Open Access Article
Open Access ArticleProtein-induced fluorescence enhancement for a simple and universal detection of protein/small molecule interactions†
Hansol Kima,
Chang Yeol Leea,
Jayeon Songa,
Junhyeok Yoona,
Ki Soo Park *b and
Hyun Gyu Park
*b and
Hyun Gyu Park *a
*a
aDepartment of Chemical and Biomolecular Engineering (BK 21+ Program), KAIST, Daehak-ro 291, Yuseong-gu, Daejeon 305-701, Republic of Korea. E-mail: hgpark@kaist.ac.kr; Fax: +82-42-350-3910; Tel: +82-42-350-3932
bDepartment of Biological Engineering, College of Engineering, Konkuk University, Seoul 05029, Republic of Korea. E-mail: kskonkuk@gmail.com; Fax: +82-2-450-3742; Tel: +82-2-450-3742
First published on 29th November 2018
Abstract
We herein describe a novel and efficient method for the detection of protein/small molecule (SM) interactions, which relies on the protein-induced fluorescence enhancement (PIFE). In this method, a duplex probe is designed to position Cy3 and SM at the optimal distance to maximize the effect of PIFE, which is utilized as the key component. In the presence of target proteins that bind to SM, the Cy3 is guided close to the target proteins, which significantly enhances the fluorescence signal through a process of PIFE. With this approach, we successfully analyzed a model target protein, streptavidin (STV) that interacts with biotin (BTN) in less than 10 min without any washing steps. In addition, the practical applicability of this method was demonstrated by reliably determining STV in human serum. Finally, the universal applicability of this method was demonstrated by monitoring the interaction between folate and folate receptors.
Introduction
Research on the interaction between proteins and small molecules (SMs) has played important roles in unveiling mechanisms of various biological processes including the cell cycle, cell development and cell signaling.1–3 In addition, it has made significant progress in the field of drug discovery, molecular diagnostics, and therapeutics. For example, the investigation of the interaction between folate (FA) and folate receptor (FR) has contributed to the early diagnosis of cancers and the effective monitoring of the drug response.4–6 Until now, to monitor the interaction between proteins and small molecules, several analytical methods, such as affinity chromatography,7 kinetic capillary electrophoresis,8,9 electrochemistry10,11 and fluorometry,12–16 have been utilized, most of which rely on the terminal protection mechanism in which SM-linked DNA that binds to the target protein is protected from exonuclease-catalyzed degradation.17–20Notably, fluorescent methods have gained considerable attention due to their intrinsic advantages including simplicity, robustness, and high sensitivity. The representative example in this type of methods is based on Förster resonance energy transfer (FRET) that exhibits the nanoscale distance-dependent fluorescence properties.21–24 Although promising, these methods require target protein labeling, which could cause conformational changes of target protein. Moreover, the requirement of two different fluorophores and either exonuclease-catalyzed digestion or relevant processes to differentiate target protein-bound DNA probe from unbound one, increases the assay complexity, cost, and time (Table S1†). For these reasons, it is highly desirable to devise a simple but efficient detection method for protein/SM interactions.
Protein-induced fluorescence enhancement (PIFE) is a phenomenon in which several organic dyes, especially cyanine dyes, show the increased fluorescence when placed in close proximity to a protein.25,26 It is known that PIFE is more flexible than FRET in that PIFE employs only a single fluorophore and it exhibits the higher spatial resolution. Based on this phenomenon, the interaction of nucleic acids with proteins such as polymerase, helicases, and restriction enzymes has been monitored.27–29 For instance, Valuchova et al. applied PIFE in order to quantify the interaction of nucleic acids with restriction enzyme BamHI, and the DNA repair complexes Ku and XPF/ERCC1.27 In recent years, DNA aptamer has been employed in conjunction with PIFE to devise the sensitive detection methods.30
In this work, we adopted the unique features of PIFE for the detection of protein/SM interactions. As a key component, a duplex probe composed of Cy3 and SM-modified DNAs, respectively, is rationally designed to precisely control the distance between target protein and Cy3. In the presence of target proteins that bind to SM, Cy3 is placed close to target proteins, which leads to the significant fluorescence enhancement through a process of PIFE. With the developed strategy, we successfully detected two model interactions, streptavidin/biotin (STV/BTN) and FR/FA, verifying its general applicability. In addition, we confirmed that the proposed assay works fine even in human serum.
Experimental
Materials
All oligonucleotides used in this study were synthesized from Bioneer® (Daejeon, Korea) and purified by HPLC. The sequences of oligonucleotides are listed in Table S2.† Lysozyme, papain and human serum were purchased from Sigma-Aldrich (St Louis, MO, USA). STV and exonuclease I (Exo I) were purchased from Thermo Fisher Scientific (Waltham, MA, USA) and New England Biolabs Inc. (Beverly, MA, USA), respectively. All other chemicals were of analytical grade and used without further purification. Ultrapure DNase/RNase-free distilled water (DW) purchased from Bioneer® (Daejeon, Korea) was used throughout all the experiments.Procedure for the detection of STV/BTN and FR/FA interaction
The duplex probe (CS3/SSBTN; 5 μM) composed of CS3 (5 μM) and SSBTN (5 μM) was first prepared in a 1× protein/SM interaction buffer (20 mM Tris–HCl, 50 mM KCl, pH 8.4). Next, 21 μL of DW, 3 μL of CS3/SSBTN (5 μM), 3 μL of 10× protein/SM interaction buffer, and 3 μL of STV at varying concentrations or other proteins were mixed in a total volume of 30 μL, which were then incubated for 10 min at room temperature (RT). The final concentration of duplex probe was 500 nM. Finally, the resulting fluorescence signals were measured at an excitation wavelength of 530 nm. For the detection of FR/FA interaction, the duplex probe (CS3/SSFA; Table S2†) was used and subjected to the same procedure as described above.Gel electrophoresis analysis for the STV/BTN interaction
The electrophoresis analysis on the reaction products was carried out with a 15% polyacrylamide gel in 1× TBE for 100 min at 120 V. After staining with GelRed (KOMA BIOTECH, Seoul, Korea), a gel image was acquired with Gel Doc™ EZ manager and Image Lab software (Bio-Rad, Hercules, USA).31,32Determination of STV in human serum
Varying concentrations of STV were spiked into undiluted human serum. Each sample was then analyzed by following the same procedure to detect the STV/BTN interaction as described above. The final concentration of human serum in the reaction mixture is 10%.Instrumentation
Fluorescence intensities were measured using a Tecan Infinite M200 pro-microplate reader (Mnnedorf, Switzerland) and black, 384-well Greiner Bio-One microplates (ref: 781077, Courtaboeuf, France).Results and discussion
Principle for the PIFE-based detection of protein/SM interactions
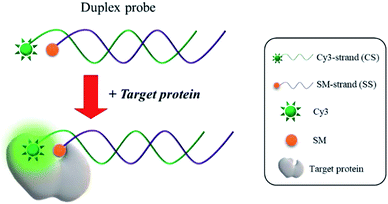
The fundamental principle of a new strategy for the detection of protein/SM interactions is illustrated in Scheme 1. The duplex probe composed of a 3′-Cy3-modified DNA (CS3) and a 5′-SM-modified complement (SSSM) is rationally designed to have the optimal distance between Cy3 and SM to maximize the effect of PIFE, which is utilized as a key component in the proposed detection system. In the presence of target protein that binds to SM, Cy3 in the duplex probe is guided close to proteins, which significantly enhances the fluorescence of Cy3 via a process of PIFE. Based on the fluorescence signal-on response, the protein/SM interaction can be detected in a single step without any washing steps.Feasibility for the PIFE-based detection of protein/SM interactions
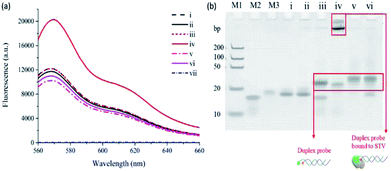
As a model target, STV/BTN interaction, which exhibits the exceptionally high binding affinity (Kd = 10−15 M),23 was selected and the detection feasibility of the proposed method was demonstrated by measuring fluorescence emission spectra. As shown in Fig. 1(a), CS3 (i) incubated with either STV (ii) or SSBTN (iii) produced the negligible fluorescence enhancement. On the other hand, the addition of STV to the duplex probe composed of CS3 and SSBTN significantly increased the fluorescence signal (iv) through a process of the PIFE. We also prepared the control duplex probe composed of CS3 and a 5′-FA-modified DNA (SSFA) instead of SSBTN (Table S2†). As envisioned, PIFE was not observed in the presence of STV since the FA does not bind to STV (v and vi in Fig. 1(a) and (b)). Importantly, STV itself gave no fluorescence (vii) and did not have any interfering effect on the fluorescence of CS3 (i, ii, v and vi), confirming that this system is not directly interrupted by STV itself. In addition, the fluorescence results were supported by the polyacrylamide gel electrophoresis, which demonstrated the formation of duplex probe and the complexation of BTN with STV (Fig. 1(b)). Overall, these results confirm that the target protein binds to SM and thus induces the significant increase of fluorescence signal by PIFE, ensuring the detection feasibility of this strategy.Optimization of reaction conditions
We optimized the reaction conditions required for the efficient analysis of STV/BTN interaction by examining the degree of fluorescence increase ((F − F0)/F0) under different conditions. First, the distance (d) between BTN and Cy3 in the duplex probe was varied by adjusting d values to find the optimal, spatial proximity for the effective PIFE. As shown in Fig. S1,† the most significant fluorescence increase was observed at d = −3, which matched well with the previous reports.33 Next, the incubation time and temperature for STV/BTN interaction were also optimized. The results in Fig. S2† show that the developed assay is not very sensitive to the incubation time and temperature, and thus 10 min and room temperature were selected for the further experiments.Detection sensitivity and selectivity
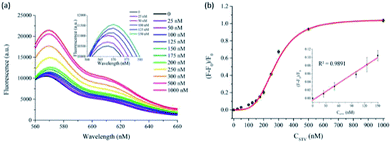
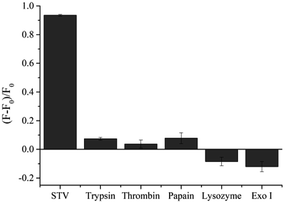
Under the optimized conditions, the analytical sensitivity of the present system was determined by measuring the fluorescence intensities at 570 nm (F570), an emission maximum of Cy3, as a function of STV concentration. As shown in Fig. 2, the fluorescence intensities increased with increasing concentrations of STV and an excellent linear relationship (R2 = 0.9891) was established in the range from 0 to 150 nM by the linear equation, (F − F0)/F0 = 0.0007 × CSTV/nM − 0.0045, where (F − F0)/F0 is the degree of fluorescence increase and CSTV is the concentration of STV. Based on the definition of the limit of detection (LOD),34 3σ/S, where σ and S are the standard deviation of blank sample and the slope of the linear relationship, respectively, LOD was calculated to be 2.93 nM, which is comparable or higher than those from previous fluorescent methods (Table S1†). It should be noted that the developed methods do not require the target protein labeling, organic synthesis of probes, and the treatment of additional enzyme reactions (e.g., exonuclease), which significantly decreases the assay time and simplifies the assay procedure. In addition, the proposed approach can be universally applied to the detection of other protein/SM interactions by simply changing the SM that matches the target proteins, which was demonstrated by the detection of FR/FA interactions (Fig. S3†). The selectivity of this method was then evaluated by testing other proteins such as trypsin, thrombin, papain, lysozyme, and exonuclease I. The results in Fig. 3 show that STV only leads to the significant fluorescence enhancement while no noticeable fluorescence changes are observed in the presence of the other proteins. These results prove that this system is highly selective to STV, indicating that PIFE originates from the specific binding of STV with BTN that places Cy3 in close proximity to STV.Practical applicability
Finally, the practical applicability of the developed system was verified by determining STV in human serum. As shown in Fig. S4,† an excellent linear relationship is made between (F − F0)/F0 and CSTV in human serum. Based on this calibration curve, we determined the concentrations of STV in human serum with high reproducibility and precision, as evidenced by the coefficients of variation (CV) (3.45–9.39%) and the recovery rates (95.17–103.35%) (Table 1). These results suggest that the proposed method would enable the reliable determination of STV in real biological samples containing a number of interfering agents.| Added STV (nM) | Measured STVb (nM) | SDc | CVd (%) | Recoverye (%) |
|---|---|---|---|---|
a To measure the concentration of STV in human serum, a calibration curve was first created by using standards containing a known concentration of STV spiked in human serum. Based on this calibration curve, the concentration of STV in human serum was determined by measuring the fluorescence at 570 nm from unknown samples.b Mean of three measurements.c Standard deviation of three measurements.d 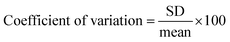 .e .e 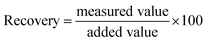 . . |
||||
| 50 | 47.58 | 2.74 | 5.76 | 95.17 |
| 150 | 155.03 | 5.35 | 3.45 | 103.36 |
| 250 | 248.54 | 23.35 | 9.39 | 99.42 |
Conclusions
In this work, we developed a simple and efficient strategy for the detection of protein/SM interactions based on the PIFE. The developed system employes a duplex probe rationally designed to maximize the effect of PIFE in response to the target protein. With this approach, we detected a model interaction, STV/BTN in less than 10 min. In addition, its general and practical applicability were verified by analyzing FR/FA interactions and reliably determining STV in human serum. Importantly, this method does not require the organic synthesis of probes, labeling of target proteins, and additional enzyme treatment, which overcomes the drawbacks such as complexity, high-cost, and long analysis time in the previous ones. To the best of our knowledge, this is the first attempt to adopt the novel phenomenon of PIFE for the analysis of protein/SM interactions, which would pave the way for the simple and efficient detection of various biological entities such as small molecules, proteins, and cells.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Bio-Synergy Research Project (NRF-2015M3A9C4070484) and Advanced Biomass R&D Center (ABC) of Global Frontier Project (ABC-2010-0029728) of the Ministry of Science, ICT and Future Planning through the National Research Foundation.Notes and references
- L. R. Coney, A. Tomassetti, L. Carayannopoulos, V. Frasca, B. a. Kamen, M. I. Colnaghi and V. R. Zurawski, Cancer Res., 1991, 51, 6125–6132 CAS.
- N. Ohoka, K. Nagai, T. Hattori, K. Okuhira, N. Shibata, N. Cho and M. Naito, Cell Death Dis., 2014, 5, e1513–10 CrossRef CAS PubMed.
- D. Leung, C. Hardouin, D. L. Boger and B. F. Cravatt, Nat. Biotechnol., 2003, 21, 687–691 CrossRef CAS PubMed.
- G. Toffoli, C. Cernigoi, A. Russo, A. Gallo, M. Bagnoli and M. Boiocchi, Int. J. Cancer, 1997, 74, 193–198 CrossRef CAS PubMed.
- P. Y. Li, S. Del Vecchio and V. Maria, J. Nucl. Med., 1996, 37, 665–672 CAS.
- P. Garin-Chesa, I. Campbell, P. E. Saigo, J. L. Lewis, L. J. Old and W. J. Rettig, Am. J. Pathol., 1993, 142, 557–567 CAS.
- J. Taumton, C. A. Hassig and S. L. Schreiber, Science, 1996, 272, 408–411 CrossRef.
- A. Petrov, V. Okhonin, M. Berezovski and S. N. Krylov, J. Am. Chem. Soc., 2005, 127, 17104–17110 CrossRef CAS PubMed.
- A. P. Drabovich, M. V. Berezovski, M. U. Musheev and S. N. Krylov, Anal. Chem., 2009, 81, 490–494 CrossRef CAS PubMed.
- Z. Wu, Z. Zhen, J. H. Jiang, G. L. Shen and R. Q. Yu, J. Am. Chem. Soc., 2009, 131, 12325–12332 CrossRef CAS PubMed.
- G. Wang, X. He, L. Wang and X. Zhang, Biosens. Bioelectron., 2013, 42, 337–341 CrossRef CAS PubMed.
- H. B. Wang, H. D. Zhang, Y. Chen and Y. M. Liu, Biosens. Bioelectron., 2015, 74, 581–586 CrossRef CAS PubMed.
- H. Shi, X. Mao, X. Chen, Z. Wang, K. Wang and X. Zhu, Biosens. Bioelectron., 2017, 91, 136–142 CrossRef CAS PubMed.
- X. Zhou, Z. Cui, L. Liu, Z. Sun, M. Lin, Q. Hu, H. Wang and X. Xiao, Analyst, 2018, 143, 2755–2759 RSC.
- C. Y. Lee, H. Y. Kim, S. Kim, K. S. Park and H. G. Park, Analyst, 2018, 143, 2023–2028 RSC.
- Z. Zhang, C. Hejesen, M. B. Kjelstrup, V. Birkedal and K. V. Gothelf, J. Am. Chem. Soc., 2014, 136, 11115–11120 CrossRef CAS PubMed.
- X. Li, X. Ding, Y. Li, L. Wang and J. Fan, Nanoscale, 2016, 8, 9852–9860 RSC.
- G. Zhou, X. Zhang, X. Ji and Z. He, Chem. Commun., 2013, 49, 8854 RSC.
- Y. He and B. Jiao, Microchim. Acta, 2016, 3183–3189 CrossRef.
- J. Chen, C. Gao, A. K. Mallik and H. Qiu, J. Mater. Chem. B, 2016, 4, 5161–5166 RSC.
- Q. Li and S. Seeger, J. Phys. Chem. B, 2011, 115, 13643–13649 CrossRef CAS PubMed.
- N. Lecat-Guillet, C. Monnier, X. Rovira, J. Kniazeff, L. Lamarque, J. M. Zwier, E. Trinquet, J. P. Pin and P. Rondard, Cell Chem. Biol., 2017, 24, 360–370 CrossRef CAS PubMed.
- D. Tu, L. Liu, Q. Ju, Y. Liu, H. Zhu, R. Li and X. Chen, Angew. Chem., Int. Ed., 2011, 50, 6306–6310 CrossRef CAS PubMed.
- S. Zhang, J. Wang, W. Xu, B. Chen, W. Yu, L. Xu and H. Song, J. Lumin., 2014, 147, 278–283 CrossRef CAS.
- E. M. S. Stennett, M. A. Ciuba, S. Lin and M. Levitus, J. Phys. Chem. Lett., 2015, 6, 1819–1823 CrossRef CAS PubMed.
- G. Luo, M. Wang, W. H. Konigsberg and X. S. Xie, Proc. Natl. Acad. Sci., 2007, 104, 12610–12615 CrossRef CAS PubMed.
- S. Valuchova, J. Fulnecek, A. P. Petrov, K. Tripsianes and K. Riha, Sci. Rep., 2016, 6, 1–10 CrossRef PubMed.
- E. Ploetz, E. Lerner, F. Husada, M. Roelfs, S. Chung, J. Hohlbein, S. Weiss and T. Cordes, Sci. Rep., 2016, 6, 1–24 CrossRef PubMed.
- H. Hwang, H. Kim and S. Myong, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 7414–7418 CrossRef CAS PubMed.
- S. Umrao, V. Jain, Anusha, B. Chakraborty and R. Roy, Sens. Actuators, B, 2018, 267, 294–301 CrossRef CAS.
- C. Y. Lee, K. S. Park and H. G. Park, Chem. Commun., 2015, 51, 13744–13747 RSC.
- K. S. Park, C. Y. Lee, K. S. Kang and H. G. Park, Biosens. Bioelectron., 2017, 88, 48–54 CrossRef CAS PubMed.
- G. Li, Y. Liu, Y. Liu, L. Chen, S. Wu, Y. Liu and X. Li, Angew. Chem., Int. Ed., 2013, 52, 9544–9549 CrossRef CAS PubMed.
- K. S. Park, M. Il Kim, M. A. Woo and H. G. Park, Biosens. Bioelectron., 2013, 45, 65–69 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra08515c |
| This journal is © The Royal Society of Chemistry 2018 |