Open Access Article
Open Access ArticleControllable extension of hairpin-structured flaps to allow low-background cascade invasive reaction for a sensitive DNA logic sensor for mutation detection†
Yun-Long
Liu‡
ac,
Hai-Ping
Wu‡
b,
Qiang
Zhou
d,
Qin-Xin
Song
d,
Jian-Zhong
Rui
a,
Xiao-Xiang
Guan
a,
Guo-Hua
Zhou
 *a and
Bing-Jie
Zou
*a
*a and
Bing-Jie
Zou
*a
aDepartment of Pharmacology, Jinling Hospital, Medical School of Nanjing University, Nanjing 210002, China. E-mail: ghzhou@nju.edu.cn; zbj523@163.com
bHuadong Research Institute for Medicine and Biotechnics, Nanjing 210002, China
cState Key Laboratory of Natural Medicines, Department of Biomedical Engineering, School of Engineering, China Pharmaceutical University, Nanjing 210009, China
dKey Laboratory of Drug Quality Control and Pharmacovigilance of Ministry of Education, China Pharmaceutical University, Nanjing 210009, China
First published on 14th December 2017
Abstract
A DNA logic sensor was constructed for gene mutation analysis based on a novel signal amplification cascade by controllably extending a hairpin-structured flap to bridge two invasive reactions. The detection limit was as low as 0.07 fM, and the analytical specificity is high enough to unambiguously pick up 0.02% mutants from a large amount of wild-type DNA. Gene mutations related to the personalized medicine of gefitinib, a typical tyrosine kinase inhibitor, were analyzed by the DNA logic sensor with only a 15 minute response time. Successful assay of tissue samples and cell-free plasma DNA indicates that the new concept we proposed here could benefit clinicians for straightforward prescription of a mutation-targeted drug.
Introduction
Gene mutations play important roles in personalized medicine.1 An increasing number of gene mutations have been found to be associated with therapeutic effects or drug resistance.2–4 For example, the clinical response of gefitinib (a tyrosine kinase inhibitor, TKI) was highly correlated with the somatic mutation status of the epidermal growth factor receptor (EGFR) gene.5EGFR L858R mutation or deletions in exon 19 confer sensitivity to gefitinib;6 in contrast, T790M mutation in EGFR would cause drug resistance.7,8 In personalized medicine, it is preferable for a method to directly give a final decision to a doctor as to which drug should be used rather than which mutation exists in the sample.The DNA logic gate is a newly developed technology for DNA computing.9–13 The main merit is to give a simple output with multiple inputs. In order to simplify the process, we tried to use the concept of the DNA logic gate to construct a sensor to deal with the relationship of multiple mutations. Different gene mutations can be employed as various inputs of the logic gate, and the output arising from the detection result of these mutations is the therapeutic regimen of a drug. Currently, there are many well-designed DNA logic gates based on different strategies including DNA hybridization reaction,14–20 functional DNA structures,21,22 or enzyme-catalyzed reaction.23–25 However, no data have shown that DNA logic gates based on the hybridization reaction and functional DNA structures could accurately sense a single-base mismatch in a target of interest. Although the enzyme-interfaced DNA logic gate achieved single-base mismatch detection,25 a high background from the mismatched target indicated that the specificity of this type of DNA logic gate is not enough to pick up ultra-low levels of personalized medicine-related somatic mutations from a large amount of wild-type DNA. In addition, with this enzyme-interfaced DNA logic gate, it is very difficult to detect mutations out of the enzyme-recognition sequence. In order to demonstrate our assumption, the DNA logic sensor must be sufficiently sensitive and specific to pick up a small amount of single-base mutants from a large amount of genomic DNA backgrounds.
Invasive reaction, which is catalyzed by flap endonuclease 1 (FEN1), can specifically recognize a one-base overlapping structure formed by two probes hybridizing to adjacent sequences in a target DNA, and trigger the cleavage of the 5′ flap of the downstream probe.26–29 The serial invasive reaction can achieve single-base difference recognition and signal amplification up to 107 fold, and is an ideal method for DNA mutation detection.30–32 However, we observed that the reported non-specific substrate (called the “X”-structure substrate) formed between the downstream probe and fluorescence resonance energy transfer (FRET) probe generates a time-dependent background signal in the cascade invasive reaction, leading to a false positive result.33,34
Here, we tried to integrate the invasive reaction and the concept of the DNA logic gate to construct a DNA logic sensor for multiple mutation analysis. To solve the issue of the “X”-structure substrate-caused background signal, we proposed a low-background invasive reaction cascade using a hairpin-structured downstream probe coupled with the controllable-extension reaction (Fig. 1). To block the “X”-structure between the downstream probe and the FRET probe, we proposed to introduce a gap sequence in the downstream probe. However, the gap would also block the formation of an overlapping structure between the cleaved flap and the FRET probe. To solve this issue, a downstream probe is designed to contain a hairpin structure (see Fig. 1 for details). In contrast to the intact downstream probe, the cleaved flap has an extendable 3′ end, hence the gap could be filled in by the polymerase-catalyzed extension reaction. To construct a one-base overlapping structure between the extended flap and FRET probe, the gap sequence was designed to contain only three kinds of base (A, G, and T), and the extension reaction of the cleaved flaps would stop at base C (artificially designed in the flap) as only three kinds of complementary dNTP (without dGTP) were added in the reaction. The controllable-extension product triggers the second invasive reaction to generate a fluorescence signal. The formation of the “X”-structure substrate is thereby efficiently suppressed by this design, and the background signal is expected to be very low.
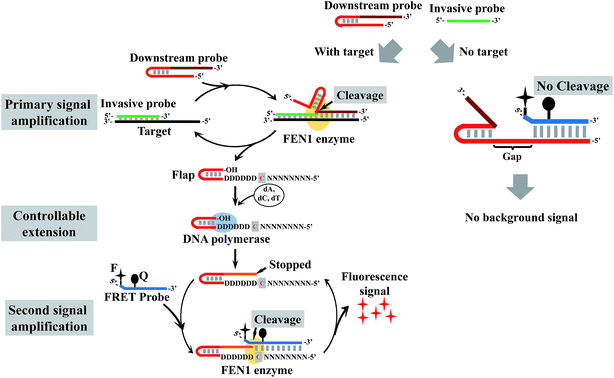 | ||
| Fig. 1 Schematic illustration of controllable-extension bridged cascade signal amplification with a hairpin downstream probe. D = A, G, or T; N = A, C, G, or T. | ||
This novel signal amplification cascade with an ultra-low background allowed us to construct sensitive DNA logic sensors for gene mutation detection, including an OR gate and an INHIBIT gate. Four DNA mutations (four inputs) in the EGFR gene were simultaneously detected in a single tube by the controllable-extension bridged cascade invasive reaction, and the output is the therapeutic regimen of gefitinib for a given patient. We have successfully applied this method to the assay of various specimens, such as tissue DNA and cell-free DNA (cfDNA).
Results and discussion
Introduction of a gap sequence into the downstream probe to suppress the formation of the “X”-structure substrate
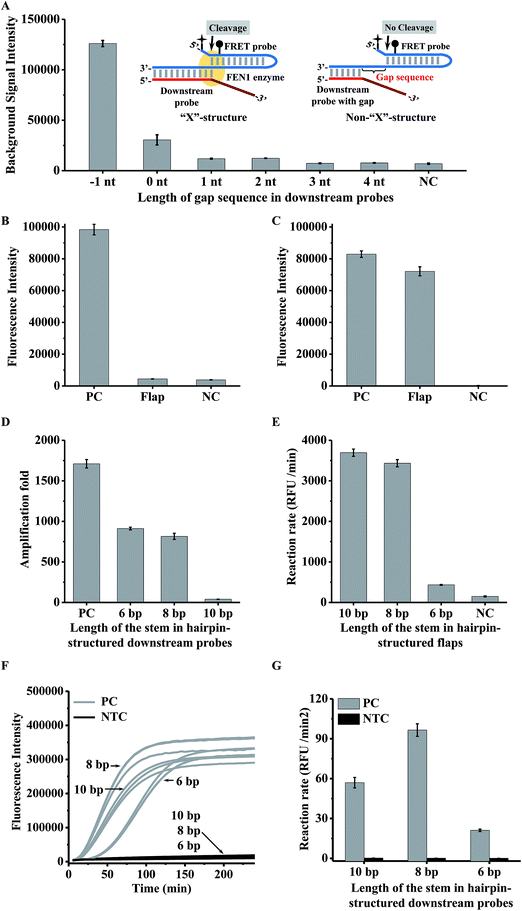
In a conventional cascade invasive reaction, the main problem is the high background signal from non-specific cleavage of the FRET probe due to the “X”-structure substrate formed by an intact downstream probe with the FRET probe. To block the formation of this “X”-structure substrate, we proposed to introduce a gap sequence into the heterodimer between a downstream probe and a FRET probe (Fig. 2A). The background signal from a series of downstream probes with different numbers of gap bases (from 0 to 4 nt) is shown in Fig. 2A. It was found that the background signal decreased with an enlarged gap sequence. When the gap length is over 3 nt, the background signal is close to the signal from the control without a downstream probe (NC in Fig. 2A). Therefore, the gap artificially inserted in the downstream probe is an effective way to suppress the background signal from the “X”-structure substrate in the cascade invasive reaction.Use of a hairpin structure as the flap sequence in the downstream probe to increase the extension efficiency
As the gap sequences also block the formation of the one-base-overlapping structure between the cleaved flaps and the FRET probe, we should extend the cleaved flaps to allow the invasive reaction to occur. However, it was found that the efficiency of the extension reaction is very low for a conventional single-stranded flap (Fig. S1† & 2B). Although the extension efficiency could be improved by decreasing the annealing temperature (25 °C) , it is still insufficient to bridge the two invasive reactions effectively (Fig. S2†). To increase the efficiency of the polymerase-catalyzed extension reaction, we proposed to design a hairpin-structured flap sequence to replace the conventional flap (Fig. 1). As shown in Fig. 2C, the efficiency of the extension reaction increased significantly although it is about 10% lower than that from the positive control (artificially synthesized extended flaps). One important thing we should point out is that the background signal from the proposed single-stranded FRET probe (negative control in Fig. 2C) is much lower than that from the conventional hairpin FRET probe (negative control in Fig. 2B), indicating that the non-specific cleavage of the hairpin FRET probe is also greatly suppressed in the proposed FRET probe.To check whether or not the gap sequences introduced in the hairpin-structured downstream probe could efficiently avoid the formation of the “X”-structure substrate, we used the intact length of the proposed hairpin-structured downstream probes with and without a gap sequence for the invasive reaction (Fig. S3A†). As shown in Fig. S3B,† the background signal from the hairpin-structured downstream probe with a gap sequence dramatically decreased to a level close to that from a negative control (without any downstream probe).
Optimization of DNA polymerase for the extension-based secondary invasive reaction
The hairpin probe extension is a bridge of the two invasive reactions, thus the extension efficiency is very important for sensitive detection. Therefore, we should choose an optimal DNA polymerase to extend the hairpin probe. On the other hand, FEN1 may compete with DNA polymerase to bind the same substrate hairpin probe, hence the concentration of DNA polymerase should also be optimized. We tested two kinds of DNA polymerase (rTaq polymerase and KlenTaq polymerase) with different concentrations for the extension-based secondary invasive reaction. As shown in Fig. S4,† the fluorescence intensities of the flap extended with rTaq polymerase were lower than those of the flap extended with KlenTaq polymerase, indicating that the extension efficiency of KlenTaq polymerase is higher than that of rTaq polymerase. Therefore, KlenTaq polymerase is preferable in our proposed assay. On the other hand, the fluorescence intensities of the artificially synthesized extended hairpin-structured flaps (Flap Ex in Fig. S4†) from the reactions containing rTaq polymerase and KlenTaq polymerase decreased with increasing amounts of polymerases, indicating that higher amounts of polymerase would affect the efficiency of the invasive reaction. The optimal amount of KlenTaq polymerase was 0.005 U.Optimization of the stem length in the hairpin region for downstream probes
The use of the hairpin structure in the downstream probe improved the extension reaction efficiency. However, it is not clear whether the primary invasive reaction could efficiently occur, because the hairpin structure in the 5′ terminal region of the flap may affect FEN1 enzyme recognition.26,35 On the other hand, the stem length of the hairpin region may affect the extension efficiency. Therefore we tried to find an optimal stem length of the hairpin for the downstream probe. Three types of downstream probe with a stem length of 10 bp, 8 bp, and 6 bp were individually employed for the primary invasive reaction, second invasive reaction triggered by the controllable-extension reaction, and controllable-extension bridged cascade invasive reaction. The amplification fold of the primary reaction was defined as the concentration ratio of the target DNA and cleaved flaps. To obtain the concentrations of the cleaved flaps, dilution of the primary invasive reaction products was employed to perform the secondary invasive reaction. The concentration of the cleaved flaps can be calculated according to a standard curve, which was plotted with different concentrations of synthesized flaps and the corresponding reaction rates by carrying out the secondary invasive reaction (Fig. S5†).As shown in Fig. 2, the hairpin structure in the downstream probe did affect the signal amplification capability. The increase in the stem length caused a decrease in the amplification folds of the primary invasive reaction (Fig. 2D) but an increase in the efficiency of the second invasive reaction triggered by the controllable-extension reaction (Fig. 2E). The controllable-extension bridged cascade invasive reaction indicated that the initial reaction rate of a downstream probe with an 8 bp stem length was the highest among the three downstream probes (Fig. 2F & G). Therefore, the downstream probe with an 8 bp stem length in the hairpin structure was selected for the controllable-extension bridged cascade invasive reaction.
Analytical sensitivity and specificity of the controllable-extension bridged cascade invasive reaction
To investigate the detection limit of the assay, a series of artificially synthesized target DNAs with different concentrations (5 pM to 1 fM) were detected by the controllable-extension bridged cascade invasive reaction. As shown in Fig. 3A, the signal from the 1 fM target could be clearly distinguished from the negative control (NTC in Fig. 3A). The detection limit (taken to be 3 times the standard deviation in NTC) was 0.07 fM, which was about 40 fold lower than that of the conventional cascade invasive reaction (the detection limit was 2.86 fM, as shown in Fig. S6A & B†). A good linear relationship was obtained between the initial reaction rates and target concentrations (Fig. 3B).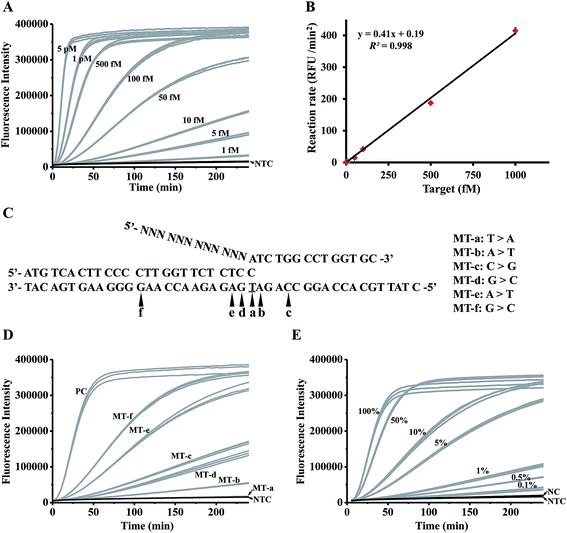 | ||
| Fig. 3 Analytical sensitivity and specificity of the controllable-extension bridged cascade invasive reaction for DNA detection. (A) The time-course curves for detecting targets with various concentrations. (B) The linear relationship between the target concentrations and the initial reaction rates. (C) Sequences of targets with single-base mutation at different positions. (D) The time-course curves for detecting the targets in panel (C). (E) The time-course curves for detecting a series of samples with different fractions of single-base mutants (the target in Table S3†) in wild-type targets (MT-a in Table S3†). PC: positive control, the reaction with 1 pM complementary targets. NC: negative control, the reaction with 1 pM wild-type targets. NTC: no target control, the reaction without a target. N = 3. | ||
In order to investigate the specificity of the assay, a set of synthesized targets with single base variation at different positions (Fig. 3C) were detected by the controllable-extension bridged cascade invasive reaction. The target with a single mismatched base in the invasive site (MT-a) gave a negative signal (MT-a in Fig. 3D). Other targets gave signals with different intensities (MT-b to MT-f in Fig. 3D), but these were lower than that of the positive control (a perfectly complementary target). As the specificity of the invasive reaction depends on the formation of an invasive structure recognized by FEN1, the lack of signal from MT-a in Fig. 3 is due to there being no formation of an invasive structure, but the appearance of signals from MT-b to MT-f is because of the presence of different amounts of invasive structure. A mismatched base in the target (MT-f and MT-e) weakened the formation of an invasive structure, but still yielded a large amount of the invasive structure, giving high signals. So it is necessary to assign the mismatched base of interest just at the position of the overlapping structure.
The specificity of the assay was further investigated through the detection of a series of samples prepared by spiking various amounts (100%, 50%, 10%, 5%, 1%, 0.5%, and 0.1%) of single-base mutated targets (the target in Table S3†) into wild-type targets (MT-a in Table S3†). As shown in Fig. 3E, samples spiked with 0.1% mutant gave recognizable signals; therefore, the proposed method is sensitive to pick up as low as 0.02% mutants (taken to be 3 times the standard deviation in NC) from a large amount of wild-type background. A good linear relationship between the mutant fractions and the initial reaction rates (Fig. S7†) suggested that our proposed method is quantitative to detect the mutants in a sample. To further investigate the feasibility of our method, two different types of mutation (EGFR L858R, c.2573T>G in exon 21 and T790M, c.2369C>T in exon 20) with various mutation fractions were tested (Fig. S8†). Similarly, 0.1% mutation fractions were accurately detected, indicating that our system is applicable to detect different types of mutation with a level as low as 0.1%.
Construction of logic gates by the proposed method
With the benefits of the low background and high sensitivity of the controllable-extension-based cascade invasive reaction, we tried to construct DNA logic gates, such as AND, OR, and INHIBIT gates.As the present assay needs multiple components to finish the whole reactions, in principle we can construct an AND gate using each of the components necessary to invasive reactions as an input. Here, a target DNA and the invasive probe were employed as inputs (inputs 1 and 2 in Fig. S9A†). As shown in Fig. S9B,† when the target DNA (input 1) and the invasive probe (input 2) were simultaneously present, the cascade invasive reaction occurs by the controllable-extension reaction, giving a high fluorescence signal, defined as the “1” output. However, neither the target DNA nor the invasive probe could trigger the cascade reaction, showing a background fluorescence, defined as the “0” output. A truth table of this AND gate is shown in Fig. S9C.† We can get output after only 15 minutes since the inputs. The output is straightforward due to the high signal-to-noise ratio (60–120).
As the primary invasive reaction generates flaps independent of the target sequence, it is very convenient to construct an OR gate by our proposed method. The OR gate could be readily achieved by employing the two different DNA targets as inputs (inputs 1 and 3 in Fig. S10A†). The key to the gate is to design two downstream probes with an identical flap sequence for sensing the two DNA targets. The two targets would produce the same flaps to trigger the subsequent invasive reaction. As shown in Fig. S10B,† when either or both of the two inputs were present, high fluorescence signals appeared leading to the “ON” state; on the other hand, no signal appeared in the absence of both DNA targets, leading to the “OFF” state. A truth table of this OR gate with two inputs is shown in Fig. S10C.† As the flap sequence was independent of the target DNA, basically there is no limitation for the number of inputs of this OR gate.
The key to our proposed method is the controllable extension of the flaps, and full extension of the flaps would not trigger the cascade invasive reaction. So the component causing full extension could be used as the input of an INHIBIT gate. As the controllable extension of the flaps was achieved by adding only three of the dNTPs (dATP, C, and T), dGTP would cause full extension of the flaps. Consequently, the target DNA and dGTP could be the inputs of the INHIBIT gate (input 1 and input 4 in Fig. S11A†). As shown in Fig. S11B,† when the target DNA (input 1) was present in the reaction, the cascade invasive reaction would be triggered, producing a high fluorescence signal (“1” output). On the other hand, when both the target DNA (input 1) and dGTP (input 4) were present, no signal was obtained, resulting in the “0” output. A truth table of this INHIBIT gate is shown in Fig. S11C.†
Construction of a DNA logic sensor for mutation detection
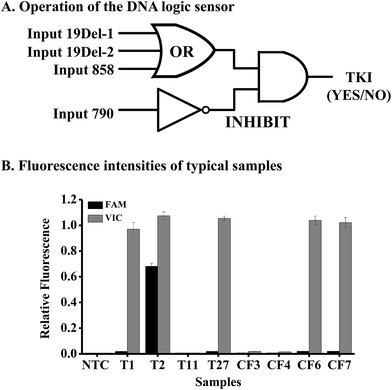
To achieve personalized medicine based on mutation detection, we tried to use the above method to design various DNA logic gates for constructing a DNA logic sensor to detect different mutations simultaneously.Mutations in the EGFR gene were taken as an example for the study. As the mutations of exon 19 deletion and L858R are responsible for the sensitivity of gefitinib, we should design an OR gate for these mutations. We should also design an INHIBIT logic gate for the mutation T790M in the EGFR gene, because this mutation causes resistance to the drug. Hence the DNA logic sensor should contain an OR logic gate and an INHIBIT logic gate (Fig. 4A). The inputs of the OR logic gate are mutations of 19Del-1 (c.2235-2249del15 in exon 19), 19Del-2 (c.2236-2250del15 in exon 19), and L858R (c.2573T>G in exon 21). We can add other mutations to the gate if necessary. In the absence of all of these three targets, no signal was obtained, and the logic gate was in the “OFF” state. However, when any of the three inputs was present alone or together, a high fluorescence signal appears, leading to the “ON” state.
The inputs of the INHIBIT logic gate are the output of the OR logic gate and T790M (c.2369C>T in exon 20). The OR gate produces a VIC fluorescence signal, defined as “1” when the fluorescence signal is positive and as “0” when the fluorescence signal is negative. The input 790 generates the FAM fluorescence signal (“1” for positive and “0” for negative). The output of the logic sensor is the difference of VIC and FAM (VIC-FAM), and value “1” means the “ON” state to give a positive result (a recommendation for the prescription of gefitinib) and “0” means the “OFF” state to give a negative result (no recommendation for the prescription of gefitinib).
To evaluate the feasibility of the proposed logic sensor, 30 tissue samples and 7 plasma cfDNA samples from patients suffering from lung cancer were analyzed (Table S5†). Results from 8 typical samples are shown in Fig. 4B, indicating that both the FAM and VIC fluorescence signals were negative for samples #T11, #CF3, and #CF4, hence there was no recommendation for the use of gefitinib. On the other hand, only the VIC fluorescence signal was positive for samples #T1, #T27, #CF6, and #CF7, giving the “1” outputs of the logic gate, hence these patients were sensitive to gefitinib. As to sample #T2, both the FAM and VIC fluorescence signals were positive, giving the “0” output of the logic gate based on FAM-VIC, hence the patient would not benefit from gefitinib. The results were verified by ARMS-PCR for tissue samples and by NGS for cfDNA samples, respectively.
Conclusions
To construct a sensitive DNA logic sensor for mutation detection, we improved the conventional cascade invasive reaction by employing a hairpin-structured downstream probe coupled with the controllable-extension reaction. The non-specific “X”-structure substrate between the downstream probe and the FRET probe was efficiently blocked by introducing a gap sequence in the downstream probe. The gap sequence was artificially designed so that the extension reaction of the cleaved flaps stopped just at the position to form an invasive structure for the subsequent invasive reaction. The background signal was significantly decreased to allow a detection limit of as low as 0.07 fM. The analytical specificity is high enough to unambiguously pick up 0.02% mutants from a large amount of wild-type DNA background (e.g., genomic DNA). This low-background cascade invasive reaction was thus used to construct various DNA logic gates, such as AND, INHIBIT, and OR gates, giving a response time of 15 min for 10 pM target DNA as inputs, much faster than most of the conventional DNA logic gates. Based on the mutations related to the sensitivity and resistance of the drug gefitinib, a DNA logic sensor was constructed for personalized medicine. Although the present study is preliminary, we believe that the new concept we proposed here should be a cost-effective tool for a doctor to make a straightforward prescription of a mutation-targeted drug for a specific patient.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (21475151, 21405176, 81673390, and 31200638), the State Key Basic Research Program of the PRC (2014CB744501 and 2016YFA0201204), Jiangsu Provincial Key Research and Development Program (No. BE2016745), Jiangsu Provincial Natural Science Foundation (No. BK20151445), Six Talent Peaks Project in Jiangsu Province (2015-WSN-085), and Jiangsu Provincial Medical Youth Talent program (No. QNRC2016889 and No. QNRC2016845).Notes and references
- L. Chin, J. N. Andersen and P. A. Futreal, Nature Medicine, 2011, 17, 297–303 CrossRef CAS PubMed.
- J. A. Ludwig and J. N. Weinstein, Nat. Rev. Cancer, 2005, 5, 845–856 CrossRef CAS PubMed.
- H. Schwarzenbach, D. S. Hoon and K. Pantel, Nat. Rev. Cancer, 2011, 11, 426–437 CrossRef CAS PubMed.
- R. Rosell, T. G. Bivona and N. Karachaliou, Lancet, 2013, 382, 720–731 CrossRef CAS.
- J. G. Paez, P. A. Janne, J. C. Lee, S. Tracy, H. Greulich, S. Gabriel, P. Herman, F. J. Kaye, N. Lindeman, T. J. Boggon, K. Naoki, H. Sasaki, Y. Fujii, M. J. Eck, W. R. Sellers, B. E. Johnson and M. Meyerson, Science, 2004, 304, 1497–1500 CrossRef CAS PubMed.
- M. Maemondo, A. Inoue, K. Kobayashi, S. Sugawara, S. Oizumi, H. Isobe, A. Gemma, M. Harada, H. Yoshizawa, I. Kinoshita, Y. Fujita, S. Okinaga, H. Hirano, K. Yoshimori, T. Harada, T. Ogura, M. Ando, H. Miyazawa, T. Tanaka, Y. Saijo, K. Hagiwara, S. Morita, T. Nukiwa and G. North-East Japan Study, N. Engl. J. Med., 2010, 362, 2380–2388 CrossRef CAS PubMed.
- S. Kobayashi, T. J. Boggon, T. Dayaram, P. A. Janne, O. Kocher, M. Meyerson, B. E. Johnson, M. J. Eck, D. G. Tenen and B. Halmos, N. Engl. J. Med., 2005, 352, 786–792 CrossRef CAS PubMed.
- K. Uchibori, N. Inase, M. Araki, M. Kamada, S. Sato, Y. Okuno, N. Fujita and R. Katayama, Nat. Commun., 2017, 8, 14768 CrossRef CAS PubMed.
- E. Katz and V. Privman, Chem. Soc. Rev., 2010, 39, 1835–1857 RSC.
- J. H. Reif, Science, 2011, 332, 1156–1157 CrossRef CAS PubMed.
- G. Chatterjee, N. Dalchau, R. A. Muscat, A. Phillips and G. Seelig, Nat. Nanotechnol., 2017, 12, 920–927 CrossRef CAS PubMed.
- D. Fan, E. Wang and S. Dong, Mater. Horiz., 2017, 4, 924–931 RSC.
- A. J. Thubagere, C. Thachuk, J. Berleant, R. F. Johnson, D. A. Ardelean, K. M. Cherry and L. Qian, Nat. Commun., 2017, 8, 14373 CrossRef CAS PubMed.
- B. M. Frezza, S. L. Cockroft and M. R. Ghadiri, J. Am. Chem. Soc., 2007, 129, 14875–14879 CrossRef CAS PubMed.
- S. Bi, M. Chen, X. Jia, Y. Dong and Z. Wang, Angew. Chem., Int. Ed. Engl., 2015, 54, 8144–8148 CrossRef CAS PubMed.
- Y. Guo, J. Wu and H. Ju, Chem. Sci., 2015, 6, 4318–4323 RSC.
- W. Engelen, L. H. Meijer, B. Somers, T. F. de Greef and M. Merkx, Nat. Commun., 2017, 8, 14473 CrossRef CAS PubMed.
- J. Zhu, L. Zhang, T. Li, S. Dong and E. Wang, Adv. Mater., 2013, 25, 2440–2444 CrossRef CAS PubMed.
- D. Y. Tam, Z. Dai, M. S. Chan, L. S. Liu, M. C. Cheung, F. Bolze, C. Tin and P. K. Lo, Angew. Chem., Int. Ed. Engl., 2016, 55, 164–168 CrossRef CAS PubMed.
- N. V. DelRosso, S. Hews, L. Spector and N. D. Derr, Angew. Chem., Int. Ed. Engl., 2017, 56, 4443–4446 CrossRef CAS PubMed.
- T. Li, E. Wang and S. Dong, J. Am. Chem. Soc., 2009, 131, 15082–15083 CrossRef CAS PubMed.
- K. He, Y. Li, B. Xiang, P. Zhao, Y. Hu, Y. Huang, W. Li, Z. Nie and S. Yao, Chem. Sci., 2015, 6, 3556–3564 RSC.
- S. Bi, B. Ji, Z. P. Zhang and J. J. Zhu, Chem. Sci., 2013, 4, 1858–1863 RSC.
- Y. Chen, Y. Song, F. Wu, W. Liu, B. Fu, B. Feng and X. Zhou, Chem. Commun., 2015, 51, 6980–6983 RSC.
- L. Ma and A. Diao, Chem. Commun., 2015, 51, 10233–10235 RSC.
- V. Lyamichev, M. A. Brow and J. E. Dahlberg, Science, 1993, 260, 778–783 CAS.
- V. Lyamichev, A. L. Mast, J. G. Hall, J. R. Prudent, M. W. Kaiser, T. Takova, R. W. Kwiatkowski, T. J. Sander, M. de Arruda, D. A. Arco, B. P. Neri and M. A. Brow, Nat. Biotechnol., 1999, 17, 292–296 CrossRef CAS PubMed.
- Y. Chen, M. R. Shortreed, M. Olivier and L. M. Smith, Anal. Chem., 2005, 77, 2400–2405 CrossRef CAS PubMed.
- H. Zou, H. Allawi, X. Cao, M. Domanico, J. Harrington, W. R. Taylor, T. Yab, D. A. Ahlquist and G. Lidgard, Clin. Chem., 2012, 58, 375–383 CAS.
- B. Zou, Y. Ma, H. Wu and G. Zhou, Angew. Chem., Int. Ed. Engl., 2011, 50, 7395–7398 CrossRef CAS PubMed.
- B. Zou, Y. Ma, H. Wu and G. Zhou, Analyst, 2012, 137, 729–734 RSC.
- B. Zou, Q. Song, J. Wang, Y. Liu and G. Zhou, Chem. Commun., 2014, 50, 13722–13724 RSC.
- J. G. Hall, P. S. Eis, S. M. Law, L. P. Reynaldo, J. R. Prudent, D. J. Marshall, H. T. Allawi, A. L. Mast, J. E. Dahlberg, R. W. Kwiatkowski, M. de Arruda, B. P. Neri and V. I. Lyamichev, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 8272–8277 CrossRef CAS PubMed.
- V. I. Lyamichev, M. W. Kaiser, N. E. Lyamicheva, A. V. Vologodskii, J. G. Hall, W. P. Ma, H. T. Allawi and B. P. Neri, Biochemistry, 2000, 39, 9523–9532 CrossRef CAS PubMed.
- J. J. Harrington and M. R. Lieber, J. Biol. Chem., 1995, 270, 4503–4508 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: The experimental section, sequences of oligonucleotides, the fluorescence intensities of clinical samples, the illustration of controllable-extension bridged cascade invasive reactions by the conventional single-stranded flaps, the background signals from different downstream probes, the sensitivity of the conventional cascade invasive reaction, the linear relationship between the mutant fractions and the initial reaction rates, and the operation of the AND, OR, and INHIBIT logic gates. See DOI: 10.1039/c7sc04210h |
| ‡ Y. L. Liu and H. P. Wu contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2018 |