Open Access Article
Open Access ArticleImproving nutrient removal performance of surface flow constructed wetlands in winter using hardy submerged plant-benthic fauna systems†
Ying Guoa,
Huijun Xieb,
Jian Zhang *ac,
Wengang Wangd,
Huu Hao Ngoe,
Wenshan Guo
*ac,
Wengang Wangd,
Huu Hao Ngoe,
Wenshan Guo e,
Yan Kanga and
Bowei Zhangb
e,
Yan Kanga and
Bowei Zhangb
aShandong Key Laboratory of Water Pollution Control and Resource Reuse, School of Environmental Science and Engineering, Shandong University, Jinan, Shandong 250100, China. E-mail: zhangjian00@sdu.edu.cn; Fax: +86-531-8836978; Tel: +86-531-88361185
bEnvironmental Research Institute, Shandong University, Jinan 250100, China
cState Key Laboratory of Microbial Technology, Shandong University, Jinan 250100, China
dShandong Academy of Environmental Science, Broadway, Jinan 250100, PR China
eSchool of Civil and Environmental Engineering, University of Technology Sydney, Broadway, NSW 2007, Australia
First published on 18th December 2018
Abstract
Constructed wetlands (CWs) have been widely used as an ecological technology for removing nutrients from aquatic ecosystems. However, the treatment efficiency of surface flow constructed wetlands (SFCWs) in winter is generally low. To enhance the nutrient removal performance of SFCWs in winter, we developed a novel hardy submerged plant-benthic fauna system by adding Chironomus riparius (C. riparius) larvae and planting Potamogeton crispus L. in SFCWs. Compared to a system without C. riparius, the paired system greatly enhanced TN and TP removal with the average removal efficiencies of 54.73% and 94.76%, respectively. Furthermore, the paired system improved NO3−–N removal efficiency by 29.51% and reached NH4+–N removal efficiency as high as 86.20% simultaneously. The mass balance analysis indicated that C. riparius larvae enhanced substrate absorption and plant uptake in the CWs. The results of microbial analysis agreed with the nutrient removal performance, showing that C. riparius larvae influence the abundance and community structure of microbes related to N removal. As a whole, this study provides a promising ecological strategy for performance intensification of SFCWs in winter.
1. Introduction
Nutrient loads in aquatic systems have increased notably, leading to eutrophication and harm to ecosystems.1 Constructed wetlands (CWs), an effective ecological technology for wastewater treatment, simulate the structure and function of natural wetlands to remove superfluous nutrients. They have been widely used due to their low cost, low operation and maintenance requirements, and landscaping function.2,3 CWs could remove nutrients by utilizing substrate adsorption and precipitation, plant uptake, and microbial degradation.4,5 However, due to withered plants and inhibited microorganism activity at low temperatures, the treatment performance of CWs decline in winter.6,7Several studies have been conducted to enhance the treatment efficiency of CWs at low temperatures. Some researchers concentrated on design innovation of CWs to achieve performance intensification, and various types of CWs were developed such as vertical subsurface flow wetlands and two-stage subsurface flow wetlands, but these CWs were expensive and prone to clogging risk.8,9 Using artificial aeration in winter can also benefit pollutants removal in CWs,10 but this approach has high costs and wastes energy. The need for a low-cost and environmentally friendly method has prompted some researchers to use ecological engineering methods, such as plant selection.11,12 A few kinds of plants can survive at low temperatures and benefit CWs in terms of removing pollutants in winter.11 Fan et al. compared some hardy plants and found that the submerged plant Potamogeton crispus L. (P. crispus) could improve the nutrient removal performance of CWs in winter especially ammonium (NH4+–N) removal.12 Nevertheless, the research they conducted showed limited nitrate (NO3−–N) removal efficiency, which was due to high NH4–N conversion efficiency to NO3−–N and insufficient denitrification.12 Thus, a more effective ecological engineering method is required.
Benthic fauna is a crucial segment of natural wetland,13,14 but it is not valued in CWs. Recently, the function of some kinds of benthic fauna attracts interest for their special performance in NO3−–N consumption with the bioturbation and the stimulation on related microbes.15,16 In a previous study, benthic fauna were added in CWs planted with emerged plant in winter and achieved the enhancement of NO3− removal.17 However, the NH4+–N removal efficiency was only 56.93% in the previous system due to the excretions of benthic fauna and insufficient nitrification.17 In the present study, we tried to add benthic fauna to submerged plant CWs and hope to achieve synergic removal of NO3−–N and NH4+–N at low temperature. On the one hand, the bioturbation of benthic fauna can create burrows in the substrate.18,19 Then more NO3−–N and carbon would move to the anaerobic zone via the burrows, and denitrification can be facilitated.15,20 On the other hand, with the prominent NH4+–N removal ability of submerged plant,21,22 the NH4+–N released by benthic fauna would be removed.
Additionally, benthic fauna may stimulate microbial metabolism,23,24 which may influence N circulation and conversion further. Besides nitrogen (N), previous studies indicated that benthic fauna also contributed to the phosphorus (P) flux between sediments and the overlying water.19,25 Based on these, microbial abundance and P removal performance were also involved in this study.
For submerged plant, hardy plant P. crispus was selected in the present study. For the benthic fauna, Chironomus riparius (C. riparius) larvae, the larvae of one of the common chironomid specie, were chosen due to its cold resistance and distinct behavioural modality. In natural wetland and other aquatic ecosystem, C. riparius larvae molt into their adult form when temperature rise and the adults will oviposit in the water when the following autumn coming.16,26,27 There are a great number of C. riparius larvae distributed in natural wetland and other aquatic ecosystem every winter.28 They roam at the sediment surface and burrow downwards, which would influence both the superficial and deeper substrate and probably affect plant thereby. Then the N and P removal could also be impacted by these behavioural modality.
So, a novel hardy submerged plant-benthic fauna system, specifically a P. crispus–C. riparius larva CW, was developed creatively to enhance the nutrient removal performance of surface flow constructed wetlands (SFCWs) in winter. Dynamic measurements were performed on the N and P concentrations in the overlying water. The fate of nutrients in this system was evaluated based on calculations of mass balance. The parameters of the microorganisms were also determined by quantitative real-time polymerase chain reaction (qPCR) and Illumina high-throughput sequencing.
2. Materials and methods
2.1 P. crispus and C. riparius larvae preparation
P. crispus were transferred from Nansi Lake, Shandong Province, China in October 2016, and were washed to remove any adhering impurities. Then they were transplanted into the laboratory-scale CWs microcosms at a density of 100 rhizomes per square meter.12 10% Hoagland solution was used to cultivate sprouted P. crispus, which were approximately 10 cm high, for three weeks.C. riparius larvae were acquired from a commercial breeding facility in Jinan, Shandong Province, China. Then the larvae were bred in the laboratory (10 cm of sediment, the same as experimental sediment; 10 °C; constant aeration; in darkness) for two weeks, feeding with synthetic water that was similar to the microcosms used. Larvae of 7 to 12 mm length were singled out and washed with purified water. After that, they were introduced into the experimental CWs microcosms at a density of 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 individuals per m2, according to the typical density observed in natural environments.15
000 individuals per m2, according to the typical density observed in natural environments.15
2.2 Experimental setup and operation
The experimental microcosms were established outdoors under a transparent rain shelter in Jinan, northern China (36°40′N, 117°03′E). Six polyethylene barrels of 50 cm depth and 43 cm inner diameter were used to develop laboratory-scale SFCWs. All the systems had an outlet at the bottom to drain water away and had a vertical perforated PVC pipe in the centre of the barrel to measure temperature and dissolved oxygen (DO). Two layers of substrate were added to each CW microcosm: the 5 cm superincumbent substrate layer (SS layer) was sediment which obtained from Baiyun Lake natural wetland (Jinan, Shandong Province) and was sieved through a 2 mm mesh as a suitable habitat for C. riparius larvae; the 15 cm underlying substrate layer (US layer) was washed sand (particle size <2 mm; mainly SiO2, Al2O3, and Fe2O3). The six CWs were divided into two groups (each group had three parallels): CWs with P. crispus larvae and C. riparius were named CWs-PC; CWs with only P. crispus were named CWs-P and served as controls.Synthetic wastewater was used for experimental influent to simulate sewage treatment plant effluent (Grade I-B Discharge Standard of Pollutants for Municipal Wastewater Treatment Plant of China (GB 18918-2002))29 by mixing the following components in tap water: 53.46 mg L−1 sucrose, 37.75 mg L−1 (NH4)2SO4, 4.39 mg L−1 KH2PO4, 86.66 mg L−1 KNO3, 10 mg L−1 MgSO4, 10 mg L−1 CaCl2, and a nutrient solution of micronutrients containing Fe, Zn, Cu, Mn, B, and Mo. The CWs were fed the synthetic wastewater in a sequencing fill-and-draw batch mode. They were filled with wastewater for treatment at the beginning of a cycle and drained from the outlet at the last day of a cycle, then refilled immediately for the next cycle. Each CW held 36 L synthetic wastewater when filled, maintained at approximately 25 cm above the substrate. According to technical specification of constructed wetlands for wastewater treatment engineering of China (HJ 2005-2010) and the actual environmental condition, a hydraulic retention time (HRT) of 8 d was adopted. The entire experimental period included eight cycles, lasting from December 2016 to the following February with the average ambient temperature ranging from −4 °C to 7 °C. When the temperature was below zero, the microcosms were wrapped by the thermal insulation materials to avoid freezing. The details about the temperature of air and water during the experimental period were shown in Fig. S1.†
2.3 Sampling and analyses
Water samples were collected from the middle water depth of all units every two days and filtered through a 0.45 mm cellulose acetate membrane. Then the samples were analysed immediately for NO3−–N, NH4+–N, TP, TN, and chemical oxygen demand (COD), following the standard methods.30 DO and water temperature were measured at the sediment–water interface in situ every two days using a DO meter (HQ40d 53LED™, Hach, USA). Potential of hydrogen (pH) was measured at the middle water depth of all units in situ using a portable pH meter (PHS-3D, Rex Instrument, Shanghai, China).The substrate samples, including SS layer samples and US layer samples, were separately collected at the beginning and the end of the experimental session to estimate N and P storage in the substrate and to analyse microbial characteristics. SS layer samples were taken from the sediment layer at the same height in each unit, and five individual samples of equal amounts were collected. For US layer sampling, the whole sand layer (depth 15 cm) was divided into three increments: 0 to 5 cm, 5 to 10 cm, and 10 to 15 cm. Five individual samples from each depth increment were collected and mixed, and sand samples from all three depth increments in each CW were blended to obtain a representative sample for each system. Then, the SS layer samples and US layer samples were both dried at −60 °C using a freeze dryer (Unicryo MC 2 L freeze dryer, Germany) for 36 h. After desiccation, the SS layer samples and US layer samples were ground separately and passed through 0.2 mm and 1.0 mm sieves, respectively. All the substrate samples were stored at −20 °C for further analysis. The TN contents in the SS layer samples and US layer samples were measured in Xinpu Environmental Technology Company (Shanxi, China) using the semi-micro Macro Kjeldahl method,31 while the TP contents were determined by molybdenum blue method after digestion with perchloric acid and sulfuric acid.32
Plant samples were collected from all parts of the plants in each unit at the beginning and the end of the experiment to estimate N and P storage. The harvested plants were washed to remove impurities and dried in an oven at 65 °C for 72 h before being ground to pass through 0.425 mm sieves. All the C. riparius larvae in each larvae-added CW system were sorted at the end of the experiment. The animal samples were then dried at −60 °C using a freeze dryer (Unicryo MC 2 L freeze dryer, Germany) for 36 h and homogenised. Plant samples and animal samples were both stored at −20 °C. The TN and TP contents in the plant samples were measured using the same methods as substrate samples measurement in Xinpu Environmental Technology Company. The TN content in animal samples was determined using an elemental analyser in the School of Chemistry and Chemical Engineering, Shandong University (Jinan, China), while the TP content was measured using an Inductively Coupled Plasma Optical Emission Spectrometry (ICP-OES) in the Science Spectrum R & D Center (Qingdao, China).
2.4 Microbial analysis
Microbial analysis was performed on the SS layer samples, US layer samples, and animal samples. The genomic DNA was extracted using MOBIO PowerSand™ DNA Isolation Kits and analysed by qPCR and Illumina high-throughput sequencing.Quantitative real-time polymerase chain reaction was conducted using a LightCycler® 480 II (Roche, USA) to detect the absolute abundance of bacterial 16S rRNA, and the key functional genes connected with nitrification and denitrification including ammonia monooxygenase (amoA), cd1-containing nitrite reductase (nirS), copper containing nitrite reductase (nirK) using primers: 338F/518R, amo598f/amo718r, nirK583F/nirK909R, nirScd3aF/nirSR3cd, respectively (Table S1†). The reaction mixture of qPCR was 20 μL, composed of 10 μL of SYBR Premix Ex Taq™, 7.2 μL of RNess-free water, 0.8 μL of the forward and reverse primers (0.4 μL of each), and 2 μL of template DNA. Each DNA sample was performed in triplicate and then averaged to get a representative result. A 10-fold dilution series of standard DNA was used to calculate a standard curve. The standard curves for the bacterial and functional genes had R2 values of 0.991–0.999 and the amplification efficiencies ranged from 80.66% to 98.52%.
In order to detect the microbial communities in the CWs, Illumina high-throughput sequencing was performed on a HiSeq 2500 platform (Illumina, USA) at the Novogene Institute (Beijing, China). Before the Illumina sequencing was performed, the DNA concentration and purity were assessed through agarose gel electrophoresis (1%) and PCR reactions were carried out using Phusion® High-Fidelity PCR Master Mix (New England Biolabs) to amplify the V4–V5 hypervariable regions of the 16S rRNA gene with primers 515F and 907R. The PCR products were assessed by electrophoresis on 2% agarose gel and the ones with the bright strip between 400 to 450 bp were chosen for further experiments. Eligible products were pooled together and purified with Qiagen Gel Extraction Kit (Qiagen, Germany), then 250 bp paired-end sequencing of 16S rRNA gene amplicons were performed using the Illumina HiSeq instrument at the sequencing institutions. After sequencing, FLASH (version 1.2.7) was used to merge paired-end reads and QIIME (version 1.7.0) was used to filter the spliced data. Sequence reads were filtered to exclude low quality reads, chimeric sequence reads and those containing ambiguous base (‘N’). Then, high quality 16S rRNA genes were classified into operational taxonomic units (OTUs) under 97% identities. Alpha diversity was calculated using Chao, Shannon, Simpson, ACE, and Good-coverage indices. All these indices were calculated with QIIME (version 1.7.0) and displayed with R software (version 2.15.3).
2.5 Mass balance calculations
The only nutrient source considered in the present system was influent (in), while the nutrients sinks were the effluent (ef), plant uptake (pu), substrate storage involving sediment storage and sand storage (ss), animal assimilation (aa) and other losses (ol) such as microbial metabolism and volatilization.| NUin = NUef + NUpu + NUss + NUaa + NUol | (1) |
The mass balance in the CWs was calculated using equations as follows:
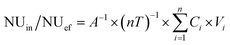 | (2) |
| NUpu/NUss/NUaa = A−1 × nT−1 × (Wend × Cend − Winitial × Cinitial) | (3) |
| NUol = NUin − NUef − NUpu − NUss − NUaa | (4) |
NUin/NUef (mg m−2 d−1) is the nutrient load for the influent or effluent, respectively; NUpu/NUss/NUaa/NUol (mg m−2 d−1) is the nutrient concentration in the different removal pathways (plant uptake, substrate storage, animal assimilation and other losses, respectively) per unit area and days of operation; i is the number of batches in sequence (i = 1, 2, 3…8); n is the total number of batches; Ci is the nutrient concentration in the influent or effluent of batch i (mg L−1); Vi is the volume of influent or effluent (L; V = 36 L); A is the area of the CW (m2; A = 0.15 m2); and T is the retention time for every batch (d; T = 8 d); Winitial/Wend (g) is the weight of the different removal agents at the beginning or end, respectively; Cinitial/Cend (mg g−1) is the nutrient concentration in the different removal agents at the beginning or end, respectively.
2.6 Statistical analysis
Data analyses were performed by Microsoft® Office Excel 2010 (Microsoft Corporation, USA), with the results expressed as mean ± standard deviation. The mean removal efficiencies of the CWs groups were calculated using the results of four typical cycles, while other mean values were calculated using the results of three replicates. One-way analysis of variance (ANOVA) tests was used to test the significance of results utilizing the statistical program SPSS 19.0 (SPSS, Chicago, USA), and differences were considered to be significant at P < 0.05.3. Results and discussion
3.1 Nitrogen and phosphorus removal performance
Because the performance of the CWs during the first four cycles was unstable (Fig. S2†), the last four experimental cycles were selected as typical cycles. The concentrations of NO3−–N, NH4+–N, TN, and TP during four typical experimental cycles for the CWs are shown in Fig. 1.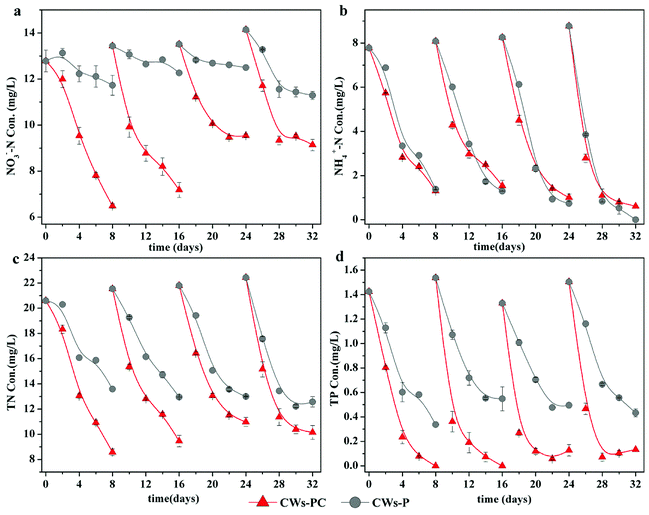 | ||
| Fig. 1 Treatment performance on NH4+–N, NO3−–N, TN and TP for each group during four typical operating cycles (n = 3). | ||
Fig. 1a shows that CWs-PC had much better performance in terms of NO3−–N removal than CWs-P (P < 0.05, n = 4), indicating C. riparius larvae addition can enhance NO3−–N removal. In the four typical experimental cycles, the final NO3−–N removal efficiencies ranged from 29.37 to 49.24% in CWs-PC while they ranged from 7.52 to 20.20% in CWs-P. The similar improvement was also reported by Lu et al. with the average NO3−–N removal efficiency increased from 10% to 30% by enhancing denitrification in winter.33 It is generally accepted that denitrification is the main mechanism of removing NO3−–N in CWs.34 In the present study, denitrification was promoted by C. riparius larvae in three aspects: O2 condition, reactants in substrate and the denitrification in C. riparius larvae body. On the one hand, the average DO concentration during the typical operating cycle was 1.87 ± 0.53 mg L−1 in CWs-PC (n = 4), while it was as high as 4.71 ± 1.07 mg L−1 in CWs-P (n = 4) (Fig. S3†), demonstrating that C. riparius larvae consumed O2 and then provided more microscopic anoxic zone in the substrate,15 which could serve as suitable habitat for denitrification microbes.35,36 On the other hand, many previous studies also illuminated that C. riparius larvae led to higher NO3−–N penetration and more particulate organic matter sedimentation into substrate, facilitating denitrification.15,16,20 Furthermore, C. riparius larvae body were distinct microsites with low O2 concentrations, high NO3−–N concentrations and plenty of denitrifiers, where denitrification could happen.20 Interestingly, no significant difference (P > 0.05, n = 4) was observed in COD removal between CWs-PC and CWs-P (Fig. S3†), indicating that carbon sources in the water might were not used for denitrification by C. riparius larvae, which should be further studied. Fig. 1a also shows differences between experimental cycles. The final NO3−–N concentrations in CWs-PC in the last two cycles were higher than those in the preceding cycles, which were related with the eclosion of C. riparius larvae.
The performance of NH4+–N removal was consistent with our expectation and no excessive accumulation of NH4+–N was observed with the addition of C. riparius larvae (P > 0.05, n = 4). As Fig. 1b shows, the treatment efficiency of NH4+-N was 86.20 ± 4.49% in CWs-PC and 89.31 ± 7.01% in CWs-P, and there was no significant difference observed between them (P > 0.05, n = 4). Compared with previous similar studies, the NH4+–N removal efficiency in the present study was much higher. In a previous study, Gao et al. achieved NH4+–N removal efficiency at 62.1 ± 8.8% in CWs by planting Iris sibirica in winter.37 In another study, Zhi et al. reported that the NH4+–N removal efficiency was 76 ± 3.9% in winter using tidal flow CWs.38 The better performance in the present study is mostly due to the effect of P. crispus. Firstly, submerged in the water, P. crispus could release oxygen produced by photosynthesis directly into the water and supply more DO to the CW system,39 which was beneficial to aerobic nitrification. Although C. riparius larvae consumed DO in CWs-PC, they could periodically ventilate the burrows created by them to supply DO to substrate,15 so the nitrification was not inhibited in CWs-PC. Secondly, P. crispus absorbed NH4+–N from the substrate and water through its roots and leaves.12 A previous study showed prominent NH4+–N removal rates by utilizing P. crispus, with NH4+–N absorption rates of 1.59 mg N min−1 g DW−1 in winter.39 Furthermore, the treatment efficiency of NH4+–N in CWs-PC and CWs-P improved in the later experimental cycles as shown in Fig. 1b, which was related to the growth of P. crispus.
Better NO3−–N removal performance and comparable NH4+–N removal performance resulted in better TN removal performance in CWs-PC than CWs-P (P < 0.05, n = 4). Fig. 1c showed that the average removal efficiency of TN reached 54.73 ± 3.19% in CWs-PC, while it was 39.57 ± 3.55% in CWs-P. In previous studies, Zhang et al. improved the TN removal efficiency of SFCWs to 44.7% in winter by plant collocation.39 Wang et al. achieved 47.5% TN removal efficiency by the subsurface flow constructed wetlands planted with hardy plant.40 Compared with these CW systems treating similar wastewater, CWs-PC had comparable TN removal efficiency and even higher by combing with C. riparius larvae and P. crispus.
As for TP removal, the performance of the two purification systems during four typical experimental cycles is shown in Fig. 1d. The TP removal efficiency was 94.76 ± 4.10% in CWs-PC, which was much higher than CWs-P (P < 0.05, n = 4). In fact, the TP removal efficiency in CWs-PC was even higher than some subsurface flow CWs. Yan and Xu mentioned the TP removal efficiency of subsurface flow CWs in north area of China ranged from 19% to 70% in winter.11 The TP removal mechanisms in CWs-PC are mainly related to substrate adsorption and plant assimilation,4 which would be discussed in Section 3.2.
3.2 Quantification and evaluation of nitrogen and phosphorus balance
In the present study, the contributions of different removal pathways to removing N and P from different wetland microcosms were quantified using mass balance. Based on areal loads of the influent (NUin), effluent (NUef), and removal rates of plant uptake (NUpu), substrate storage (NUss), animal assimilation (NUaa), and other losses (NUol), the results of the mass balance are displayed in Fig. 2. The TP content of larvae body changed from 4.54 mg g−1 to 4.72 ± 0.24 mg g−1 (P > 0.05, n = 3) during the experimental period, which was statistically insignificant, so the animal assimilation was not taken in into account in Fig. 2b.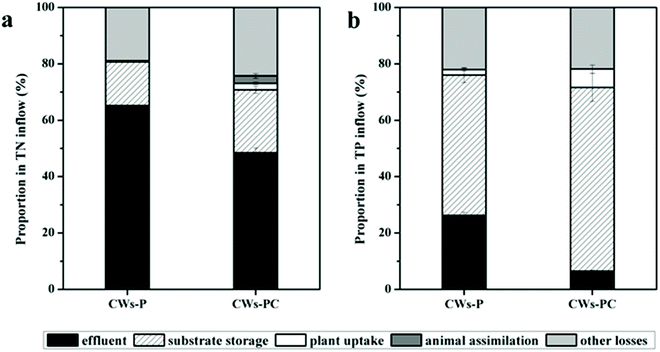 | ||
| Fig. 2 Proportion of N and P removed by different pathways among different wetlands during the experimental period (n = 3). | ||
For N removal, other losses and substrate absorption were the dominant pathway in both CWs-PC and CWs-P (Fig. 2a). Other losses include volatilization and gas emission via microbial reactions such as nitrification–denitrification, accounting for 18.89–24.24% of the TN input in all the CWs. In the present study, the volatilization of ammonia is negligible because both CWs-PC and CW-P were slightly alkaline.41 Therefore, microbial degradation might play an important role in N removal in the two systems. The results were consistent with some previous studies which suggested the significance of microbial reactions for N removal and showed a high proportion of nitrification–denitrification for N removal.42,43 It is noteworthy that the proportion of other losses in the CWs-PC accounted in TN input was 5.35% higher than in the CWs-P (P < 0.05, n = 3), which suggest that adding C. riparius larvae might influence microbial N removal and we would discuss this part in Section 3.3. Substrate absorption accounted for 15.45–22.39% of the TN input. The proportions of substrate absorption were comparable to the proportions of 20.5–34.4% found in the previous study.44 Furthermore, the proportion of plant uptake accounted in TN input varied from 0.44–2.24% in all the CWs. These values are lower than those seen in previous studies that also assess the contribution of submerged plant uptake on N removal.45,46 The minor contribution of plant uptake in the present study was mainly because of the lower biomass production of P. crispus. Besides, a previous study compared four aquatic plant treatment systems and found the P. crispus system had lowest NH4+–N uptake rate but highest NH4+–N removal efficiency,47 suggesting P. crispus mostly removed N by nitrification rather than absorption by itself. In this study, C. riparius larvae also assimilated N in CWs-PC, and the TN content of larvae body increased from 406.4 mg g−1 to 433.05 ± 2.65 mg g−1 (P < 0.05, n = 3). Due to the eclosion of C. riparius larvae and the limit of manual sorting from substrate at the end of the experiment, the larvae mass (shown in Table S2†) did not increase over the course of the experiment (P > 0.05, n = 3). However, although some larvae mass was not taken into account, animal assimilation in CWs-PC accounted for 2.71% of the TN input due to the increased TN content of larvae body, indicating direct assimilation by C. riparius larvae also contribute to N removal.
On the other hand, the proportion of TN input stored in the substrate was 6.92% higher in the CWs-PC than in the CWs-P (P < 0.05, n = 3), indicating that C. riparius larvae enhanced substrate absorption, which also contributes to TN removal. In the present study, C. riparius larvae dug downwards and irrigated the burrows they produced.15 A mass of C. riparius larvae were found in the substrate at a depth of 15 cm (Fig. S4†). Fluxes of N between the deep substrate and water could be enhanced using the burrows in the deeper substrate so that fresh inorganic N could bind to the deeper substrate and be adsorbed.48 Moreover, the burrows were rich in NH4+–N excreted by C. riparius larvae and this portion of the NH4+–N would precipitate in the deep substrate following excretion.49 As can be seen in Table 1, the TN content of the US layer was higher in the CWs-PC than in the CWs-P (P < 0.05), although there was no obvious difference in the SS layer. Thus, C. riparius larvae could improve the TN storage of US layers and increase TN removal. Besides, plant uptake in the CWs-PC removed 1.80% of the TN input higher than in the CWs-P (P < 0.05), suggesting that the contributions of plant uptake to N removal were strengthened by adding C. riparius larvae. This is because C. riparius larvae increase the biomass and N content of P. crispus. Table S2† shows the total dry weight of P. crispus in CWs-PC was 2.15 fold higher than in the CWs-P at the end of the experiment (P < 0.05), suggesting that P. crispus in CWs-PC grew better. The better growth condition was related to the higher TN storage in the substrate stimulated by C. riparius. Table 1 shows P. crispus in CWs-PC has higher TN content than CWs-P (P < 0.05), which might be related to the regulation of nitric oxide produced by larvae-based denitrification on plant assimilation.50 As a result, the plant uptake in CWs-PC was promoted by C. riparius and TN removal was promoted thereby.
| CWs | Items | Parameters | |
|---|---|---|---|
| TN (mg g−1) | TP (mg g−1) | ||
| a Indicate significant differences (P < 0.05). | |||
| CWs-P | SS layer | 1.033 ± 0.035 | 0.065 ± 0.001 |
| US layer | 0.028 ± 0.002 | 0.015 ± 0.002 | |
| Plant | 23.520 ± 1.470 | 5.944 ± 0.427 | |
| CWs-PC | SS layer | 0.915 ± 0.056 | 0.062 ± 0.001a |
| US layer | 0.128 ± 0.010a | 0.026 ± 0.001a | |
| Plant | 32.620 ± 0.770a | 6.321 ± 0.050 | |
The contributions of different pathways in different wetland microcosms to TP input were shown in Fig. 2b. The results showed that substrate storage removed 49.75–65.11% of the TP input in CWs-PC and CWs-P, while P removal through plant uptake was quantified as 1.96–6.50% and other losses accounted for 21.86–22.03%. The substrate storage of P was much higher in the CWs-PC than in the CWs-P (P < 0.05), indicating that C. riparius larvae had a great influence on TP storage. By adding C. riparius larvae, more soluble and labile P was pumped into the deeper substrate layer through bioturbation and the burrows created.25 Metal ions such as aluminium (Al), iron (Fe), and calcium (Ca), and organic matter in the substrate were then able to take up soluble and labile P, binding them in the substrate. As can be seen in Table 1, the TP contents of US layer were significantly higher with the addition of C. riparius larvae. Table 1 also showed the TP content of the SS layer was lower, which suggested that the P in the overlying wastewater and released from the SS layer would be absorbed by the US layer in CWs-PC. Furthermore, the pH value was 8.70 ± 0.10 in CWs-PC while the pH value was 8.41 ± 0.08 in CWs-P (P < 0.05, n = 3), and the higher pH value could increase P sorption capacity of alkaline CW.51 Therefore, the effect of substrate adsorption on TP removal can be enhanced in CWs-PC. Similar to the effect of C. riparius on plant uptake in terms of removing TN, C. riparius improved the performance of P. crispus on TP removal and a higher proportion of plant uptake can be obtained in CWs-PC (P < 0.05, n = 3). As to other losses, they mainly included microbe assimilation and algal component removal and no differences were observed between the CWs-PC and CWs-P (P > 0.05, n = 3). In previous studies, researchers found that plant uptake removed 36% of TP input and the P load due to adsorption, desorption, precipitation, and exchange with groundwater was estimated to be approximately 26%.52 Another study used SFCWs to treat the eutrophicated waters of a lake and showed that the TP in plants was 19.2% of TP removed.53 It seems that the contributions of different pathways to TP removal vary between different plant species and substrates.
3.3 Microbial abundance and structure
There were obvious differences in the percentage of other losses accounted in TN input observed between the CWs-PC and CW-P (as described in Section 3.2), which may be related to microbial degradation. Based on the well-founded impact of C. riparius larvae on microbial N removal,23,24,54 the abundance and structure of N-related microbes were detected in this study.It is accepted that nitrite reductase genes (nirK and nirS) and ammonia monooxygenase gene (amoA) can be used to detect denitrifying bacteria and nitrifying bacteria in environmental samples, respectively.55,56 Table 2 shows the copy numbers of these functional genes and bacterial 16S rRNA in the substrate and C. riparius larvae samples of wetland microcosms, reflecting the absolute abundance of related bacteria in the two systems. From Table 2, it can be seen that the copy number of nirS in the SS layer of the CWs-PC was higher than in the CWs-P (P < 0.05, n = 3) and the mean copy number of nirK and nirS in the US layer of CWs-P and CWs-PC were at different magnitudes, indicating that C. riparius larvae increased the abundance of denitrifying bacteria in substrate. The copy number of nirK gene and nirS gene in larval bodies were (7.90 ± 2.90) × 106 copies per g and (5.62 ± 1.73) × 106 copies per g, respectively, suggesting that denitrification could occur in the larval body. These results were consistent with the NO3−–N removal performance of CWs-PC. Moreover, the copy number of amoA in the SS layer of the CWs-PC was higher than in the CWs-P (P < 0.05), which was due to the stimulation of C. riparius larvae on nitrifying bacteria. The results agree with a previous study which showed C. riparius larvae could facilitate nitrification by concomitant ventilation and ammonium excretion in the burrows.15 The probable effect of C. riparius larvae on nitrification may be a reason for NH4+–N removal in CWs-PC. As for the absolute abundance of bacterial 16S rRNA, no obvious difference was detected between the two layers in either CWs-PC or CWs-P (P > 0.05, n = 3), indicating that C. riparius larvae did not alter the total bacterial abundance in CWs, probably changed the bacterial abundance of associated pollutants only.
| Genes | CWs | SS layer | US layer | Larvae body |
|---|---|---|---|---|
| (copies per g) | ||||
| a Indicate significant differences (P < 0.05); – means no data. | ||||
| nirK | CWs-P | (1.23 ± 0.33) × 108 | (3.43 ± 2.56) × 105 | — |
| CWs-PC | (1.36 ± 0.16) × 108 | (4.62 ± 3.42) × 106 | (7.90 ± 2.90) × 106 | |
| nirS | CWs-P | (3.47 ± 0.33) × 107 | (5.66 ± 0.00) × 104 | — |
| CWs-PC | (6.01 ± 0.53) × 107a | (4.79 ± 3.75) × 105 | (5.62 ± 1.73) × 106 | |
| amoA | CWs-P | (2.29 ± 0.32) × 105 | (9.85 ± 0.03) × 103 | — |
| CWs-PC | (3.53 ± 0.17) × 105a | (1.64 ± 0.16) × 104 | (3.27 ± 0.62) × 104 | |
| 16S rRNA | CWs-P | (7.67 ± 0.74) × 109 | (4.76 ± 0.01) × 107 | — |
| CWs-PC | (9.16 ± 0.99) × 109 | (4.87 ± 3.66) × 108 | (1.36 ± 0.16) × 109 | |
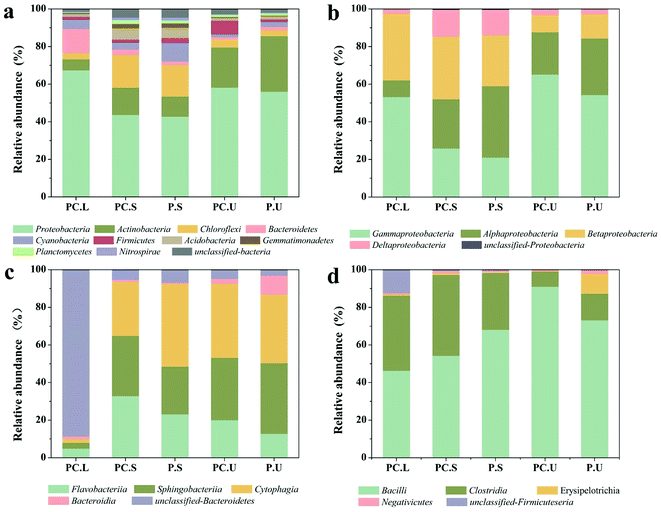
Also, the microbial communities were investigated in the present study using high-throughput sequencing. The diversity index and species richness of samples from CWs-PC and CWs-P (Table S3†) elucidated that the species diversity and richness increased imperceptibly in response to the addition of C. riparius larvae. Fig. 3a shows the bacterial composition of different samples at the phylum level. In the C. riparius larvae bodies, Proteobacteria and Bacteroidetes were the two predominant phyla, which include many types of denitrifiers.57 In the substrates of both CWs-PC and CWs-P, Proteobacteria, Actinobacteria, and Chloroflexi dominated, comprising 70.20–88.80% of all detected OTUs, and they are related to N and C cycling.58 Kang et al. also indicated Proteobacteria and Actinobacteria predominated in their benthic fauna added CWs.2
Previous studies have reported that the three typical phyla of Proteobacteria, Bacteroidetes, and Firmicutes strains play a crucial role in N-transformation.59,60 To detect the effect of C. riparius larvae on microbial communities that relate to N removal, subdivisions of these three phyla were all further analysed at class level (Fig. 3). From Fig. 3b, it can be seen that the relative abundance of gamma-Proteobacteria in the SS layer of the CWs-PC was 4.86% higher than that of the CWs-P, while in the US layer the relative abundance was 10.96% higher in the CWs-PC than in the CWs-P. Fig. 3c shows that Flavobacteriia, within the phylum Bacteroidetes, in the SS layer of the CWs-PC was 9.71% higher than that of the CWs-P, and US layer was 7.26% higher. For Firmicutes phyla (Fig. 3d), the relative abundance of Bacillus in the SS layer was 13.78% lower in the CWs-PC than in the CWs-P, but in the US layer the relative abundance of Bacillus was 17.89% higher in the CWs-PC. The relative abundance of Clostridia in the SS layer of the CWs-PC was 12.74% higher than in the same layer of the CWs-P, and there was no obvious difference observed between relative abundance in the US layer of the CWs-PC and CWs-P. Among these classes, gamma-Proteobacteria, Flavobacteriia and Clostridia are all related to denitrifying processes.61,62 Bacillus is reported to be an aerobic bacterium and involved in heterotrophic nitrification.63 The results of the relative abundance of Bacteroidetes, Proteobacteria, and Firmicutes subdivisions in the CWs at the class level indicated that C. riparius larvae could improve the relative abundance of denitrifiers in both the SS layer and US layer. Specifically, the results from the Bacillus class also suggested that the P. crispus–C. riparius CWs could supply oxygen to deeper substrates and could stimulate nitrifiers.
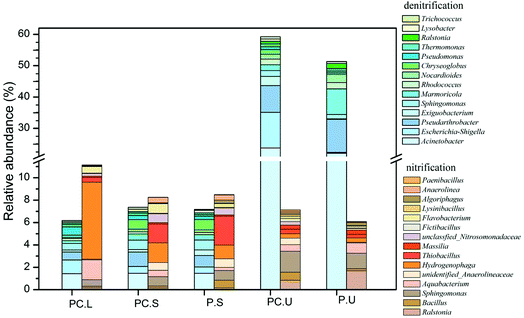
To further detect the influences of C. riparius larvae on denitrifiers and nitrifiers, the main potential functional bacteria responsible for denitrification and nitrification were identified by genus-level analysis in Fig. 4. The results showed that abundant denitrifiers and nitrifiers were observed in C. riparius larvae bodies, which provide evidence for microbial denitrification and nitrification prevailed. As for the substrate samples, no obvious difference was detected between the CWs-PC and CWs-P in SS layer. In US layer, the relative abundance of denitrifiers was higher in the CWs-PC than in the CWs-P, indicating that adding C. riparius larvae enhance denitrification in US layer, which would benefit NO3−–N removal. Besides, the relative abundance of nitrifiers was slightly higher in CWs-PC, which was related to the ventilation of C. riparius larvae burrows.
3.4 Implications for practical application of hardy submerged plant-benthic fauna systems
In the present study, we tested the performance of the P. crispus–C. riparius system on performance intensification of SFCWs in winter. Our results, based on laboratory-scale microcosms, clearly showed that the P. crispus–C. riparius system intensified the nutrient removal in winter. Because C. riparius can acclimate and maintain populations in CWs every winter with its high reproductive ability and the tolerance in varieties of environment,27 we are confident on the long-term nutrients removal performance of such ecological system. Now, further experiments are needed to validate the long-term viability of large scale P. crispus–C. riparius CWs.4. Conclusions
The P. crispus–C. riparius system could enhance the nutrient removal performance of CWs in winter. In this system, the NH4+–N removal efficiency was high with the prominent oxygen supply ability of P. crispus and the ventilation of burrows created by C. riparius larvae. The bioturbation and burrows of C. riparius larvae facilitated fluxes of nutrient between US layer and water. As a result, N transformation was enhanced and NO3−–N removal efficiency was improved with C. riparius addition. Besides, the substrate storage and plant uptake towards nutrient were enhanced, contributing to the enhanced TN and TP removal. In addition, the microbial analysis reflected that C. riparius larvae influenced the abundance and structure of N related microbes, which also beneficial to N removal. Thus, hardy submerged plant-benthic fauna systems could be a promising ecological technology to improve the performance of SFCWs in winter.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by China Major Science and Technology Program for the Water Pollution Control and Treatment (2017ZX07101003), the National Natural Science Foundation of China (51578321 and 51720105013).References
- M. Zamparas and I. Zacharias, Sci. Total Environ., 2014, 496, 551–562 CrossRef CAS PubMed.
- Y. Kang, J. Zhang, H. Xie, Z. Guo, H. H. Ngo, W. Guo and S. Liang, Bioresour. Technol., 2017, 224, 157–165 CrossRef CAS PubMed.
- Y. Li, G. Zhu, W. J. Ng and S. K. Tan, Sci. Total Environ., 2014, 468–469, 908–932 CrossRef CAS PubMed.
- I. Blanco, P. Molle, L. E. Sáenz de Miera and G. Ansola, Water Res., 2016, 89, 355–365 CrossRef CAS PubMed.
- O. Coban, P. Kuschk, U. Kappelmeyer, O. Spott, M. Martienssen, M. S. Jetten and K. Knoeller, Water Res., 2015, 74, 203–212 CrossRef CAS PubMed.
- Y. Chen, X. Wu, M. Chen, K. Li, J. Peng and P. Zhan, Front. Environ. Sci. Eng., 2013, 7, 906–912 CrossRef CAS.
- F. Wang, Y. Liu, Y. Ma, X. Wu and H. Yang, Ecol. Eng., 2012, 42, 124–129 CrossRef.
- G. F. Hua, W. Zhu, L. F. Zhao and J. Y. Huang, J. Hazard. Mater., 2010, 180, 668 CrossRef CAS PubMed.
- G. Langergraber, K. Leroch, A. Pressl, K. Sleytr, R. Rohrhofer, R. Haberl, M. Borin, M. Malagoli and M. Schiavon, Desalination, 2009, 246, 55–68 CrossRef CAS.
- C. Ouellet, Ecol. Eng., 2006, 27, 258–264 CrossRef.
- Y. Yan and J. Xu, Wetlands, 2014, 34, 243–253 CrossRef.
- J. Fan, J. Zhang, H. H. Ngo, W. Guo and X. Yin, Bioresour. Technol., 2016, 218, 1257–1260 CrossRef CAS PubMed.
- J. W. Day, A. Westphal, R. Pratt, E. Hyfield, J. Rybczyk, G. Paul Kemp, J. N. Day and B. Marx, Ecol. Eng., 2006, 27, 242–257 CrossRef.
- A. Gezie, W. Anteneh, E. Dejen and S. T. Mereta, Environ. Monit. Assess., 2017, 189, 152 CrossRef PubMed.
- S. Lagauzère, L. Pischedda, P. Cuny, F. Gilbert, G. Stora and J. M. Bonzom, Environ. Pollut., 2009, 157, 1234–1242 CrossRef PubMed.
- J. Shang, L. Zhang, C. Shi and C. Fan, J. Environ. Sci., 2013, 25, 978–985 CrossRef CAS.
- Y. Kang, J. Zhang, H. Xie, Z. Guo, P. Li, C. Cheng and L. Lv, RSC Adv., 2016, 6, 34841–34848 RSC.
- F. Hölker and P. Stief, Behav. Ecol. Sociobiol., 2005, 58, 256–263 CrossRef.
- J. K. Biswas, S. Rana and J. N. Bhakta, Ecol. Eng., 2009, 35, 1444–1453 CrossRef.
- M. Poulsen, M. V. W. Kofoed, L. H. Larsen, A. Schramm and P. Stief, Syst. Appl. Microbiol., 2014, 37, 51–59 CrossRef CAS PubMed.
- Y. Dai, C. Jia, W. Liang, S. Hu and Z. Wu, Ecol. Eng., 2012, 40, 113–116 CrossRef.
- Z. Wang, Z. Zhang, Y. Zhang, J. Zhang, S. Yan and J. Guo, Chemosphere, 2013, 92, 177–183 CrossRef CAS PubMed.
- P. Stief, Biogeosciences, 2013, 10, 7829–7846 CrossRef.
- S. Bonaglia, F. J. Nascimento, M. Bartoli, I. Klawonn and V. Brüchert, Nat. Commun., 2014, 5, 5133 CrossRef CAS PubMed.
- M. Chen, S. Ding, L. Liu, D. Xu, C. Han and C. Zhang, Environ. Pollut., 2015, 204, 241–247 CrossRef CAS PubMed.
- A. P. Mackey, Oikos, 1977, 28, 270–275 CrossRef.
- L. C. V Pinder, Annu. Rev. Entomol., 1986, 31, 1–23 CrossRef.
- Z. Xie, J. Zhang, K. Cai, Z. Xu, D. Wu and B. Wang, Acta Ecol. Sin., 2016, 36, 16–22 CrossRef.
- Q. H. Zhang, W. N. Yang, H. H. Ngo, W. S. Guo, P. K. Jin, M. Dzakpasu, S. J. Yang, Q. Wang, X. C. Wang and D. Ao, Environ. Int., 2016, 12, 11–12 CrossRef PubMed.
- APHA, AWWA and WEF, Standard Methods for the Examination of Water and Wastewater, 2005 Search PubMed.
- L. A. Douglas, A. Riazi and C. J. Smith, Soil Sci. Soc. Am. J., 1980, 44, 431–433 CrossRef CAS.
- X. Shi, J. Fan, J. Zhang and Y. Shen, Environ. Sci. Pollut. Res., 2017, 24, 22524–22534 CrossRef CAS PubMed.
- S. Lu, H. Hu, Y. Sun and J. Yang, J. Environ. Sci., 2009, 21, 1036–1043 CrossRef CAS.
- T. Sirivedhin and K. A. Gray, Ecol. Eng., 2006, 26, 167–181 CrossRef.
- Z. Fu, F. Yang, F. Zhou and Y. Xue, Bioresour. Technol., 2009, 100, 136–141 CrossRef CAS PubMed.
- S. A. Ong, K. Uchiyama, D. Inadama, Y. Ishida and K. Yamagiwa, Bioresour. Technol., 2010, 101, 7239–7244 CrossRef CAS PubMed.
- J. Gao, W. Wang, X. Guo, S. Zhu, S. Chen and R. Zhang, Ecol. Eng., 2014, 70, 351–361 CrossRef.
- W. Zhi, L. Yuan, G. Ji and C. He, Environ. Sci. Technol., 2015, 49, 4575–4583 CrossRef CAS PubMed.
- J. Zhang, H. Sun, W. Wang, Z. Hu, X. Yin, H. H. Ngo, W. Guo and J. Fan, Bioresour. Technol., 2016, 224, 222 CrossRef PubMed.
- P. Wang, H. Zhang, J. Zuo, D. Zhao, X. Zou, Z. Zhu, N. Jeelani, X. Leng and S. An, Sci. Rep., 2016, 6, 33600 CrossRef CAS PubMed.
- M. A. Valero and D. Mara, Water Sci. Technol., 2007, 55, 87–92 CrossRef CAS.
- Y. Chen, Y. Wen, Q. Zhou and J. Vymazal, Water Res., 2014, 63, 158–167 CrossRef CAS PubMed.
- A. K. Søvik and P. T. Mørkved, Sci. Total Environ., 2008, 392, 157–165 CrossRef PubMed.
- H. Wu, J. Zhang, R. Wei, S. Liang, C. Li and H. Xie, Environ. Sci. Pollut. Res., 2013, 20, 443–451 CrossRef CAS PubMed.
- P. Luo, F. Liu, S. Zhang, H. Li, R. Yao, Q. Jiang, R. Xiao and J. Wu, Bioresour. Technol., 2018, 258, 247–254 CrossRef CAS PubMed.
- S. Zhang, F. Liu, R. Xiao, Y. He and J. Wu, J. Sci. Food Agric., 2017, 97, 505–511 CrossRef CAS PubMed.
- J. Li, X. Yang, Z. Wang, Y. Shan and Z. Zheng, Bioresour. Technol., 2015, 179, 1–7 CrossRef CAS PubMed.
- N. Risgaard-Petersen, S. L. Nielsen, G. T. Banta and M. F. Pedersen, Aquatic Ecology Series, 2004, 263–280 Search PubMed.
- J. M. Svensson, Hydrobiologia, 1997, 346, 157–168 CrossRef CAS.
- L. Mur, K. Hebelstrup and K. Gupta, Front. Plant Sci., 2013, 4, 288 CAS.
- D. Xu, J. Xu, J. Wu and A. Muhammad, Chemosphere, 2006, 63, 344–352 CrossRef CAS PubMed.
- S. Y. Lu, F. C. Wu, Y. F. Lu, C. S. Xiang, P. Y. Zhang and C. X. Jin, Ecol. Eng., 2009, 35, 402–409 CrossRef.
- M. Martín, N. Oliver, C. Hernández-Crespo, S. Gargallo and M. C. Regidor, Ecol. Eng., 2013, 50, 52–61 CrossRef.
- P. Stief, Freshwater Biol., 2007, 52, 1807–1819 CrossRef CAS.
- G. Angnes, R. S. Nicoloso, M. L. B. Da Silva, P. A. V. De Oliveira, M. M. Higarashi, M. P. Mezzari and P. R. M. Miller, Bioresour. Technol., 2013, 140, 368–375 CrossRef CAS PubMed.
- K. Enwall, I. N. Throbäck, M. Stenberg, M. Söderström and S. Hallin, Appl. Environ. Microbiol., 2010, 76, 2243–2250 CrossRef CAS PubMed.
- M. Yu, R. Liao, X. X. Zhang, W. Yuan, W. Zhu, S. Peng, L. Bo and A. Li, Water Res., 2015, 76, 43–52 CrossRef PubMed.
- A. Cydzik-Kwiatkowska, Bioresour. Technol., 2015, 181, 312–320 CrossRef CAS PubMed.
- J. Luo, H. Liang, L. Yan, J. Ma, Y. Yang and G. Li, Bioresour. Technol., 2013, 148, 189 CrossRef CAS PubMed.
- Y. Xia, Y. Kong, T. R. Thomsen and N. P. Halkjaer, Appl. Environ. Microbiol., 2008, 74, 2229–2238 CrossRef CAS PubMed.
- D. Chen, H. Wang, B. Ji, K. Yang, L. Wei and Y. Jiang, Process Biochem., 2015, 50, 1904–1910 CrossRef CAS.
- H. Wang, Q. He, D. Chen, L. Wei, Z. Zou, J. Zhou, K. Yang and H. Zhang, Appl. Microbiol. Biotechnol., 2015, 99, 10829–10837 CrossRef CAS PubMed.
- K. K. Joong, J. P. Kyoung, S. C. Kyoung, S. W. Nam, T. J. Park and R. Bajpai, Bioresour. Technol., 2005, 96, 1897–1906 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra06451b |
| This journal is © The Royal Society of Chemistry 2018 |