Open Access Article
Open Access ArticleLong-lived protein expression in hydrogel particles: towards artificial cells†
Xiaoyu
Zhou
,
Han
Wu
,
Miao
Cui
,
Sze Nga
Lai
and
Bo
Zheng
 *
*
Department of Chemistry, The Chinese University of Hong Kong, Shatin, Hong Kong, P. R. China. E-mail: bozheng@cuhk.edu.hk
First published on 16th April 2018
Abstract
Herein we report a new type of cell-mimic particle capable of long-lived protein expression. We constructed the cell-mimic particles by immobilizing the proteinaceous factors of the cell-free transcription and translation system on the polymer backbone of hydrogel particles and encapsulating the plasmid template and ribosome inside the hydrogel. With the continuous supply of nutrients and energy, the protein expression in the cell-mimic particles remained stable for at least 11 days. We achieved the regulation of protein expression in the cell-mimic particles by the usage of lac operon. The cell-mimic particles quickly responded to the concentration change of isopropyl β-D-1-thiogalactopyranoside (IPTG) in the feeding buffer to regulate the mCherry expression level. We also constructed an in vitro genetic oscillator in the cell-mimic particles. Protein LacI provided a negative feedback to the expression of both LacI itself and eGFP, and the expression level change of eGFP presented an oscillation. We expect the cell-mimic particles to be a useful platform for gene circuit engineering, metabolic engineering, and biosensors.
Introduction
Artificial cells are an essential tool in the research of the origin of life,1–4 synthetic biology,5–7 drug delivery,8–11 gene therapy,12,13etc. Artificial cells are generally capable of some functions or behaviors of natural cells, such as protein expression,14–17 communication,18–21 and self-replication.1,22 These artificial cells are mostly liposomes,23,24 polymersomes,25,26 hydrogels,27,28 or simply aqueous droplets29 encapsulating the cell-free expression system. Cell-free expression systems are usually cell extracts from Escherichia coli (E. coli), wheat embryo and rabbit reticulocytes30,31 supplemented with energy components and free amino acids.32–34 In addition, a purified cell-free transcription and translation system, termed the protein synthesis using recombinant elements (PURE) system was developed by Ueda and co-workers.35 The PURE system was reconstituted from the essential elements of the E. coli transcription and translation system.Cell-free protein expression in an artificial cell in the batch format mostly terminated within a few hours due to the depletion of energy and nutrients and the accumulation of waste products.33,35 To solve this problem, many efforts were devoted to building artificial cells capable of long-lived protein expression through continuous energy and nutrient uptake and waste elimination. Dialysis membranes encapsulating the cell-free protein expression system inside were introduced, and the protein expression in the artificial cells could last tens of hours with higher protein yield.14 However, dialysis membranes presented a molecular weight cut-off, making it difficult for macromolecules including protein products to pass through the pores. Recently, Maerkl and co-workers built a micro-reactor that supported regulated protein expression up to 30 hours.36 Mass exchange and mixing were actively controlled by the integrated pneumatic valves. Noireaux et al. reported vesicle-based artificial cells which expressed and anchored the pore proteins to the vesicle membrane.37 Small molecules such as the energy source and nutrients can diffuse through the nanopores in the membrane, thus facilitating continuous protein expression up to 4 days. However, vesicle-based artificial cells required a delicate balance of osmotic pressure to prevent vesicle bursting. In contrast to the microencapsulation of the cell free expression system, micro-compartmentation of DNA templates, e.g., DNA gel27 and DNA brushes,21,38 allowed continuous protein expression when the cell free expression system was supplied together with energy and nutrients. DNA brush based artificial cells provide a useful platform for studying synthetic networks.21 However, DNA brushes present characteristics more of a biochip than an artificial cell, and functions such as multi-compartments, motility and chemotaxis39 are difficult to achieve.
Herein, we aimed to construct hydrogel-based cell-mimic particles capable of long-lived protein expression. Instead of physical entrapment of the transcription and translation system in polymeric particles as in previous studies40–42 to achieve cell-free protein expression, we immobilized the proteinaceous factors of the PURE system on the polymer backbone of hydrogel particles through chelating bonds. We achieved selective nutrient, waste, and energy exchange by the diffusion process between the surrounding buffer and the cell-mimic particles. The resulting cell-mimic particles were an open system that supported not only long-lived protein expression, but also expression regulation and genetic oscillators.
Results and discussion
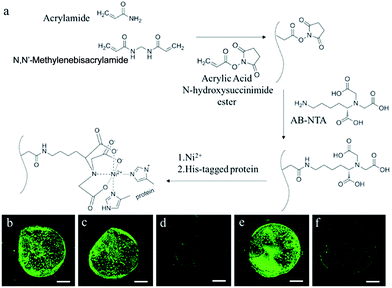
We first prepared a functionalized polyacrylamide (PA) hydrogel that presented a nickel(II)–nitrilotriacetic acid (Ni2+–NTA) moiety on the polymer backbone (Fig. 1a). First, acrylic acid N-hydroxysuccinimide ester was copolymerized with the hydrogel precursors. Then Nα,Nα-bis(carboxymethyl)-L-lysine hydrate (AB-NTA) was incorporated into the polymer backbone through the reaction with the N-hydroxysuccinimide (NHS) group. After the formation of the Ni2+–NTA complex, the resulting functionalized PA hydrogel can be used for the immobilization of His-tagged proteins.43 We evaluated the immobilization efficiency of the functionalized hydrogel by using the His-tagged eGFP. The PA hydrogel particles were treated with the solution of His-tagged eGFP, followed by washing with water several times. A control experiment was performed by treating the unfunctionalized PA hydrogel with the same solution of His-tagged eGFP. The His-tagged eGFP remained in the functionalized PA hydrogel after washing with water (Fig. 1b and c), on the other hand, the His-tagged eGFP in the functionalized PA hydrogel could be washed away with imidazole (Fig. 1d), further indicating that the His-tagged eGFP was immobilized through the interaction between the His-tag and the Ni2+–NTA complex.We started the construction of cell-mimic particles by preparing uniform functionalized PA hydrogel particles with a size of 50 μm using the microfluidic T-junction platform.44 The hydrogel particles were heated in air at 100 °C to be dehydrated. The PURE system proteinaceous factors, ribosome and plasmid templates were loaded into the PA hydrogel particles when the predried PA hydrogel particles were rehydrated in the PURE system solution.45 The enzyme mixture of the PURE system includes 20 tRNA synthetases, 3 initiation factors (IF1, IF2, and IF3), 3 elongation factors (EF-G, EF-Tu, and EF-Ts), 3 release factors (RF1, RF2, and RF3), ribosome recycling factor, myokinase, creatine kinase, nucleoside-diphosphate kinase, peptidylprolyl-isomerase, T7 RNA polymerase, and 70S ribosome. The proteinaceous factors of the PURE system were all labelled with the His-tag and immobilized through the interaction of the His-tag and the Ni2+–NTA complex. The ribosomes were non-His-tagged with a relatively large size of around 20 nm, while the average pore size of the PA hydrogel was around 12 nm. The ribosomes were encapsulated during the rehydration of the predried PA hydrogel particles, which facilitated convective transport induced by expanding the gel network.45 After the rehydration, the ribosomes were entrapped inside the cell-mimic particles due to the larger size than the hydrogel pores. We also observed that the plasmid templates were encapsulated inside the hydrogel, probably due to both the interaction between the Ni2+ and the DNA phosphates46 and the entanglement of the DNA strands with the hydrogel polymer backbone (Fig. S1†).
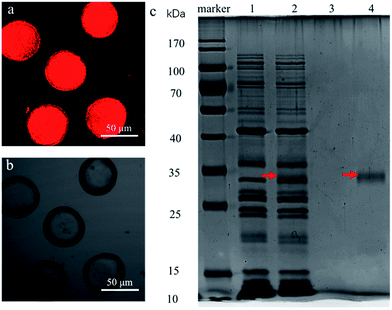
The expression of non-His-tagged mCherry was performed in the cell-mimic particles to verify the capability of protein expression. A red fluorescence was observed in the cell-mimic particles when the plasmid containing the non-His-tagged mCherry coding gene was encapsulated into the cell-mimic particles (Fig. 2a and b). The SDS-PAGE results (Fig. 2c) confirmed that non-His-tagged mCherry was expressed inside the cell-mimic particles and then diffused out to the surroundings. By contrast, no proteinaceous factors or ribosome bands could be found in the surroundings, indicating that the proteinaceous factors and ribosome of the PURE system remained inside the cell-mimic particles. The non-His-tagged mCherry observed in the surroundings indicated that the protein product freely diffused out of the cell-mimic particles, providing a potential means of communication with nearby cell-mimic particles. The results proved that the cell-mimic particle was (i) a self-maintaining system that kept the proteinaceous factors, ribosome and plasmid template inside the cell-mimic particle; and (ii) an open system that allowed exchange of nutrients, energy, and expression products between the cell-mimic particle and the surroundings.
The expression yield of non-His-tagged eGFP was measured to evaluate the efficiency of the cell-mimic particles. A group of ten cell-mimic particles in 5 μL feeding buffer reached the expression equilibrium after 4 hours, with the final non-His-tagged eGFP concentration in the surrounding buffer at 197 μg mL−1. In the control experiment, we prepared 5 μL feeding buffer with the same composition. The buffer solution also contained the same amount of proteinaceous factors and ribosome of the PURE system as in the combined ten cell-mimic particles. The yield of non-His-tagged eGFP in the solution of the control experiment reached equilibrium after 2 hours, with the final non-His-tagged eGFP concentration of 87 μg mL−1. By comparison, the in vivo expression in E. coli yielded eGFP with the concentration on the scale of tens of μg mL−1.47,48 The results showed that the protein yield and the expression efficiency were higher in the cell-mimic particles than in the aqueous solution and in the E. coli cells.47–49 The high expression efficiency of the cell-mimic particles was an unexpected observation. In E. coli, on which the PURE system is based, the transcription and translation processes are coupled, which require a close proximity of ribosome, RNA polymerase, and other related factors. The efficient expression of non-His-tagged eGFP in the cell-mimic particles suggested that the polymer backbone was flexible enough to support the diffusive movement of the immobilized factors and facilitate the coupled transcription and translation processes.50 The better performance of the cell-mimic particles in protein expression is probably because the proteinaceous factors were confined inside the cell-mimic particles with a much higher local concentration.27 We expect that smaller or thinner cell-mimic particles with higher surface-area-to-volume ratio will have higher expression yield, as the exchange of nutrients, energy, and expression products by diffusion will take less time.27
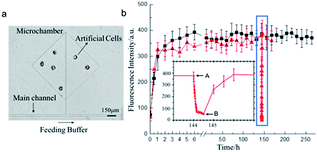
Next, we tested the capability of long-lived protein expression of the cell-mimic particles. The cell-mimic particles were placed in a microchamber which was connected to the main channel through a side channel (Fig. 3a). Due to the sub-millimeter dimension of the microchannel and the low flow rate, the exchange of nutrients, energy, and expression products between the cell-mimic particles and the solution in the main channel was achieved by diffusion through the side channel. The openness of the cell-mimic particles was crucial to the long-lived protein expression in the cell-mimic particles for the efficient energy and nutrient supply and waste discharge. During the experiment, the nutrients and the energy agents continuously diffused into the cell-mimic particles to support the protein expression, while the protein products and by-products diffused out of the cell-mimic particles. When continuously feeding the cell-mimic particles, we observed stable expression of non-His-tagged mCherry for at least 11 days (Fig. 3b). In the control experiment, the temperature was decreased to room temperature to pause the expression. The associated decrease of the fluorescence signal indicated the loss of non-His-tagged mCherry in the cell-mimic particles by diffusion (Fig. S2†). Once the temperature returned to 37 °C, the expression resumed and the fluorescence signal in the cell-mimic particles was recovered (Fig. 3b). We expect the protein expression to continue until the immobilized proteinaceous factors of the PURE system and the ribosomes degrade. The long-lived protein expression of the cell-mimic particles facilitated more complex functions of the cell-mimic particles such as protein expression regulation and gene networks.
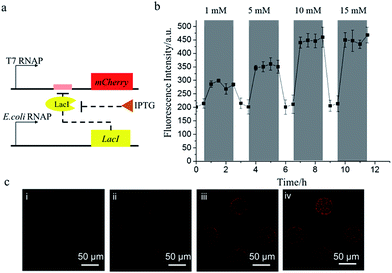
The cell-mimic particles were then used to achieve the regulation of the gene expression. E. coli RNA polymerase was added to the PURE system to initiate the transcription and translation of the protein LacI, which would impede the transcription of the non-His-tagged mCherry gene by binding with the Lac operator linked to the mCherry gene. When isopropyl β-D-1-thiogalactopyranoside (IPTG) was added into the feeding buffer, LacI would bind to IPTG, releasing the Lac operator to activate the mCherry expression (Fig. 4a). After the addition of IPTG into the feeding buffer, the mCherry expression level would be enhanced. The mCherry expression level increased with the increase of the IPTG concentration until the IPTG concentration reached 10 mM (Fig. 4b). When IPTG was removed from the feeding buffer, the mCherry expression returned to the initial low level as the LacI protein would bind to the Lac operator again. The cell-mimic particles' response to IPTG showed their potential to be a biosensor platform in addition to being a useful cell model for the research of gene networks and cell–cell communication.
We further demonstrated a negative feedback gene oscillator in the cell-mimic particles. With the transcription action of T7 RNA polymerase, protein LacI and non-His-tagged eGFP were expressed simultaneously in the cell-mimic particles. The accumulation of protein LacI would inhibit the expression of eGFP and LacI as a negative feedback, leading to the decreased expression level of LacI and eGFP (Fig. 5a).51 Once the accumulated LacI diffused out of the cell-mimic particles, the promoters became free of bound LacI and the cycle began anew. As expected, the fluorescence signal of eGFP showed an oscillation, with a period of about 2 minutes (Fig. 5b). The period of the oscillator in the cell-mimic particles was much shorter than the period of the same oscillator in natural cells.51 The difference is likely because the oscillator period was related to the removal rate of the proteins.51 In natural cells, the protein products were degraded by catabolic enzymes, while in the cell-mimic particles, the decrease of the protein level was dependent on the diffusion process, which was much quicker than the degradation process.51 With the demonstration of the gene oscillator, we expect that cell-mimic particles possess great potential as a platform to test the dynamic behavior of the gene network.
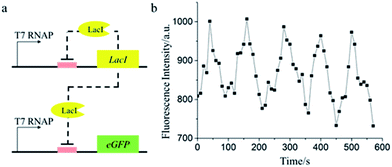 | ||
| Fig. 5 (a) The schematic of negative feedback oscillation. (b) The fluorescence trajectory of a single cell-mimic particle containing the gene network with negative feedback. | ||
Conclusions
In conclusion, we built a new type of self-maintaining and self-regulating cell-mimic particle. The prominent features of the cell-mimic particles are the long-lived and efficient protein expression in the particles. The features were achieved through the immobilization of the cell-free transcription and translation system on the polymer backbone of the cell-mimic particles, which rendered the particles capable of continuous uptake of nutrients and energy from the surroundings and the diffusion of the protein expression products out of the particles.With the long-lived and efficient protein expression, the cell-mimic particles supported the gene regulation and gene circuit engineering. We envision that the cell-mimic particles will also be a useful platform for biosensors such as toehold-based sensors for viruses52 and for the research of cell–cell communication such as biofilm disruption.53 In addition, with a coating of a lipid bilayer, the cell-mimic particles provide a potential starting point to the construction of artificial cells and drug delivery vehicles.54
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge the financial support by the Research Grants Council of Hong Kong (GRF 14303917), The Chinese University of Hong Kong (Direct Grant 4053158), and the Hong Kong Ph.D. Fellowship (H.·W.) from the Research Grants Council of Hong Kong.Notes and references
- J. W. Szostak, D. P. Bartel and P. L. Luisi, Nature, 2001, 409, 387–390 CrossRef CAS PubMed.
- D. Deamer, Trends Biotechnol., 2005, 23, 336–338 CrossRef CAS PubMed.
- P. L. Luisi, F. Ferri and P. Stano, Naturwissenschaften, 2006, 93, 1–13 CrossRef CAS PubMed.
- S. Rasmussen, L. Chen, D. Deamer, D. C. Krakauer, N. H. Packard, P. F. Stadler and M. A. Bedau, Science, 2004, 303, 963–965 CrossRef CAS PubMed.
- C. Lartigue, J. I. Glass, N. Alperovich, R. Pieper, P. P. Parmar, C. A. Hutchison III, H. O. Smith and J. C. Venter, Science, 2007, 317, 632–638 CrossRef CAS PubMed.
- A. Pohorille and D. Deamer, Trends Biotechnol., 2002, 20, 123–128 CrossRef CAS PubMed.
- C. Chiarabelli, P. Stano and P. L. Luisi, Curr. Opin. Biotechnol., 2009, 20, 492–497 CrossRef CAS PubMed.
- C. E. Ashley, E. C. Carnes, G. K. Phillips, D. Padilla, P. N. Durfee, P. A. Brown, T. N. Hanna, J. Liu, B. Phillips, M. B. Carter, N. J. Carroll, X. Jiang, D. R. Dunphy, C. L. Willman, D. N. Petsev, D. G. Evans, A. N. Parikh, B. Chackerian, W. Wharton, D. S. Peabody and C. J. Brinker, Nat. Mater., 2011, 10, 389–397 CrossRef CAS PubMed.
- Y. Zhang, W. C. Ruder and P. R. LeDuc, Trends Biotechnol., 2008, 26, 14–20 CrossRef CAS PubMed.
- K. Morigaki and P. Walde, Curr. Opin. Colloid Interface Sci., 2007, 12, 75–80 CrossRef CAS.
- J. Liu, A. Stace-Naughton, X. Jiang and C. J. Brinker, J. Am. Chem. Soc., 2009, 131, 1354–1355 CrossRef CAS PubMed.
- S. Prakash and M. L. Jones, J. Biomed. Biotechnol., 2005, 2005, 44–56 CrossRef PubMed.
- T. M. S. Chang, Nat. Rev. Drug Discovery, 2005, 4, 221–235 CrossRef CAS PubMed.
- A. S. Spirin, V. I. Baranov, L. A. Ryabova, S. Y. Ovodov and Y. B. Alakhov, Science, 1988, 242, 1162–1164 CAS.
- W. Yu, K. Sato, M. Wakabayashi, T. Nakaishi, E. P. Ko-Mitamura, Y. Shima, I. Urabe and T. Yomo, J. Biosci. Bioeng., 2001, 92, 590–593 CrossRef CAS PubMed.
- S. M. Nomura, K. Tsumoto, T. Hamada, K. Akiyoshi, Y. Nakatani and K. Yoshikawa, ChemBioChem, 2003, 4, 1172–1175 CrossRef CAS PubMed.
- T. Oberholzer, K. H. Nierhaus and P. L. Luisi, Biochem. Biophys. Res. Commun., 1999, 261, 238–241 CrossRef CAS PubMed.
- P. M. Gardner, K. Winzer and B. G. Davis, Nat. Chem., 2009, 1, 377–383 CrossRef CAS PubMed.
- R. Lentini, S. P. Santero, F. Chizzolini, D. Cecchi, J. Fontana, M. Marchioretto, C. Del Bianco, J. L. Terrell, A. C. Spencer and L. Martini, Nat. Commun., 2014, 5, 4012 CAS.
- K. P. Adamala, D. A. Martin-alarcon, K. R. Guthrie-honea and E. S. Boyden, Nat. Chem., 2016, 9, 431–439 CrossRef PubMed.
- E. Karzbrun, A. M. Tayar, V. Noireaux and R. H. Bar-Ziv, Science, 2014, 345, 829–832 CrossRef CAS PubMed.
- K. Kurihara, M. Tamura, K. Shohda, T. Toyota, K. Suzuki and T. Sugawara, Nat. Chem., 2011, 3, 775–781 CrossRef CAS PubMed.
- P. Walde, BioEssays, 2010, 32, 296–303 CrossRef CAS PubMed.
- N.-N. Deng, M. Yelleswarapu, L. Zheng and W. T. S. Huck, J. Am. Chem. Soc., 2017, 139, 587–590 CrossRef CAS PubMed.
- N. P. Kamat, J. S. Katz and D. A. Hammer, J. Phys. Chem. Lett., 2011, 2, 1612–1623 CrossRef CAS PubMed.
- M. Li, X. Huang, T.-Y. D. Tang and S. Mann, Curr. Opin. Chem. Biol., 2014, 22, 1–11 CrossRef CAS PubMed.
- N. Park, S. H. Um, H. Funabashi, J. Xu and D. Luo, Nat. Mater., 2009, 8, 432–437 CrossRef CAS PubMed.
- M. Bayoumi, H. Bayley, G. Maglia and K. T. Sapra, Sci. Rep., 2017, 7, 45167 CrossRef CAS PubMed.
- C. Martino and A. J. DeMello, Interface Focus, 2016, 6, 20160011 CrossRef PubMed.
- M. W. Nirenberg and J. H. Matthaei, Proc. Natl. Acad. Sci. U. S. A., 1961, 47, 1588–1602 CrossRef CAS.
- E. D. Carlson, R. Gan, C. E. Hodgman and M. C. Jewett, Biotechnol. Adv., 2012, 30, 1185–1194 CrossRef CAS PubMed.
- T. Sawasaki, T. Ogasawara, R. Morishita and Y. Endo, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 14652–14657 CrossRef CAS PubMed.
- A. Zemella, L. Thoring, C. Hoffmeister and S. Kubick, ChemBioChem, 2015, 16, 2420–2431 CrossRef CAS PubMed.
- D. M. Kim and J. R. Swartz, Biotechnol. Bioeng., 1999, 66, 180–188 CrossRef CAS PubMed.
- Y. Shimizu, A. Inoue, Y. Tomari, T. Suzuki, T. Yokogawa, K. Nishikawa and T. Ueda, Nat. Biotechnol., 2001, 19, 751–755 CrossRef CAS PubMed.
- H. Niederholtmeyer, V. Stepanova and S. J. Maerkl, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 15985–15990 CrossRef CAS PubMed.
- V. Noireaux and A. Libchaber, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 17669–17674 CrossRef CAS PubMed.
- D. Gerber, S. J. Maerkl and S. R. Quake, Nat. Methods, 2009, 6, 71–74 CrossRef CAS PubMed.
- B. C. Buddingh' and J. C. M. van Hest, Acc. Chem. Res., 2017, 50, 769–777 CrossRef PubMed.
- J. Thiele, Y. Ma, D. Foschepoth, M. M. K. Hansen, C. Steffen, H. A. Heus and W. T. S. Huck, Lab Chip, 2014, 14, 2651–2656 RSC.
- D. Yang, S. Peng, M. R. Hartman, T. Gupton-Campolongo, E. J. Rice, A. K. Chang, Z. Gu, G. Q. Lu and D. Luo, Sci. Rep., 2013, 3, 3165 CrossRef PubMed.
- K.-H. Lee, K.-Y. Lee, J.-Y. Byun, B.-G. Kim and D.-M. Kim, Lab Chip, 2012, 12, 1605–1610 RSC.
- R. L. Schnaar and Y. C. Lee, Biochemistry, 1975, 14, 1535–1541 CrossRef CAS PubMed.
- Z. Han, W. Li, Y. Huang and B. Zheng, Anal. Chem., 2009, 81, 5840–5845 CrossRef CAS PubMed.
- Y. Li, D. Guo and B. Zheng, RSC Adv., 2012, 2, 4857–4863 RSC.
- T. Pawłowski, J. Swiatek, K. Gasiorowski and H. Kozłowski, Inorg. Chim. Acta, 1987, 136, 185–189 CrossRef.
- P. Lu, C. Vogel, R. Wang, X. Yao and E. M. Marcotte, Nat. Biotechnol., 2007, 25, 117–124 CrossRef CAS PubMed.
- C. Albano, L. Randers-Eichhon, Q. Chang, W. Bentley and G. Rao, Biotechnol. Tech., 1996, 10, 953–958 CrossRef CAS.
- Y. Chen, L.-N. Wei and J. D. Muller, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 15492–15497 CrossRef CAS PubMed.
- R. A. Sheldon, Adv. Synth. Catal., 2007, 349, 1289–1307 CrossRef CAS.
- J. Stricker, S. Cookson, M. R. Bennett, W. H. Mather, L. S. Tsimring and J. Hasty, Nature, 2008, 456, 516–519 CrossRef CAS PubMed.
- S. Slomovic, K. Pardee and J. J. Collins, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 14429–14435 CrossRef CAS PubMed.
- R. Lentini, N. Y. Martín, M. Forlin, L. Belmonte, J. Fontana, M. Cornella, L. Martini, S. Tamburini, W. E. Bentley, O. Jousson and S. S. Mansy, ACS Cent. Sci., 2017, 3, 117–123 CrossRef CAS PubMed.
- P. F. Kiser, G. Wilson and D. Needham, Nature, 1998, 394, 459–462 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details and results. See DOI: 10.1039/c8sc00383a |
| This journal is © The Royal Society of Chemistry 2018 |