Open Access Article
Open Access ArticleProtein scaffolded DNA tetrads enable efficient delivery and ultrasensitive imaging of miRNA through crosslinking hybridization chain reaction†
Du-Juan
Huang‡
,
Zhi-Mei
Huang‡
,
Hu-Yan
Xiao
,
Zhen-Kun
Wu
*,
Li-Juan
Tang
and
Jian-Hui
Jiang
 *
*
Institute of Chemical Biology & Nanomedicine, State Key Laboratory of Chemo/Biosensing & Chemometrics, College of Chemistry & Chemical Engineering, Hunan University, Changsha 410082, China. E-mail: jianhuijiang@hnu.edu.cn; tomwu@hnu.edu.cn
First published on 5th May 2018
Abstract
Efficient intracellular delivery of nucleic acids to achieve sensitive detection and gene regulation is essential for chemistry and biology. Here we developed a novel protein scaffolded DNA tetrad, a four-arm DNA nanostructure constructed using streptavidin (SA) protein and four biotinylated hairpin DNA probes for efficient nucleic acid delivery and ultrasensitive miRNA imaging through crosslinking hybridization chain reaction (cHCR). DNA tetrads were easy to prepare and allowed precise control of the structure of the probes. DNA tetrads showed rapid intracellular delivery of DNA probes and high efficiency in lysosome escape by using confocal images for individual cells and flow cytometry for a large population of cells. cHCR allowed generating clumps of crosslinked hydrogel networks specifically to target miRNA, affording high sensitivity and spatial resolution for imaging. To our knowledge, this is the first time that HCR amplification has been realized in situ on nanostructures. Moreover, the FRET based design of cHCR conferred improved precision with the use of dual-emission ratiometric imaging to avoid false signals in biological systems. Intracellular imaging experiments further showed that DNA tetrad based cHCR could realize ultrasensitive and accurate miRNA imaging in living cells. Moreover, DNA tetrad based cHCR provided a potential tool for quantitative measurement of intracellular miRNA. The results suggested that this developed strategy provided a useful platform for nucleic acid delivery and low level biomarker imaging.
Introduction
Nucleic acids play critical roles in many fundamental cellular processes involving genetic information carriers, intracellular regulatory molecules, and catalytic enzymes. Because of their ability to interact with various molecules via canonical base-pairing or conformation-fitted binding, nucleic acids have become a valuable regulatory and analytical tool with exquisite specificity for cell biology and medicine.1–3 A prime quest to use nucleic acids for biomedical applications is the transfection system that allows their efficient delivery into different cells. DNA/RNA nanostructures represent an attractive option for intracellular delivery of nucleic acids. Prominent examples are spherical nucleic acids (SNAs),4–6 and self-assembled nucleic acid nanostructures.7–9 These nucleic acid nanostructures have demonstrated their ability to enter different cells without the aid of cationic carriers, affording a powerful platform for developing diagnostic assays,10–12 therapeutic drugs,13,14 and gene regulation agents15,16 for live cell and in vivo applications. Nevertheless, major challenges still exist in facile synthesis or purification of DNA/RNA nanostructures, high-efficiency delivery of nucleic acid reagents into cells and limited sensitivity for intracellular detection.Proteins provide an alternative promising scaffold for constructing DNA/RNA nanostructures. In contrast to synthetic nanoparticles,17,18 proteins are characterized by a surface with unevenly distributed reactive sites and a core with well-defined conformation and diverse oligomeric structures. Motivated by this advantage, protein-based SNAs, which use proteins as the core and utilize their surface amine and thiol groups for dense functionalization of DNA, have been engineered.19–21 This particular type of SNAs is shown to not only enhance cellular uptake of functional proteins, but also offer building blocks for precisely modulating the nanoparticle superlattice architecture. However, protein-scaffolded DNA nanostructures that are facile in synthesis and purification and efficient for delivery of nucleic acid reagents have rarely been explored.
To allow facile synthesis of the protein-scaffolded DNA nanostructure, we turn to employing high-affinity non-covalent interactions for assembling DNA on the protein surface. Streptavidin (SA) is known for its exceptionally tight binding of biotin with the dissociation constant down to 10−14 M,22 and fusion to SpyTag has generated assemblies of SA subunits capable of binding a varying number of biotin conjugates.23 Hence, SA and its recombinant protein fusion have the potential to provide a modular scaffold for assembling biotinylated DNA into nanostructures with precise control of the number and position of DNA. Motivated by this hypothesis, we choose SA as the model scaffold and develop a simple DNA nanostructure, a DNA tetrad, in which the surface of SA is provided with four biotinylated DNA oligonucleotides via high-affinity SA–biotin interactions. Additionally, nanostructures reported so far had dense DNA probes on their surface, which may generate high steric hindrance and inhibit the DNA-assembly based amplification; the tiny DNA tetrad developed here with only four DNA probes resulted in a relatively loose structure, offering the possibility of HCR in situ on nanostructures. Motivated by this assumption and based on previous work on DNA circuits for signal amplification by our group and others,24–29 we further demonstrated the utility of DNA tetrads for ultrasensitive imaging of miRNA using a novel crosslinking hybridization chain reaction (cHCR), as shown in Scheme 1. As a proof-of-principle, we chose miR-21, a known biomarker overexpressed in many cancers,30–32 as a case of study to demonstrate the design of DNA tetrads for intracellular cHCR imaging. In cHCR, two biotinylated hairpin probes H1 and H2 are designed, one labeled with a Cy3 fluorescence donor and the other with a Cy5 fluorescence acceptor. Then, DNA tetrads with H1 and H2 probes separately displayed on SA are synthesized. In the presence of target miRNA, it opens the hairpin probe H1, which in turn initializes HCR with a cascade of alternating hybridization between H1 and H2 and induces crosslinking of the tetrads and form 3D crosslinked hydrogel networks. In the cHCR products, Cy3 fluorescence donors and Cy5 acceptors are drawn into close proximity, which activates a FRET signal as an indicator for the expression of target miRNA. In the absence of target miRNA, cHCR between H1 and H2 probes does not occur and no FRET signal from Cy3 to Cy5 is observed. It is worth noting that, compared to current DNA circuit based imaging strategies,25–29 this DNA tetrad based strategy shows remarkable advantages. First, DNA tetrads provide a more robust, convenient and modular platform because of their fast and easy preparation, and precise control of the structure and concentration of the probes. It is also demonstrated that DNA tetrads are able to enter live cells quickly and efficiently without the aid of transfection carriers. Most importantly, DNA tetrads showed desirable efficiency in lysosome escape. Second, because each tetrad has four reactive sites, HCR can induce crosslinking of the tetrads and form 3D crosslinked hydrogel networks, affording high sensitivity and spatial resolution for imaging. To our knowledge, this is the first time that HCR amplification has been realized in situ on nanostructures, providing the first integrated system for DNA delivery and signal amplified imaging in living cells. Finally, the Föster resonance energy transfer (FRET) based design confers improved precision because it allows the use of dual-emission ratiometric imaging to avoid false signals in complex biological environments. Therefore, the DNA tetrad enabled cHCR strategy can offer a highly robust, accurate, and ultrasensitive approach for RNA based diagnostics.
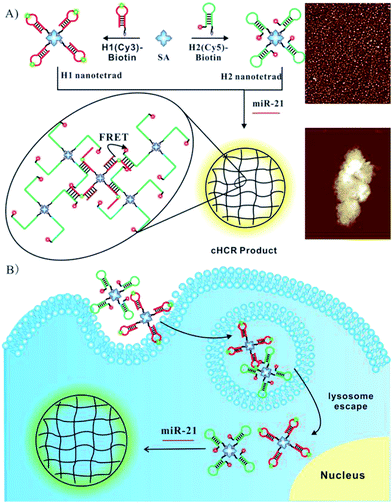 | ||
| Scheme 1 Illustration of the (A) DNA tetrad enabled cHCR and (B) ultrasensitive intracellular miRNA imaging strategy. | ||
Results and discussion
Characterization and cellular permeability of DNA tetrads
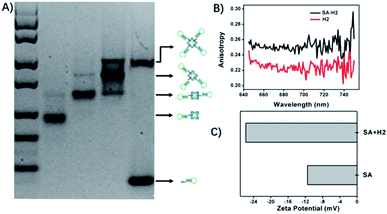
To study the cellular permeability of DNA tetrads, we synthesized a DNA tetrad by incubating SA with probe H3. As SA is a tetramer protein with four biotin-binding sites, four types of SA–DNA complexes can be obtained, corresponding to the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 SA–DNA complex to the 1
1 SA–DNA complex to the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 SA–DNA complex. These SA–DNA complexes were readily characterized by gel electrophoresis (Fig. 1A). As anticipated, at different concentration ratios of H3 to SA, the gel image showed one or two bands with varying mobility in each lane (2–4), and four bands with different molecular weights were obtained in these lanes. This result gave clear evidence for the formation of four types of SA–DNA complexes. Because of the well-separated bands for SA-complexes, the DNA tetrads were readily purified by cutting the corresponding DNA band followed by DNA isolation from the gel. The purified product was also conformed in the gel image (Fig. S1†). Fluorescence anisotropy analysis and zeta potential analysis were both used to verify the interaction between DNA probes and SA (Fig. 1B and C).
4 SA–DNA complex. These SA–DNA complexes were readily characterized by gel electrophoresis (Fig. 1A). As anticipated, at different concentration ratios of H3 to SA, the gel image showed one or two bands with varying mobility in each lane (2–4), and four bands with different molecular weights were obtained in these lanes. This result gave clear evidence for the formation of four types of SA–DNA complexes. Because of the well-separated bands for SA-complexes, the DNA tetrads were readily purified by cutting the corresponding DNA band followed by DNA isolation from the gel. The purified product was also conformed in the gel image (Fig. S1†). Fluorescence anisotropy analysis and zeta potential analysis were both used to verify the interaction between DNA probes and SA (Fig. 1B and C).
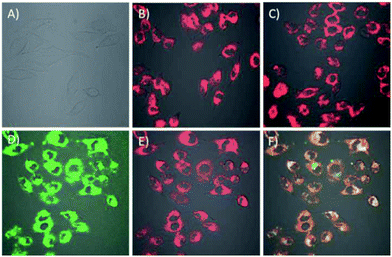
The cell permeability of the DNA tetrads was investigated. To visualize the DNA tetrads, we labelled the DNA tetrads on SA protein with Cy3 or on the H2 probe with Cy5. After 60 min of incubation with the DNA tetrads, the cells displayed bright fluorescence with typical cytosolic localization (Fig. 2B and C). The results suggested that DNA tetrads were capable of entering the cells without the aid of transfection reagents. By contrast, in a control experiment in which cells were incubated with free SA protein labelled with Cy3, no appreciable fluorescence was observed (Fig. 2A). According to this control, it was clear that surface-immobilized DNA probes were essential for the cell permeability of DNA tetrads. A further interrogation of cells incubated using DNA tetrads with SA protein labelled using Cy3 and the H2 probe tagged using Cy5 showed very intense and highly colocalized fluorescence signals in the cells at both Cy3 and Cy5 channels (Fig. 2D–F). This finding evidenced that DNA tetrads retained their structural integrity in cells with no dissociation of the biotinylated DNA probes from SA protein.
Further investigation of cellular uptake of the DNA tetrads in HeLa cells was performed (Fig. S2†). The confocal microscopy images showed a bright fluorescence when cells were incubated with DNA tetrads at 37 °C for 1 h. However, for cells pretreated at 4 °C before adding DNA tetrads, a dramatic reduction of Cy5 fluorescence intensity was observed, which suggested that the reaction with DNA tetrads involved a temperature-dependent cellular uptake process. Another experiment by pretreating cells with NaN3, an inhibitor of ATPase which is involved in all energy dependent endocytic pathways,29 also displayed significantly reduced cellular uptake of DNA tetrads.
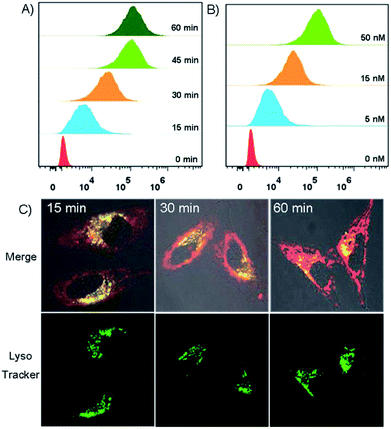
The trafficking dynamics of DNA tetrad uptake were monitored using both flow cytometry for a large population of cells and confocal fluorescence microscopy for individual cells. As shown in Fig. 3A, after incubating the cells with DNA tetrads, a substantial fluorescence peak shift was observed within 15 min, which saturated at ∼45 min, demonstrating an efficient delivery approach using DNA tetrads. Confocal microscopy imaging further verified these results (Fig. S3†). The dependency of uptake efficiency on the concentration of DNA tetrads was also investigated. With the concentration of DNA tetrads increased from 0 to 50 nM, the fluorescence intensity was significantly increased (Fig. 3B). To elucidate whether the internalization of DNA tetrads is dependent upon the conformation of the DNA probe, a control experiment using DNA tetrads with single strand DNA L1 was performed (Fig. S4†). A large peak red-shift was also observed for cells incubated with DNA tetrads carrying L1, verifying that DNA tetrads afford an efficient intracellular delivery approach for either linear or hairpin DNA probes. We next studied the subcellular localization of DNA tetrads. With the incubation time increased from 15 to 60 min, the Cy5 fluorescence gradually increased and spread out from the lysosomes into the cytosol with lysosome colocalization coefficients decreasing from 69.1% to 47.2% to 26.8% (Fig. 3C, and S5†).
Additionally, a Z-axis optical sectioning study further verified the efficiency of DNA tetrads in lysosome escape (Fig. S6 and S7†). These results suggested that DNA tetrads could be a useful platform for nucleic acid delivery with efficiency in lysosome escape.
In vitro response of DNA tetrads
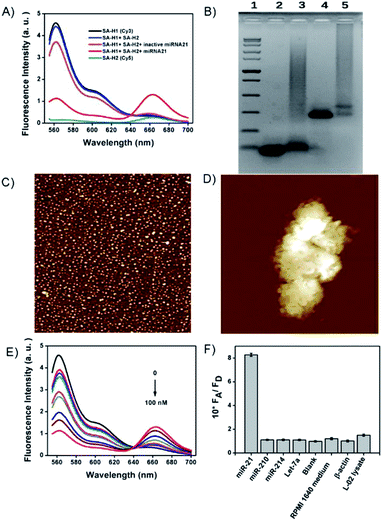
To our knowledge, HCR has not been realized in situ on nanostructures. Presumably, existing nanostructures with dense DNA probes may generate huge steric hindrance that prevents the growth of HCR polymers. Because of the tiny size of tetrads and sparsely tethered DNA probes, we reasoned that DNA tetrads offer the possibility of HCR amplification in situ on the nanostructures. As a result, we then investigated the feasibility of DNA tetrads for HCR amplification. As shown in Fig. 4A, almost no FRET occurred without target miR-21 due to the stable intramolecular hybridization of H1 and H2 on the tetrads, but a strong FRET signal was observed in the presence of target miR-21. A negative control experiment using inactive miR-21 merely displayed a low FRET signal. Agarose gel electrophoresis analysis was also performed to verify the design (Fig. 4B). The formation of crosslinked hydrogel networks was further demonstrated by atomic force microscopy. In contrast to the monomers of DNA tetrads with a ∼5 nm diameter (Fig. 4C), clumps with a size in the micron order for the cHCR products was observed (Fig. 4D); this result clearly evidenced the formation of crosslinked hydrogels in cHCR assay.The FRET signals showed dynamic responses to miR-21 with concentrations ranging from 0.05 to 100 nM with an estimated detection limit of 6 pM (Fig. 4E, and S8†). This detection limit was much better than those of the existing miRNA imaging methods,26,32 indicating the advantage of our strategy in sensitivity enhancement. We also made a comparison between this cHCR and conventional HCR using dimer probes of H1 and H2 (Fig. S9†). Interestingly, cHCR displayed a higher signal for Cy5 emission and a lower peak for Cy3 emission than those for conventional HCR. We ascribed the improved FRET signals of cHCR to the proximity of probes in DNA tetrads, which facilitates the reaction kinetics of HCR in the tetrads. Besides the improved sensitivity, our design also afforded excellent selectivity. As shown in Fig. 4F, target miR-21 triggered an obvious FRET signal. Control experiments using other miRNAs, a blank sample of the reaction buffer, RIPM 1640 cell culture medium or β-actin protein in place of target miRNA all displayed very low FRET signals, demonstrating the high selectivity of our method to detect miR-21. Furthermore, a lysate of negative L-02 cells with no miR-21 expression also did not produce an enhanced FRET signal, suggesting that our design had excellent resistance to false signals in complex biological environments. In other words, these results indicated that the design could improve the accurate image of miRNA in complex biological systems.
Intracellular cHCR imaging of DNA tetrads for miRNA
On the basis of preliminary results, we next applied DNA tetrads for imaging miR-21 in living cells. When HeLa cells were incubated with two DNA tetrads carrying H1 and H2, bright fluorescence was observed both in the green channel and red channel, suggesting that miR-21 initiated the cHCR reaction to generate the FRET signal (Fig. 5A, and S10†). To further verify the specificity of the fluorescence response, HeLa cells pre-treated with an miR-21 inhibitor, synthetic single-stranded RNAs designed to specifically bind to and selectively decrease the active concentration of miR-21,30,32 showed dim red fluorescence but very bright green fluorescence (Fig. 5B), indicating the specific response to miR-21. Moreover, HeLa cells pre-treated with a small chemically modified miR-21 mimic to imitate miRNA-21 high expression displayed higher red fluorescence intensity but lower green fluorescence intensity than that observed in untreated HeLa cells (Fig. 5C). These results revealed that fluorescence signals were dynamically correlated to the concentration of miR-21. The ratiometric images also showed similar results, giving direct evidence for cHCR and distinguishing different levels of intracellular miRNA while avoiding false signals. Furthermore, we transfected HeLa cells with dimer probes of H1 and H2 using the Lipofectamine 3000 reagent for conventional HCR assay. As anticipated, cells with conventional HCR assay showed much weaker FRET signals than those in DNA tetrad based cHCR assay (Fig. S11†), demonstrating that cHCR afforded higher amplification efficiency and better imaging contrast than conventional HCR. We ascribed the improved contrast of cHCR to the high delivery efficiency of DNA tetrads, facilitated HCR kinetics in DNA tetrads, and highly crosslinked HCR polymers with more localized signals.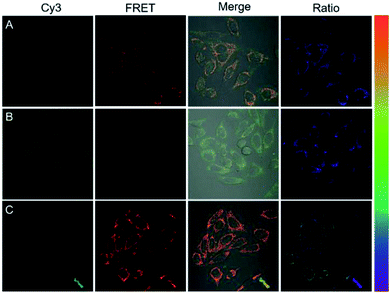 | ||
| Fig. 5 Fluorescence images of HeLa cells treated with DNA tetrads (A), 300 nM miR-21 inhibitor (B) and 300 nM miR-21 mimic (C). | ||
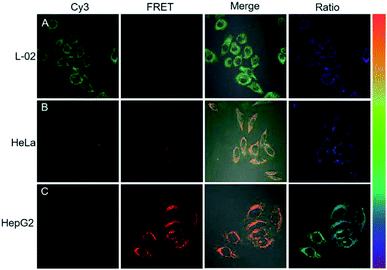
Next, the potential of the strategy for quantitative evaluation of the miR-21 expression in different cell lines was explored. Fluorescence images of varying brightness and ratiometric images were obtained (Fig. 6). HepG2 cells showed the highest miR-21 expression whereas L-02 displayed the lowest expression, which were consistent with the previous report for miRNA-21 expression levels in these cells.29 Ratiometric images were dynamically correlated to the concentration of miR-21, as analyzed by RT-PCR (Fig. S12†). These results verified the ability of the cHCR-based strategy for quantitative measurement of miRNA in living cells.
Conclusions
In summary, we have developed a novel DNA tetrad as a facile platform for efficient nucleic acid delivery and ultrasensitive miRNA imaging through cHCR. The DNA tetrads were demonstrated to be a highly robust delivery agent because of facile synthesis and purification, precise control of the structure and concentration of the probes, rapid nucleic acid delivery and efficiency in lysosome escape. cHCR amplification allows generating clumps of crosslinked hydrogel networks specifically to target miRNA, affording sensitive and spatially resolved miRNA imaging in living cells. To our knowledge, this is the first time that HCR amplification has been realized in situ on nanostructures. Moreover, the FRET based signal activation mode improved the precision of detection, avoiding false signals in complex biological systems. In virtue of these advantages, DNA tetrads may provide an effective strategy for nucleic acid delivery, low level biomarker imaging and related theranostics.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (21527810 and 21521063).Notes and references
- D. G. Spiller, C. D. Wood, D. A. Rand and M. R. White, Nature, 2010, 465, 736–745 CrossRef PubMed.
- M. R. Jones, N. C. Seeman and C. A. Mirkin, Science, 2015, 347, 1260901 CrossRef PubMed.
- Y. Ke, L. L. Ong, W. Sun, J. Song, M. Dong, W. M. Shih and P. Yin, Nat. Chem., 2014, 6, 994–1002 CrossRef PubMed.
- A. B. Chinen, C. M. Guan and C. A. Mirkin, Angew. Chem., Int. Ed., 2015, 54, 527–531 Search PubMed.
- C. H. Choi, L. Hao, S. P. Narayan, E. Auyeung and C. A. Mirkin, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 7625–7630 CrossRef PubMed.
- (a) R. J. Macfarlane, B. Lee, M. R. Jones, N. Harris, G. C. Schatz and C. A. Mirkin, Science, 2011, 334, 204–208 CrossRef PubMed; (b) H. Peng, X. F. Li, H. Zhang and X. C. Le, Nat. Commun., 2017, 8, 14378 CrossRef PubMed.
- L. He, D. Q. Lu, H. Liang, S. Xie, C. Luo, M. Hu, L. Xu, X. Zhang and W. Tan, ACS Nano, 2017, 11, 4060–4066 CrossRef PubMed.
- H. Pei, L. Liang, G. B. Yao, J. Li, Q. Huang and C. H. Fan, Angew. Chem., Int. Ed., 2012, 51, 9020–9024 CrossRef PubMed.
- J. Wang, J. Chao, H. Liu, S. Su, L. Wang, W. Huang, I. Willner and C. Fan, Angew. Chem., Int. Ed., 2017, 56, 2171–2175 CrossRef PubMed.
- D. Zheng, D. S. Seferos, D. A. Giljohann, P. C. Patel and C. A. Mirkin, Nano Lett., 2009, 9, 3258–3261 CrossRef PubMed.
- J. I. Cutler, E. Auyeung and C. A. Mirkin, J. Am. Chem. Soc., 2012, 134, 1376–1391 CrossRef PubMed.
- X. Tan, X. Lu, F. Jia, X. Liu, Y. Sun, J. K. Logan and K. Zhang, J. Am. Chem. Soc., 2016, 138, 10834–10837 CrossRef PubMed.
- X. Tan, B. B. Li, X. Lu, F. Jia, C. Santori, P. Menon, H. Li, B. Zhang, J. J. Zhao and K. Zhang, J. Am. Chem. Soc., 2015, 137, 6112–6115 CrossRef PubMed.
- T. G. W. Edwardson, K. M. M. Carneiro, C. K. Mclaughlin, C. J. Serpell and H. F. Sleiman, Nat. Chem., 2013, 5, 868–875 CrossRef PubMed.
- N. L. Rosi, D. A. Giljohann, C. S. Thaxton, A. K. Lytton-Jean, M. S. Han and C. A. Mirkin, Science, 2006, 312, 1027–1030 CrossRef PubMed.
- D. A. Giljohann, D. S. Seferos, A. E. Prigodich, P. C. Patel and C. A. Mirkin, J. Am. Chem. Soc., 2009, 131, 2072–2073 CrossRef PubMed.
- H. Liang, X. B. Zhang, Y. Lv, L. Gong, R. Wang, X. Zhu, R. Yang and W. Tan, Acc. Chem. Res., 2014, 47, 1891–1901 CrossRef PubMed.
- C. Wang, H. Zhang, D. Zeng, L. San and X. Mi, Chin. J. Chem., 2016, 34, 299–307 CrossRef.
- J. D. Brodin, A. J. Sprangers, J. R. McMillan and C. A. Mirkin, J. Am. Chem. Soc., 2015, 137, 14838–14841 CrossRef PubMed.
- P. Cigler, A. K. Lytton-Jean, D. G. Anderson, M. G. Finn and S. Y. Park, Nat. Mater., 2010, 9, 918–922 CrossRef PubMed.
- J. D. Brodin, E. Auyeung and C. A. Mirkin, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 4564–4569 CrossRef PubMed.
- D. E. Hyre, I. Le Trong, E. A. Merritt, J. F. Eccleston, N. M. Green, R. E. Stenkamp and P. S. Stayton, Protein Sci., 2006, 15, 459–467 CrossRef PubMed.
- C. Zhang, C. Tian, F. Guo, Z. Liu, W. Jiang and C. Mao, Angew. Chem., Int. Ed., 2012, 51, 3382–3385 CrossRef PubMed.
- (a) F. Wang, J. Elbaz, R. Orbach, N. Magen and I. Willner, J. Am. Chem. Soc., 2011, 133, 17149–17151 CrossRef PubMed; (b) F. Wang, C. H. Lu, X. Liu, L. Freage and I. Willner, Anal. Chem., 2014, 86, 1614–1621 CrossRef PubMed.
- C. Wu, S. Cansiz, L. Zhang, I. T. Teng, L. Qiu, J. Li, Y. Liu, C. Zhou, R. Hu, T. Zhang, C. Cui, L. Cui and W. Tan, J. Am. Chem. Soc., 2015, 137, 4900–4903 CrossRef PubMed.
- (a) Z. Cheglakov, T. M. Cronin, C. He and Y. Weizmann, J. Am. Chem. Soc., 2015, 137, 6116–6119 CrossRef PubMed; (b) L. Li, J. Feng, H. Liu, Q. Li, L. Tong and B. Tang, Chem. Sci., 2016, 7, 1940–1945 RSC.
- F. Chen, M. Bai, K. Cao, Y. Zhao, J. Wei and Y. Zhao, Adv. Funct. Mater., 2017, 27, 1702748 CrossRef.
- (a) Z. Wu, H. Fan, N. S. R. Satyavolu, W. Wang, R. Lake, J. H. Jiang and Y. Lu, Angew. Chem., Int. Ed., 2017, 56, 8721–8725 CrossRef PubMed; (b) M. Liu, Q. Zhang, D. Chang, J. Gu, J. D. Brennan and Y. Li, Angew. Chem., Int. Ed., 2017, 56, 6142–6146 CrossRef PubMed.
- Z. Wu, G. Q. Liu, X. L. Yang and J. H. Jiang, J. Am. Chem. Soc., 2015, 137, 6829–6836 CrossRef PubMed.
- J. Lu, G. Getz, E. A. Miska, E. Alvarez-Saavedra, J. Lamb, D. Peck, A. Sweet-Cordero, B. L. Ebert, R. H. Mak, A. A. Ferrando, J. R. Downing, T. Jacks, H. R. Horvitz and T. R. Golub, Nature, 2005, 435, 834–838 CrossRef PubMed.
- M. V. Joglekar, V. M. Joglekar and A. A. Hardikar, Gene Expression Patterns, 2009, 9, 109–113 CrossRef PubMed.
- Z. M. Ying, Z. Wu, B. Tu, W. Tan and J. H. Jiang, J. Am. Chem. Soc., 2017, 139, 9779–9782 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8sc01001c |
| ‡ D.-J. H. and Z.-M. H. contributed equally to the manuscript. |
| This journal is © The Royal Society of Chemistry 2018 |