (P < 0.05) (Fig. 3B). In the immunohistochemical study, only a few Gr-1+ cells existed around the hepatic portal vein area in the lean group (Fig. 3C); however, increased numbers of Gr-1+ cells were appeared around the hepatic portal vein area in the HFD group (Fig. 3D). The Gr-1+ cells were significantly reduced in HFD + PD group compared with HFD group (Fig. 3E).3.4 Effects of PD on the regulation of pro-inflammatory factors in liver tissues of HFD mice
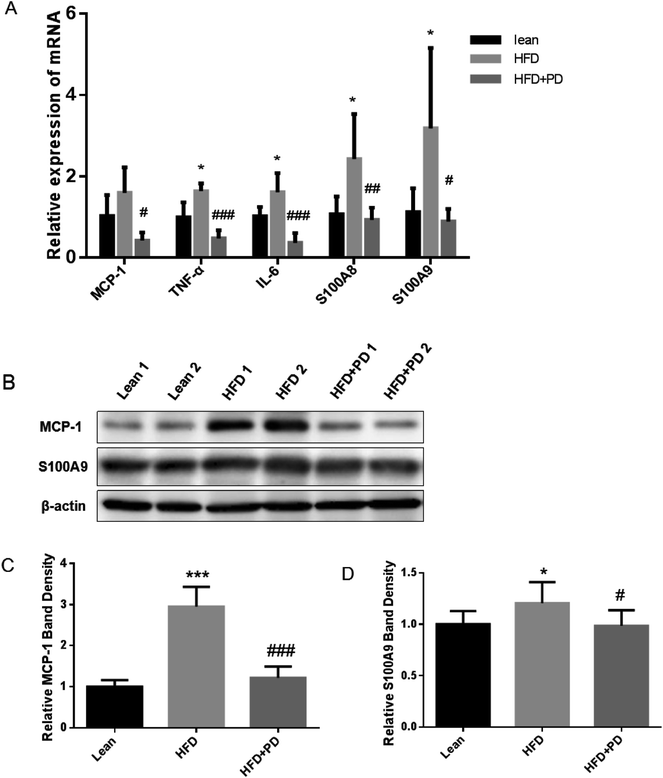
Finally, we investigated expressions of several pro-inflammatory factors in the liver tissues of HFD + PD group. The mRNA expression levels of pro-inflammatory factors, including TNF-α, IL-6, S100A8, S100A9 and MCP-1 were significantly increased in the liver of HFD group compared with the lean group (Fig. 4A). The gene expressions of these pro-inflammatory factors were significantly decreased in the HFD + PD compared with the HFD group (Fig. 4A). Western Blot analysis showed that the protein level of MCP-1 and S100A9 in HFD + PD group were markedly lower than that in the HFD group (Fig. 4B). The quantified immunoblots analyzed by Image J software showed a 2.4 fold decreased in the MCP-1 protein levels (Fig. 4C) (P < 0.001) and a 1.2 fold decreased in the S100A9 protein levels in the HFD + PD compared with the HFD group (Fig. 4D) (P < 0.05).4 Discussion
Obesity is a chronic inflammatory state associated with metabolic syndromes and NAFLD,14 including metabolic disorders, dyslipidemia, and hepatic steatosis.15 In the present study, we observed that PD treatment reduced the serum levels of TG, LDL, AST and ALT, as well as decreased lipid droplets accumulation in the liver of a high fat diet induced obese mice model, an animal model used to study obesity associated NAFLD, indicating that PD may have protective effects in NAFLD by ameliorating the severity of hepatic steatosis. According to previous reports, in high fat diet animal models, there is a a linkage between HFD and liver lipid metabolism disorder.16–18 Our results suggested that PD has the capacity of reducing lipid accumulation in hepatocytes of obese mice, and therefore may be involved in the regulation of lipid metabolism in liver.Previous studies have demonstrated that PD possessed its protective effects against liver injury and fibrosis through the inhibition of inflammation and oxidative stress in a mice model with injured liver induced by CCl4.19 PD reduced the serum levels of ALT and AST, as well as alleviated oxidative stress of the hepatic tissue in an alcohol-induced acute liver injury mice model.20 Taken together, these studies may provide indication that PD serve as a novel pharmaceutical candidate to alleviate NAFLD. Currently, several categories of medicines are approved to be used for the treatment of NAFLD, such as drugs targeting hepatic fat accumulation (elafibranor, saroglitazar),21,22 lipogenesis inhibitors (aramchol),23 bile acid-farnesoid X receptor axis (obeticholic acid),24 oxidative stress alleviation (emricasan, amlexanox, vitamin E),25–27 and anti-obesity (orlista, simtuzumab).28,29 However, there is still lack of a standard therapy with the most effective drugs for NAFLD treatment. Lifestyle modification and medications are the important treatment option for the treatment of NAFLD.30 In our study, PD which is derived from Polygonum cuspidatum, could significantly reduce blood lipid and alleviate hepatic fat deposition, although we have not done a comparison study with other traditional medicines used for NAFLD treatment, we still consider that PD is a promising compound with its capacity in the regulation of lipid metabolism.
In the subsequent study, we have explored the molecular mechanisms of PD in alleviating NAFLD. We observed that PD could significantly reduce the percentage of MDSCs in the peripheral blood of HFD mice by detecting CD11b and Gr-1, the surface markers of mice MDSCs.31 In addition, the results of immunohistochemistry showed the decreased number of cells with Gr-1 positive staining in the liver tissue of HFD mice treated with PD. MDSCs are a heterogeneous population of immune cells and have been considered to play a relevant role in a variety of hepatic diseases such as hepatic fibrosis, inflammation and hepatocellular carcinoma.32,33 In the hepatic microenvironment, MDSCs could impair T cell proliferation and effector function, that lead to a complex suppressive microenvironment.34 We hypothesized that NAFLD induced by obesity could promote the release of MDSCs from bone marrow to the circulation, and MDSCs might be further recruited into the injured liver by inflammatory cytokines. It is still not fully established what are the fundamental roles of MDSCs accumulation during liver injury induced by obesity. In our study, PD appears to reduce MDSCs both in the circulation and in the liver of obese mice, suggesting PD may be able to restore the dysregulated immune state in obesity. However, whether PD takes a direct or indirect role in the regulation of MDSCs is still unclear and extensive studies are needed in the future.
NAFLD is considered to be a chronic inflammatory state as hepatic resident cells macrophages, lymphocytes, can produce a wide range of pro-inflammatory factors such as MCP-1, TNF-α, and IL-6.35 These factors are contributed to hepatocyte apoptosis, immune cell activation and recruit immune and nonimmune cells into the inflamed sites.35 In our study, we showed that PD could reduce the liver inflammatory response by decreasing inflammatory factors locally in the liver of obese mice. The gene expression of MCP-1, TNF-α, and IL-6 were decreased in the liver of obese mice after PD treatment, further confirmed by the decreased MCP-1 protein levels in the liver of obese mice with PD treatment. In addition, the mRNA levels of S100A8 and S100A9, as well as the protein expression of S100A9 were downregulated in the liver of HFD induced obese mice after PD treatment. S100A8 and S100A9 are expressed in granulocytes, monocytes and macrophages, and the secreted S100A8 and S100A9 form a heterodimeric protein relating to pro-inflammatory mechanisms.36 Furthermore, S100A8 and S100A9 are considered to be markers of MDSCs and can also be released by MDSCs.37 S100A8/A9 can promote the expansion of MDSCs, resulting in an increase of inflammatory factors.38 The downregulation of S100A8/A9 indirectly prove MDSCs, and reduced the further accumulation of MDSCs. Taken together, these data suggested that PD may be a potential candidate to treat NAFLD by regulating immune activities and inflammatory responses systematically and locally.
In conclusion, PD has beneficial effects on decreasing triglyceride, reducing the lipids accumulation in hepatocytes, alleviating hepatocytes steatosis, and decreasing inflammatory response in the liver of NAFLD induced by obesity.
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was supported by the Natural Science Foundation of Zhejiang Province, Zhejiang, China (LY16H070007), Health Bureau of Zhejiang Province, Zhejiang, China (2016KYB292), Science and Technology Bureau of Jiaxing, Zhejiang, China (2016AY23054).References
- Q. H. Du, C. Peng and H. Zhang, Pharm. Biol., 2013, 51, 1347–1354 CrossRef PubMed.
- E. Gugliandolo, R. Fusco, F. Biundo, R. D Amico, F. Benedetto, R. Di Paola and S. Cuzzocrea, Pharmacol. Res., 2017, 123, 83–92 CrossRef PubMed.
- H. Zhang, C. Li, S. Kwok, Q. Zhang and S. Chan, J. Evidence-Based Complementary Altern. Med., 2013, 2013, 1–13 Search PubMed.
- L. Q. Xu, Y. L. Xie, S. H. Gui, X. Zhang, Z. Z. Mo, C. Y. Sun, C. L. Li, D. D. Luo, Z. B. Zhang, Z. R. Su and J. H. Xie, Food Funct., 2016, 7, 4545–4555 RSC.
- B. Liu, S. Li, X. Sui, L. Guo, X. Liu, H. Li, L. Gao, S. Cai, Y. Li, T. Wang and X. Piao, Resveratrol, and Emodin, Front. Pharmacol., 2018, 9, 347 CrossRef PubMed.
- H. Zhang, C. H. Yu, Y. P. Jiang, C. Peng, K. He, J. Y. Tang and H. L. Xin, PLoS One, 2012, 7, e46574 CrossRef PubMed.
- Q. Zhang, Y. Tan, N. Zhang and F. Yao, Mol. Med. Rep., 2015, 11, 603–610 CrossRef PubMed.
- M. V. Machado, World J. Gastroenterol., 2014, 20, 12956 CrossRef PubMed.
- C. C. Lindenmeyer and A. J. McCullough, Clin. Liver Dis., 2018, 22, 11–21 CrossRef PubMed.
- J. Ko and K. Kim, J. Exerc. Nutrition Biochem., 2013, 17, 181–188 CrossRef PubMed.
- J. Steiner and C. Lang, Alcohol, Biomolecules, 2017, 7, 16 CrossRef PubMed.
- J. Hutcheson, Cytokine, 2015, 75, 272–279 CrossRef PubMed.
- O. Massoud and M. Charlton, Clin. Liver Dis., 2018, 22, 201–211 CrossRef PubMed.
- Y. Luo, P. Abiri, S. Zhang, C. C. Chang, A. H. Kaboodrangi, R. Li, A. K. Sahib, A. Bui, R. Kumar, M. Woo, Z. Li, R. Packard, Y. C. Tai and T. K. Hsiai, Theranostics, 2018, 8, 1636–1647 CrossRef PubMed.
- E. Fabbrini, S. Sullivan and S. Klein, Hepatology, 2010, 51, 679–689 CrossRef PubMed.
- Z. G. Pan and X. S. An, Biochem. Biophys. Res. Commun., 2018, 498, 416–423 CrossRef PubMed.
- G. Fouret, S. Gaillet, J. Lecomte, B. Bonafos, F. Djohan, B. Barea, E. Badia, C. Coudray and C. Feillet-Coudray, Br. J. Nutr., 2018, 119, 368–380 CrossRef PubMed.
- S. Bashir, Y. Sharma, A. Elahi and F. Khan, J. Nutr. Biochem., 2016, 33, 82–90 CrossRef PubMed.
- X. Zhao, R. Li, Y. Liu, X. Zhang, M. Zhang, Z. Zeng, L. Wu, X. Gao, T. Lan and Y. Wang, Arch. Biochem. Biophys., 2017, 629, 1–7 CrossRef PubMed.
- M. Koneru, B. D. Sahu, S. Gudem, M. Kuncha, H. G. Ravuri, J. M. Kumar, E. K. Kilari and R. Sistla, Phytomedicine, 2017, 27, 23–32 CrossRef PubMed.
- V. Ratziu, S. A. Harrison, S. Francque, P. Bedossa, P. Lehert, L. Serfaty, M. Romero-Gomez, J. Boursier, M. Abdelmalek, S. Caldwell, J. Drenth, Q. M. Anstee, D. Hum, R. Hanf, A. Roudot, S. Megnien, B. Staels and A. Sanyal, Gastroenterology, 2016, 150, 1147–1159 CrossRef PubMed.
- M. Benedict and X. Zhang, World J. Hepatol., 2017, 9, 715–732 CrossRef PubMed.
- R. Safadi, F. M. Konikoff, M. Mahamid, S. Zelber-Sagi, M. Halpern, T. Gilat and R. Oren, Clin. Gastroenterol. Hepatol., 2014, 12, 2085–2091 CrossRef PubMed.
- H. Herrema, M. Meissner, T. H. van Dijk, G. Brufau, R. Boverhof, M. H. Oosterveer, D. J. Reijngoud, M. Muller, F. Stellaard, A. K. Groen and F. Kuipers, Hepatology, 2010, 51, 806–816 CrossRef PubMed.
- F. J. Barreyro, S. Holod, P. V. Finocchietto, A. M. Camino, J. B. Aquino, A. Avagnina, M. C. Carreras, J. J. Poderoso and G. J. Gores, Liver Int., 2015, 35, 953–966 CrossRef PubMed.
- G. Mintziori and S. A. Polyzos, Expert Opin. Pharmacother., 2016, 17, 1937–1946 CrossRef PubMed.
- K. V. Kowdley, L. A. Wilson and M. L. Van Natta, Hepatology, 2015, 62, 264A Search PubMed.
- S. Zelber-Sagi, A. Kessler, E. Brazowsky, M. Webb, Y. Lurie, M. Santo, M. Leshno, L. Blendis, Z. Halpern and R. Oren, Clin. Gastroenterol. Hepatol., 2006, 4, 639–644 CrossRef PubMed.
- A. H. Talal, Simtuzumab, J. Hepatol., 2013, 58, S532 CrossRef.
- Y. Rotman and A. J. Sanyal, Gut, 2017, 66, 180–190 CrossRef PubMed.
- A. C. Foks, G. H. Van Puijvelde, J. Wolbert, M. J. Kroner, V. Frodermann, T. Van Der Heijden, P. J. Van Santbrink, L. Boon, I. Bot and J. Kuiper, Cardiovasc. Res., 2016, 111, 252–261 CrossRef PubMed.
- H. Zhang, M. Lian, J. Zhang, Z. Bian, R. Tang, Q. Miao, Y. Peng, J. Fang, Z. You, P. Invernizzi, Q. Wang, M. E. Gershwin and X. Ma, Hepatology, 2018, 67, 232–246 CrossRef PubMed.
- B. Höchst, J. Mikulec, T. Baccega, C. Metzger, M. Welz, J. Peusquens, F. Tacke, P. Knolle, C. Kurts, L. Diehl and I. Ludwig-Portugall, PLoS One, 2015, 10, e119662 CrossRef PubMed.
- H. Chou, C. Hsieh, H. Yang, L. Wang, Y. Arakawa, K. Brown, Q. Wu, F. Lin, M. Peters, J. J. Fung, L. Lu and S. Qian, Hepatology, 2011, 53, 1007–1019 CrossRef PubMed.
- E. Seki and R. F. Schwabe, Hepatology, 2015, 61, 1066–1079 CrossRef PubMed.
- M. Ichikawa, R. Williams, L. Wang, T. Vogl and G. Srikrishna, Mol. Cancer Res., 2011, 9, 133–148 CrossRef PubMed.
- F. Zhao, B. Hoechst, A. Duffy, J. Gamrekelashvili, S. Fioravanti, M. P. Manns, T. F. Greten and F. Korangy, Immunology, 2012, 136, 176–183 CrossRef PubMed.
- P. Sinha, C. Okoro, D. Foell, H. H. Freeze, S. Ostrand-Rosenberg and G. Srikrishna, J. Immunol., 2008, 181, 4666–4675 CrossRef.
| This journal is © The Royal Society of Chemistry 2018 |