Open Access Article
Open Access ArticleDetection of nucleic acids via G-quadruplex-controlled L-cysteine oxidation and catalyzed hairpin assembly-assisted signal amplification†
Piaopiao Chena,
Pingyue Hub,
Ke Huang b,
Erica Sawyera,
Ke Suna,
Binwu Yinga,
Xiawei Weia and
Jia Geng
b,
Erica Sawyera,
Ke Suna,
Binwu Yinga,
Xiawei Weia and
Jia Geng *a
*a
aDepartment of Laboratory Medicine, State Key Laboratory of Biotherapy, West China Hospital, Sichuan University and Collaborative Innovation Center for Biotherapy, Chengdu, Sichuan 610041, China. E-mail: geng.jia@scu.edn.cn
bCollege of Chemistry and Material Science, Sichuan Normal University, Chengdu, Sichuan 610068, China
First published on 5th December 2018
Abstract
The development of simple, sensitive and cost-effective methods for specific nucleic acid detection has attracted tremendous attention due to its importance to the early diagnosis of genetic diseases and to biodefense applications. In this work, we demonstrated a fluorescent turn-off mode DNA assay based on L-cysteine-modulated synthesis of CdTe quantum dots (CdTe QDs), horseradish peroxidase-mimicking G-quadruplex–hemin–K+ complex controlled oxidation of L-cysteine to cystine, and catalyzed hairpin assembly (CHA)-assisted signal amplification. After the addition of target DNA, the CHA signal amplification reaction was triggered and numerous H1–H2 double-stranded DNA were formed, initiating the release of G-quadruplex sequences in H2 simultaneously. Thus, the degree of inhibition of the synthesis of CdTe QDs is proportional to the concentration of the G-quadruplex sequence in this method. In contrast, when the target DNA was absent, the CHA could not be triggered, and the fluorescence signal was high due to the remaining intact L-cysteine. Under optimal experimental conditions, the homogeneous fluorescence method achieved the detection of HIV DNA with a linear range from 0.1 pM to 1 nM and a detection limit of 0.12 pM. This novel biosensor exhibits excellent specificity in differentiating DNA sequences with a single-base and two-base mismatch. To the best of our knowledge, this a label-free and highly sensitive bioassay utilizing CHA-assisted signal amplification and G-quadruplex control of in situ synthesis of CdTe QDs strategy was not reported in previous. Thus, this proposed strategy is anticipated to find use in basic biochemical research and clinical diagnosis.
Introduction
Recently, sequence-specific nucleic acid detection has received more and more attention due to its extreme importance in the clinical molecular diagnosis of diseases, biomedical studies, and cancer therapy.1,2 A variety of sensitive approaches have been developed, such as colorimetric,3 fluorescent,4 chemiluminescence (CL),5 electrochemiluminescence (ECL),6 electrochemical,7 surface enhanced Raman spectroscopy (SERS),8 quartz crystal microbalance (QCM),9 atomic fluorescence spectrometry (AFS),10 inductively coupled plasma mass spectrometry (ICP-MS) methods,11 and nanopores etc.12 Among these sensing methods, the fluorescence-based is one of the most powerful and widely used approaches because of its rapidity, simplicity, high sensitivity, and accessible instrument requirements.Fluorescent inorganic nanoparticles have been generously synthesized by biomineralization processes, in which biomolecules are used as templates to mediate the formation of fluorescent nanoparticles.13 Among these biomineralized fluorescent inorganic metal nanoparticles, quantum dots (QDs) exhibit unique photophysical properties, such as high fluorescence quantum yields, large Stokes shifts, narrow and symmetric emission spectra, optically tunable light emissions, and superior stability against photobleaching. QDs have potential applications in numerous fields, such as sensing, bioimaging, and disease diagnosis, etc.14–16 Also well known, L-cysteine could serve as an excellent ligand for CdTe QDs growths.17 The G-quadruplex–hemin–K+ complex DNAzyme, which mimics horseradish peroxidase, has attracted substantial interest in the past decade due to its catalytic ability to oxidize various substrates, such as ABTS2− and L-cysteine. These properties were extensively used to develop optical sensors for DNA,18 aptamer–substrate complexes,19 and small molecule.20 Therefore, through a combination of the L-cysteine-mediated synthesis of quantum dots and the catalytic oxidation of L-cysteine to cystine (thiols to disulfides) by the G-quadruplex–hemin–K+ complex, fluorescent bioanalysis may be feasible.
Until now, various signal amplification strategies have been pursued to explore ultrasensitive detection capability, which can be classified into enzyme-assisted and enzyme-free approaches. The enzyme-assisted approach signal amplification, such as polymerase, ligase, nicking endonuclease, exonuclease, and double-strand specific nuclease.21–24 Although target cycling methods via nucleases are highly sensitive and selective, the detection cost increase greatly and applications may be limited. Compared to the enzyme-assisted approach, enzyme-free signal amplification, such as hybridization chain reaction (HCR),25 strand displacement amplification (SDA),26 and catalyzed hairpin assembly (CHA),27 possesses unique advantages in detection due to its simple, robust, and low-cost features. Among the signal amplification methods, CHA signal amplification is an isothermal amplification reaction, and it utilizes a toehold-mediated strand displacement mechanism to continuously assemble DNA duplexes and translate the input of nucleic acid targets into an amplified signal with a low detection limit.28
To further extend the range of applications of biomimetic synthesis of luminescent nanomaterials and improve their analytical performance, a simple, homogeneous fluorescent turn-off nanoplatform by taking full advantage of G-quadruplex–hemin–K+-catalyzed oxidation reaction, L-cysteine-mediated synthesis of fluorescent QDs and CHA nucleic acid amplification technique was developed for HIV DNA assay in this work. Given the high signal amplification efficiency of CHA, and the intrinsically high sensitivity of fluorescence detection, the proposed method provides a simple and highly sensitive platform for nucleic acid assay with a low detection limit.
Experimental section
Materials
All reagents used were of the highest purity available but at least of analytical grade and used without further purification. High purity sodium nitrate (NaNO3), sodium hydroxide (NaOH), potassium chloride (KCl), cadmium chloride (CdCl2·2.5H2O), sodium tellurite (Na2TeO3) and sodium borohydride (NaBH4) were purchased from Kelong Chemical Reagents (Chengdu, China). Hemin and 3-(N-morpholino) propanesulfonic acid (MOPS) were purchased from Solarbio Technology Co., Ltd. (Beijing, China). Dimethyl sulfoxide (DMSO) was obtained from Sigma-Aldrich (St. Louis, MO, USA.). L-Cysteine was obtained from Sinopharm Chemical Reagent Co., Ltd. (Beijing, China). All oligonucleotides were synthesized and purified by Sangon Biotechnology Co. Ltd. (Shanghai, China). The sequences of the employed DNA oligonucleotides are shown in Table 1. Human serum samples were provided by the Department of Laboratory Medicine, West China Hospital of Sichuan University (Chengdu, China). All work solutions were prepared with MOPS buffer solution (10 mM, 150 mM NaNO3, pH 7.6). High purity deionized water (18.2 MΩ cm) from a water purification system (Chengdu Ultrapure Technology Co., Ltd., Chengdu, China) was used throughout this work. All solutions were stored at 4 °C in refrigerator.| Name | Sequence (5′-3′) |
|---|---|
| G1 (G-quadruplex) | GGGTTTTGGGTTTTGGGTTTTGGG |
| Target DNA | AGTCAGTGTGGAAAATCTCTAGC |
| H1 (hairpin probe 1) | GCTAGAGATTTTCCACACTGACTTCTCTAGCGGGTTTTGGGTTTTAGTCAGTGTGGAAAA |
| H2 (hairpin probe 2) | CTGACTAAAACCCAAAACCCGCTAGAGAAGTCAGTGTGGAAAATCTCTAGCGGGTTTTGGGTTTTGGGTTTTGGG |
| Single-base mismatch | AGTCAGTGTGG![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) AAATCTCTAGC AAATCTCTAGC |
| Two-base mismatch | AGTCAGT![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) TGGAAA TGGAAA![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) TCTCTAGC TCTCTAGC |
| Non-complementary | GCATGTATATTCTTCATCAATAG |
Instruments
The pH of the buffer was determined by PHS-3C pH Benchtop meter (INESA, Shanghai, China). UV-visible absorption spectra were recorded on a Hitachi U-1750 UV-vis spectrophotometer (Shimadzu, Japan). The fluorescence spectrum measurements were recorded on F-7100 (Hitachi, Tokyo, Japan). Temperature was controlled with an RCT basic package magnetic stirrer (IKA, Germany). A MIX-25P shaker (Miou, Hangzhou, China) was used to mix the solutions. All the photographs in the experiment were taken by an EOS M3 Digital Single Lens Reflex (Canon, Japan) with transmitted ultraviolet irradiation. The morphology of the CdTe QDs was recorded by JEM-2100F transmission electron microscopy (TEM).Synthesis of quantum dots
The general procedure for the synthesis of CdTe quantum dots was as follows. Typically, 200 μL L-cysteine (100 μM) was added to a solution of 200 μL MOPS buffer (10 mM, 150 mM NaNO3, pH 7.6). Then, 200 μL CdCl2·2.5H2O (2 mM), 200 μL Na2TeO3 (0.4 mM) and 20 μL NaBH4 (60 mM) were added in order into the above solution. Finally, after incubating for 75 min at 100 °C, the CdTe quantum dots were obtained.Target DNA sensing
In a typical assay, 40 μL of target DNA of different concentrations, 40 μL H1 DNA (1.25 μM) and 40 μL H2 DNA (1.25 μM) were first mixed with MOPS buffer (200 μL, pH 7.4, 10 mM) and incubated at room temperature for 90 min to complete the CHA signal amplification reaction, thus releasing the G-quadruplex sequence. Then, after introducing 15 μM hemin (10 μL) and 1 M K+ (10 μL) into the above solution, it was incubated for 15 min at room temperature to form the G-quadruplex–hemin–K+ complex. Then, 200 μL of L-cysteine (100 μM) was added into the above solution to initiate the oxidation reaction, which took place over 45 min at room temperature. Finally, 200 μL CdCl2·2.5H2O (2 mM), 200 μL Na2TeO3 (0.4 mM) and 20 μL NaBH4 (60 mM) were sequentially added into the resulting solution. After incubation for 75 min at 100 °C, the final solution was cooled to room temperature. The fluorescence spectra of the mixtures were measured on the spectrophotometer with a 365 nm excitation wavelength.Pretreatment of human serum samples
Serum samples were taken and centrifuged for 15 min (8000 rpm) using an ultrafiltration centrifuge tube (MWCO = 3 kDa). Then, the ultrafiltrates were diluted 100 times by ultrapure water and stored in 4 °C before analysis.Results and discussion
Sensing principle
The principle of this G-quadruplex-controlled L-cysteine oxidation and CHA-assisted signal amplification fluorescence detection of DNA is elucidated in Scheme 1. The proposed strategy for the DNA assay included the following three steps: (1) CHA-assisted signal amplification triggered by target DNA, which releases the free G-quadruplex sequences, (2) the addition of hemin and K+ to form the G-quadruplex–hemin–K+ complex to initiate the following oxidation reaction of L-cysteine to cystine, (3) synthesis of fluorescent CdTe QDs by adding the CdCl2, Na2TeO3 and NaBH4 into the resulting aforementioned solution. Two types of DNA hairpin probes were ingeniously designed and employed in this system, including H1 and H2 (hairpin probes, shown in Table 1). The H1 and H2 probes could form a stem-loop structure due to the binding of the complementary sequences at the ends. H2 was the signal hairpin composed of a G-quadruplex sequence. When H2 retains its hairpin structure, the segment of the G-quadruplex sequence is hidden in the stem. Thus, the G-quadruplex cannot form which decreases the background CHA. Additionally, the sequence of the H1 is partially complementary to the target DNA, while the entire sequence of H1 is complementary to H2. When the H1 was confronted with target DNA, the recognition between the target DNA and H1 would break the hairpin structure, initiating the first branch migration and the opening of H1. Then, the newly exposed sequence in the opened H1 serves as a new trigger which hybridizes with H2 and initiates the second branch migration, resulting in the formation of an H1–H2 complex and the release of the G-quadruplex sequences. Finally, the released G-quadruplex has the capability to bind with hemin and act as an HRP-like DNAzyme for catalyzing L-cysteine oxidation, and L-cysteine serves as a ligand to mediate biomimetic synthesis of luminescent quantum dots. Hence, the degree of inhibition for the QDs synthesis is proportional to the concentration of target DNA. In this way, the target DNA trigger catalyzes the autonomous assembly and generation of numerous H1–H2 complexes and G-quadruplexes, which are exponentially produced. Therefore, the basis of the L-cysteine-mediated synthesis of fluorescent CdTe QDs and CHA-assisted target recycling strategy provides the possible high sensitivity for the assay of nucleic acids through the turn-off mode.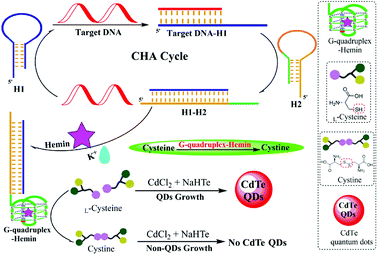 | ||
| Scheme 1 Schematic illustration of the label-free and homogeneous fluorescence DNA assay based on the G-quadruplex-controlled oxidation of L-cysteine and CHA-assisted signal amplification. | ||
Feasibility of the fluorescence DNA detection
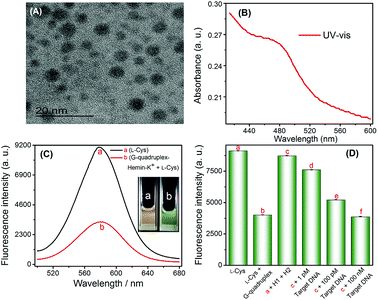
Fluorescence experiments were performed to investigate the feasibility of the G-quadruplex–hemin–K+-controlled oxidation of L-cysteine and CHA-assisted signal amplification triggered by target nucleic acids.The ability of G-quadruplex–hemin–K+ to catalyze the oxidation of L-cysteine to cystine, and L-cysteine to serve as the ligand for CdTe QDs synthesis was affirmed first. Evidently, in the absence of the G-quadruplex–hemin–K+ complex, L-cysteine could not be oxidized and still maintained the capability to assist CdTe QDs synthesis. As the TEM image shows, the CdTe QDs which used L-cysteine as a ligand are quasi-spherical and non-aggregated with an average diameter of ∼4 nm (Fig. 1A).29 As shown in Fig. 1B, the characteristic absorption peak at approximately 480 nm was found in the UV-vis spectra. Furthermore, a significant fluorescence signal was acquired (as shown in Fig. 1C-a, and inset (a)). However, in the presence of the G-quadruplex–hemin–K+ complex, L-cysteine was effectively converted into cystine via the catalyzed oxidation reaction, which led to a much lower fluorescence intensity (shown in Fig. 1C-b, and inset (b)).
To investigate the feasibility of the proposed strategy towards target HIV DNA detection, fluorescence spectra signals of the sensing system under various conditions were recorded and displayed in Fig. 1D. Obviously, a relatively weak fluorescence signal (Fig. 1D-b) was observed when G1 (G-quadruplex sequence, Table 1) was pre-incubated with L-cysteine in the presence of hemin and K+ (b vs. a). No significant fluorescence signal changes were observed when H1 and H2 DNA (in the absence of a target DNA) were added into L-cysteine (solution a), which indicated that the formation of the G-quadruplex structure is effectively suppressed by the formation of the H2 hairpin structure, and H1 and H2 can stably coexist in solution. In contrast, in the presence of the target DNA, the fluorescence intensity of CdTe QDs considerably decreased (c vs. d–f). Such a decrease in the fluorescence signal is due to the fact that the target DNA binds and opens the H1 and H2 hairpin structure, leading to the release of G-quadruplex sequence, and then causes the formation of the G-quadruplex–hemin–K+ complex; this complex oxidizes L-cysteine to cystine, thus inhibiting the formation of quantum dots. These experimental results clearly demonstrated that the fluorescence detection of HIV DNA could be readily achieved using the proposed system.
Optimization of experimental conditions
Before the standard DNA assay, some important parameters were optimized to obtain the best sensing performance, including the conditions of CdTe QDs synthesis, CHA-assisted signal amplification reaction, formation of the G-quadruplex–hemin–K+ complex, and the oxidation of L-cysteine to cystine, etc. As shown in Fig. S1A and S1B,† the maximum fluorescence signals were obtained at 100 °C, and the peak fluorescent intensity was achieved at the time of 75 min for CdTe QDs synthesis (Fig. S1C and S1D†). Therefore, the conditions of 75 min incubation at 100 °C were chosen to synthesize CdTe QDs. As shown in Fig. S2 and S3,† the maximum fluorescence value obtained at the concentrations of L-cysteine and NaBH4 were 100 μM and 60 mM, respectively. The CHA-assisted signal amplification reaction conditions mainly involve the concentrations of H1 and H2, and the total reaction time after adding target DNA into blank solution. As shown in Fig. S4 and S5,† the concentrations of H1 and H2 at 1.25 μM, and reaction time of 90 min were used in the subsequent experiments, respectively. The formation of the G-quadruplex–hemin–K+ complex and oxidation L-cysteine to cystine is crucial for the entire experiment. The concentration of K+ and hemin were investigated subsequently, respectively. As shown in Fig. S6 and Fig. S7,† the maximum fluorescence difference values were obtained at the concentrations of 10 mM K+ and 15 μM hemin. The time of 15 min for the formation the G-quadruplex–hemin–K+ complex after added the hemin and K+, and 45 min for the oxidation of L-cysteine to cystine by the G-quadruplex–hemin complex was sufficient (as shown in Fig. S8 and S9†).Analytical performance of the DNA sensor
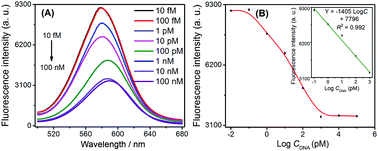
Under the optimal conditions, we used this G-quadruplex-controlled oxidation of L-cysteine and L-cysteine-mediated synthesis of fluorescent quantum dots system to detect synthetic HIV DNA by monitoring fluorescent signals. A series of fluorescence spectra are shown in Fig. 2A. The fluorescence intensity of CdTe QDs gradually decreased in conjunction with increasing concentrations of target DNA from 10 fM to 100 nM. It could be seen that the fluorescence intensity of CdTe QDs and the logarithmic concentration of HIV DNA showed a good linear relationship within the range from 0.1 pM to 1 nM (as shown in the inset of Fig. 2B). The linear equation was found to be Y = −1405![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C + 7796, with the correlation coefficient R2 = 0.992, and the detection limit was 0.12 pM (3σ). The reproducibility expressed as relative standard deviation (RSD) of 10 pM target DNA was 4.8%. This high sensitivity could be ascribed to the effective signal amplification of the developed enzyme-free CHA-assisted signal amplification strategy.
C + 7796, with the correlation coefficient R2 = 0.992, and the detection limit was 0.12 pM (3σ). The reproducibility expressed as relative standard deviation (RSD) of 10 pM target DNA was 4.8%. This high sensitivity could be ascribed to the effective signal amplification of the developed enzyme-free CHA-assisted signal amplification strategy.
The analytical performance of this assay was further compared with several other reported methods and summarized in Table S1.† Compared to the previous enzyme-involved and enzyme-free protocols, the proposed method exhibited comparable sensitivity with a simpler operation and design.
Specific discrimination for base mismatches
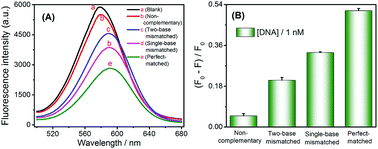
The capability to specifically detect base mismatches was investigated. The selectivity of the sensing scheme was also evaluated by monitoring the fluorescence response of CdTe QDs when this method was challenged with one base- and two base-mismatches, and the non-complementary DNA as well as perfectly complementary DNA sequences at same concentration of 1 nM. Fig. 3A displayed an obvious fluorescence signal decreased with one base- and two base-mismatches as well as fully complementary DNA sequences, respectively. In contrast to the significant reduction of fluorescence intensity caused by the target DNA, the addition of the non-complementary and base-mismatched strands triggered a less reduction in fluorescence intensity (as shown in Fig. 3B). These results demonstrated that this method can distinguish single-base mismatches in DNA sequences.Practical applicability
To validate the application of this proposed DNA assay to real biological samples, human serum samples spiked with different concentrations of target HIV DNA were analyzed. To eliminate potential interferences, the serum samples were filtered using a centrifugal filtration device (MWCO = 3 kDa), which removed small molecules (e.g. GSH) and ions (e.g. K+). The resulting filtrate was diluted 100-fold before use. As demonstrated in Table 2, the spiked recovery of HIV DNA from the serum samples was 93–104%, and the relative standard deviation (RSD) was as low as 3.3%, which showed a strong consistency between the spiked concentrations. Therefore, these results demonstrated the tolerance of the proposed method to the complicated sample matrix of serum.Conclusions
In summary, a well-designed synthetic fluorescent quantum dot-based novel turn-off mode strategy for the highly sensitive label-free detection of DNA was developed. This method mainly was based on the L-cysteine-modulated synthesis of CdTe QDs, the horseradish peroxidase-mimicking oxidative control of the L-cysteine to cystine conversion performed by G-quadruplex–hemin–K+, and the CHA-assisted signal amplification. Benefitting from the CHA-assisted signal amplification, and the intrinsically high sensitivity of fluorescence detection, this system revealed good performance in nucleic acid analysis with a detection limit of 0.12 pM. This method is more straightforward and cost-effective than other current analytical methods, and this is the first example showing the technique of biomimetic synthesis of luminescent nanomaterials coupled with nucleic acid signal amplification to directly monitor DNA.30 Also, considering the versatility of aptamers or peptides as recognition ligands, the proposed biomolecular-modulated quantum dot synthesis-based DNA bioassay platform is expected to have a broad spectrum of applications in bioanalysis.31,32Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Key Research and Development Program of China (Grant No. 2016YFA0201400 and No. 2016YFC1200300).Notes and references
- S. K. Arya and S. Bhansali, Chem. Rev., 2011, 111, 6783–6809 CrossRef CAS PubMed.
- J. Bell, Nature, 2004, 429, 453–456 CrossRef CAS PubMed.
- P. P. Liang, J. Canoura, H. X. Yu, O. Alkhamis and Y. Xiao, ACS Appl. Mater. Interfaces, 2018, 10, 4233–4242 CrossRef CAS PubMed.
- W. Li, C. X. Yang and X. P. Yan, Chem. Commun., 2017, 53, 11469–11471 RSC.
- N. X. Li, J. Zheng, C. R. Li, X. X. Wang, X. H. Ji and Z. K. He, Chem. Commun., 2017, 53, 8486–8488 RSC.
- P. Zhang, J. Jiang, R. Yuan, Y. Zhuo and Y. Q. Chai, J. Am. Chem. Soc., 2018, 140, 9361–9364 CrossRef CAS PubMed.
- X. Y. Wang, F. Chen, D. X. Zhang, Y. Zhao, J. Wei, L. H. Wang, S. P. Song, C. H. Fan and Y. X. Zhao, Chem. Sci., 2017, 8, 4764–4770 RSC.
- Y. D. Sun, P. Peng, R. Y. Guo, H. H. Wang and T. Li, Biosens. Bioelectron., 2018, 104, 32–38 CrossRef CAS PubMed.
- Y. Zhao, H. M. Wang, W. Tang, S. C. Hu, N. Li and F. Liu, Chem. Commun., 2015, 51, 10660–10663 RSC.
- P. P. Chen, P. Wu, J. B. Chen, P. Yang, X. F. Zhang, C. B. Zheng and X. D. Hou, Anal. Chem., 2016, 88, 2065–2071 CrossRef CAS PubMed.
- R. Liu, C. Q. Wang, Y. M. Xu, J. Y. Hu, D. Y. Deng and Y. Lv, Anal. Chem., 2017, 89, 13269–13274 CrossRef CAS PubMed.
- B. Y. Guo, Y. Y. Sheng, K. Zhou, Q. S. Liu, L. Liu and H. C. Wu, Angew. Chem., Int. Ed., 2018, 57, 3602–3606 CrossRef CAS PubMed.
- N. Goswami, F. X. Lin, Y. B. Liu, D. T. Leong and J. P. Xie, Chem. Mater., 2016, 28, 4009–4016 CrossRef CAS.
- Y. F. Kong, J. Chen, H. W. Fang, G. Heath, Y. Wo, W. L. Wang, Y. X. Li, Y. Guo, S. D. Evans, S. Y. Chen and D. J. Zhou, Chem. Mater., 2016, 28, 3041–3050 CrossRef CAS PubMed.
- Y. Hu, Z. Y. Miao, M. J. Zhang, X. T. Yang, Y. Y. Tang, S. Yu, C. X. Shan, H. M. Wen and D. Zhu, Anal. Chem., 2018, 90, 5678–5686 CrossRef CAS PubMed.
- K. Huang, K. L. Xu, J. Tang, L. Yang, J. R. Zhou, X. D. Hou and C. B. Zheng, Anal. Chem., 2015, 87, 6584–6591 CrossRef CAS PubMed.
- Y. F. Chen, Z. Chen, Y. J. He, H. L. Lin, P. T. Sheng, C. B. Liu, S. L. Luo and Q. Y. Cai, Nanotechnology, 2010, 21, 125502 CrossRef PubMed.
- A. Niazov-Elkan, E. Golub, E. Sharon, D. Balogh and I. Willner, Small, 2014, 10, 2883–2891 CrossRef CAS PubMed.
- H. Y. Li, J. F. Chang, P. P. Gai and F. Li, ACS Appl. Mater. Interfaces, 2018, 10, 4561–4568 CrossRef CAS PubMed.
- F. A. Wang, X. Q. Liu, C. H. Lu and I. Willner, ACS Nano, 2013, 7, 7278–7286 CrossRef CAS PubMed.
- Y. He, D. L. Chen, M. X. Li, L. Fang, W. J. Yang, L. J. Xu and F. F. Fu, Biosens. Bioelectron., 2014, 58, 209–213 CrossRef CAS PubMed.
- D. F. Wang, C. L. Vietz, T. Schroeder, G. Acuna, B. Lalkens and P. Tinnefeld, Nano Lett., 2017, 17, 5368–5374 CrossRef CAS PubMed.
- X. L. Zuo, F. Xia, Y. Xiao and K. W. Plaxco, J. Am. Chem. Soc., 2010, 132, 1816–1818 CrossRef CAS PubMed.
- B. C. Yin, Y. Q. Liu and B. C. Ye, J. Am. Chem. Soc., 2012, 134, 5064–5067 CrossRef CAS PubMed.
- J. Huang, Y. R. Wu, Y. Chen, Z. Zhu, X. H. Yang, C. Y. J. Yang, K. M. Wang and W. H. Tan, Angew. Chem., Int. Ed., 2011, 50, 401–404 CrossRef CAS PubMed.
- P. P. Chen, P. Wu, Y. X. Zhang, J. B. Chen, X. M. Jiang, C. B. Zheng and X. D. Hou, Anal. Chem., 2016, 88, 12386–12392 CrossRef CAS PubMed.
- H. M. Fang, N. L. Xie, M. Ou, J. Huang, W. S. Li, Q. Wang, J. B. Liu, X. H. Yang and K. M. Wang, Anal. Chem., 2018, 90, 7164–7170 CrossRef CAS PubMed.
- Y. Jiang, B. L. Li, J. N. Milligan, S. Bhadra and A. D. Ellington, J. Am. Chem. Soc., 2013, 135, 7430–7433 CrossRef CAS PubMed.
- J. b. Liu, X. H. Yang, K. M. Wang, X. X. He, Q. Wang, J. Huang and Y. Liu, ACS Nano, 2012, 6, 4973–4983 CrossRef CAS PubMed.
- X. W. He and N. Ma, Small, 2013, 9, 2527–2531 CrossRef CAS PubMed.
- H. M. Meng, H. Liu, H. L. Kuai, R. Z. Peng, L. T. Mo and X. B. Zhang, Chem. Soc. Rev., 2016, 45, 2583–2602 RSC.
- Y. Cheng, C. L. Sun, X. W. Ou, B. F. Liu, X. D. Lou and F. Xia, Chem. Sci., 2017, 8, 4571–4578 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra08296k |
| This journal is © The Royal Society of Chemistry 2018 |