Microfluidic bioanalytical flow cells for biofilm studies: a review†
Mohammad
Pousti
a,
Mir Pouyan
Zarabadi
a,
Mehran
Abbaszadeh Amirdehi
 a,
François
Paquet-Mercier
a and
Jesse
Greener
a,
François
Paquet-Mercier
a and
Jesse
Greener
 *ab
*ab
aDépartement de chimie, Faculté des sciences et de génie, Université Laval, Québec City, Québec G1 V 0A6, Canada
bCHU de Quebec Research Centre, Laval University, 10 rue de l'Espinay, Quebec City, (QC) G1L 3L5, Canada
First published on 17th October 2018
Abstract
Bacterial biofilms are among the oldest and most prevalent multicellular life forms on Earth and are increasingly relevant in research areas related to industrial fouling, medicine and biotechnology. The main hurdles to obtaining definitive experimental results include time-varying biofilm properties, structural and chemical heterogeneity, and especially their strong sensitivity to environmental cues. Therefore, in addition to judicious choice of measurement tools, a well-designed biofilm study requires strict control over experimental conditions, more so than most chemical studies. Due to excellent control over a host of physiochemical parameters, microfluidic flow cells have become indispensable in microbiological studies. Not surprisingly, the number of lab-on-chip studies focusing on biofilms and other microbiological systems with expanded analytical capabilities has expanded rapidly in the past decade. In this paper, we comprehensively review the current state of microfluidic bioanalytical research applied to bacterial biofilms and offer a perspective on new approaches that are expected to drive continued advances in this field.
Introduction
Living biofilms are one of the most challenging materials to study quantitatively. Analytical tools based in electrochemistry, spectroscopy and quantitative imaging are beneficial, but their full potential can only be realised with proper control over the liquid phase. Therefore, microfluidics is quickly becoming a fundamental aspect of modern bioanalytical sciences.Literature review
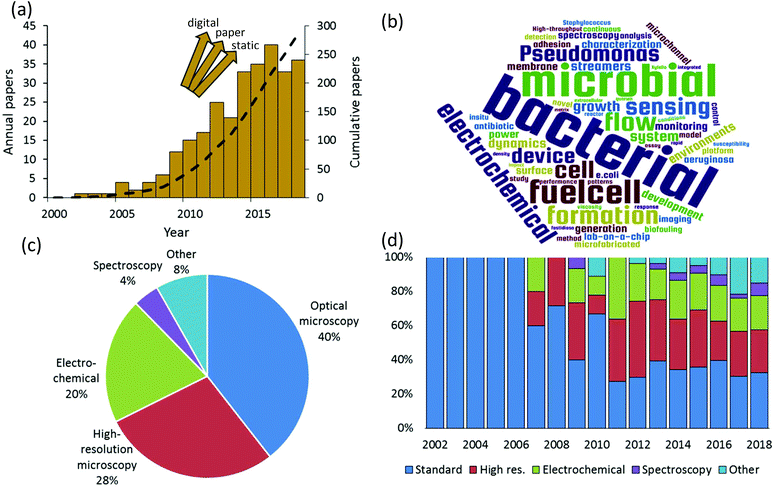
We rigorously analysed all published papers that used microfluidic devices to study bacterial biofilms. We categorised the applied measurement modalities to assemble a global overview of the state of microfluidic bioanalytical methods. Therefore, this work complements and goes beyond previous reviews of the use of microfluidics which have previously focused on certain specific phenomena such as biofilm formation, adhesion and detachment;1–3 biofilm susceptibility to antibiotics and toxicity testing;4 and electroactive biofilms including those used for microbial fuel cells.5–8Fig. 1a shows the annual numbers of manuscripts published since 2002 that report microfluidic studies of bacterial biofilms. These numbers grew rapidly between 2007 and 2016, with nearly 300 papers published by mid-2018. It is difficult to know if the apparent levelling off after 2016 is significant or if it is just an aberration related to the estimated number of papers in 2018, which was based on an extrapolation to the end of the calendar year. We note that the graphic is not comprehensive across all microfluidic domains because it does not include alternative platforms such as digital9 and paper10 microfluidics. Also excluded are “static” microscale devices, which can share microfabrication techniques and materials with channel-based microfluidics but do not involve flow. Keywords from the titles of all analysed papers are presented in graphical format in Fig. 1b. This graphic shows some of the major drivers of the field, based on title keywords (excluding “microfluidics” and “biofilms”). A text file listing the most popular title keyword is given in the ESI.† Based on a detailed reading of nearly 300 relevant papers that were published since 2002, a global breakdown of characterisation methods is given in Fig. 1c. Here multiple uses of different techniques within the same category were only counted once. For example, work using multiple spectroscopic or electrochemical measurements was considered a single instance in either category. The categories identified in this figure define the subject matter and the organization of this paper.
Optical microscopy is the historically dominant measurement modality. This includes transmission modes, which enable quantification of biofilm mass and dynamic motion via optical density and time-lapse imaging, respectively.11 Epifluorescence methods provide high-contrast imaging and can target specific biological and biochemical groups via fluorescent probe molecules. High-resolution microscopy, which we define here as either providing higher spatial resolution or higher dimensionality than transmission and fluorescence microscopy, has been the second most frequently used in published microfluidic biofilm studies. This category includes confocal laser scanning microscopy (CLSM), atomic force microscopy (AFM) and electron microscopy such as scanning and transmission electron microscopy (SEM, TEM). With few exceptions, AFM and electron microscopy are only applicable to ex situ microfluidic biofilm studies, usually after the experiment is finished. On the other hand CLSM is easily applied in situ. This almost certainly accounts for the majority use of CLSM in this category (>60%). By far, the most important non-imaging approaches are electrochemical. This has been driven by a growing interest in electroactive biofilms, though some electrochemical techniques can even be applied to non-electroactive biofilms, further increasing their relevance. Spectroscopic studies represent an important undercurrent in the literature. This category is mainly composed of vibrational techniques (Raman and infrared spectroscopy). Finally, other techniques and emerging approaches are discussed. These include important developments and trends toward on-chip implementation of specialised microscope imaging techniques, electrochemistry and spectroscopy, such as on-chip magnetic resonance imaging, as well as emerging approaches of mass spectrometry, respirometry. These methods are also compared in terms of their ability to meet a range of characterization requirements for biofilm studies.
True to the concept of lab-on-a-chip, fully one third of the published studies applied more than one characterisation category in a single study. This also supports recent conclusions that multiple analytical techniques applied to the native biofilm are required to obtain most meaningful results and continued advancement of the field.12 We also note in passing that approximately 15% of all relevant papers used numerical simulations, which were mainly applied to predict the flow properties within the microchannels. The final global characteristic extracted from this literature search was the significant diversification of the analytical modalities used over time (Fig. 1d). Although microscopy was dominant in early biofilm work, a decisive shift has occurred toward other approaches. This observation matches the trends in non-microfluidic experiments, which increasingly involve chemical and bio-chemical characterisation approaches such as respirometry,13 spectral microscopy and chemical imaging14 and electrochemistry.15 Taken together, these trends indicate that biofilm research is becoming a driver for the development of integrated multifunctional microfluidic technology. To appreciate the need for such sophistication, a short summary of bacterial biofilms is presented next.
Bacterial biofilms, grand analytical challenges
Sessile microbes, i.e., those living at surfaces, preferentially form biofilms. Contrary to their name, biofilms are three-dimensional materials with heterogeneous structural and chemical properties. In addition to living bacteria, biofilms are usually composed of an extracellular polymeric substance (EPS) consisting of biomacromolecules such as proteins, polysaccharides and nucleic acids. The EPS helps to regulate molecular diffusion, microbe-to-microbe interactions and other biochemical functions, while simultaneously maintaining the physical and mechanical integrity necessary for survival under hydrodynamic conditions.16,17 Therefore, microbial biofilms can form their own niche to achieve homeostasis in harsh or fluctuating conditions. This special property allowed microbial biofilms to become one of the first multicellular life forms on ancient Earth, approximately 3 to 4 billion years ago. Some of these ancient biofilms can still be found in certain niche environments today such caves18 and in extreme temperature, acidic and anaerobic conditions19–21 but are now joined by a diverse range of modern species that have proliferated to all corners of modern Earth.Recently, a rapid expansion in biofilm research is been driven by multiple areas of enquiry. In addition to fundamental questions focused on the relationship between biofilm structure and ecological function,22 these drivers include applied research into marine and industrial fouling,23,24 dental research,25 indoor moulds,26 virulent biofilms27–30 and biocatalysis.31,32 To complement the tremendous advances in molecular biology studies of biofilms in the last 10–15 years, greater attention has been placed on quantitative analysis under well-defined physiochemical conditions. In other words, modern bioanalytical investigations are increasingly directed toward improved control over growth conditions. This situation is especially true for biofilms, which efficiently respond to environmental cues by altering fundamental properties such as metabolic activity, biochemistry, physical structure and molecular mass transport.14,33–35
In the following section, we discuss different flow cell designs and the potential for microfluidic platforms to deliver enhanced control over key environmental parameters during biofilm studies.
Control using microfluidics
Biofilm growth cells are either static or flow-based. Static reactors are attractive because they are straightforward in design and easy to use. For example, assays conducted in microwells are common, with fluid handling usually being conducted via manual or robotic pipetting. A major drawback is that planktonic bacteria and other suspended material can contaminate biofilms due to gravitational settling. This problem can be alleviated with modified growth cell designs such as the Calgary device (Fig. 2a) in which biofilms are grown on pegs that are inserted into microwells from the top side. Periodic withdrawal of the biofilm-coated pegs for analysis leaves debris and foreign bacteria to settle at the bottom of the well.36 Drawback include changes in the biochemical environment due to continuous nutrient depletion and by-product accumulation. Frequent sample handling also limits measurement reproducibility and can introduce contamination. In contrast, flow cells maintain a constant flux of nutrient solution against the biofilm in a sealed environment, while debris and planktonic bacteria are washed downstream, which can significantly reduce all of these problems.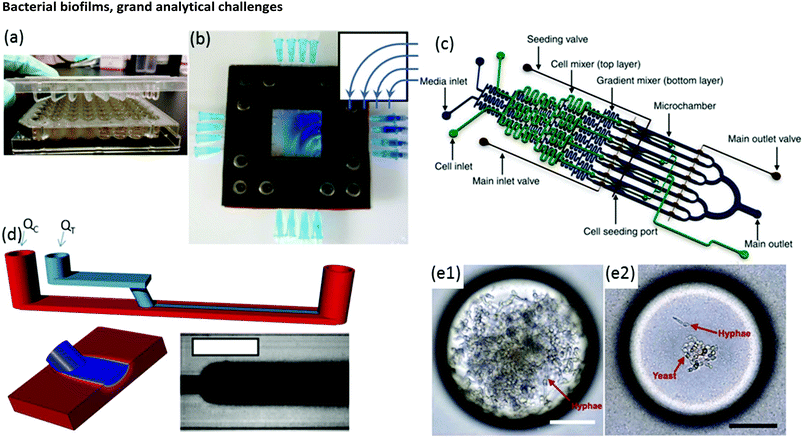 | ||
| Fig. 2 (a) Conventional Calgary device. (b) Microfluidic planar flow cell with height of 600 μm.48 Inset shows the direction of flow. (c) Multi-level device featuring complex fluidic circuitry designed to generate concentration gradients with built-in mixers, purpose-specific inlets, valves and separated growth compartments.64 (d) Laminar flow-templating device for creating linear biofilms patterns using liquid–liquid interfaces to prevent growth in microchannel corners. (e) Encapsulated microenvironments containing yeast and hyphae forms of biofilm forming Candida albicans cells after encapsulation (e1) and the more robust yeast cells when incubated in a culture of Pseudomonas aeruginosa PA14.69 | ||
Moreover, flow is among the most important factors in determining fundamental biofilm behaviour due to its effect on imposed shear stress37,38 molecular flux,39 concentration gradients in and around the biofilm40–43 and interactions with planktonic bacteria.44 As such, flow affects most observable biofilm properties, including growth kinetics,45 structure46–49 and function.50 However, due to the large liquid volumes required, standard flow cells present challenges in long-term studies over a comprehensive range of flow rates. Microfluidics can expand the benefits of standard flow cells while also offering other unique advantages. On-chip volumes at the μL scale or smaller require remarkably low flow rates, even during manipulation of hydrodynamic properties over a large dynamic range. Thus, nutrient solution utilisation can be strongly limited, which is beneficial in long-term experiments. Unlike in miniature chemostats or millifluidic flow cells, dead volumes in microchannels are vanishingly small, ensuring predictable liquid/biofilm contact times and applied concentrations. Flexibility in channel design and fluid pumping sequences enables complex multi-step experimental sequences and rapid adjustment of physiochemical parameters via on-chip dilution51,52 or integrated microvalves.53 For example, a microfluidic planar flow cell facilitated the application of a wide range of concentrations to the biofilm culture environment (Fig. 2b).48 A range of rapid prototyping techniques exists for fabrication of different microchannel geometries,54–56 enabling purpose-driven environments such as pseudo-porous and serpentine channels for the study of streamers,57,58 highly parallelised channels for high-throughput assays,59 complex fluidic circuitry60 and other moving components.61,62 For example, microfluidic concentration-gradient generators with complex circuitry for parallel biofilm assays using separate media and cell seeding inlets have been successfully applied in a number of studies (Fig. 2c).63,64 Studies on the effects of biofilms under moving three-phase interfaces can be efficiently conducted via the imposition of bubbles with well-defined sizes, velocities and wall interactions.65–67 Nevertheless, biofilms are highly efficient at modifying their environment, potentially undermining the advantages above.37,68 Solutions to these problems have been demonstrated, including templating biofilm growth using liquid–liquid (Fig. 2d)45 or liquid–gas67 interfaces, confinement in microdroplets69,70 (Fig. 2e) and isolation methods with integrated heating elements71 or via confinement by locally high shear forces.72 Isolation of electroactive biofilms at the surface of an integrated electrode can also limit channel overgrowth.73
To build on the previous achievements summarised above, the challenge for future biofilm researchers consists of merging microfluidic biofilm flow cells with powerful in situ analytical tools. The resulting synergy is expected to enable future discoveries and drive continued growth of microfluidics applied to biofilm research and bioanalytical chemistry generally.
Microscopy
Optical microscopy
Transmission microscopy and fluorescence microscopy were the most popular analytical tools for early microfluidic studies of bacterial biofilms. This is unsurprising because the combination of clear planar surfaces and the small internal dimensions of most microfluidic devices offer an excellent environment for undistorted, high-resolution imaging with no special requirements imposed on device fabrication or operation. In addition to supplying structural information at the micron to centimetre scale,74 optical microscopy also enables quantitative optical density measurements.11 Biofilm structural heterogeneity can also be quantified from microscope images via the coefficient of variance, fractal dimension and other image analysis techniques.74,75 Because microscope-based analysis of biofilms has become well understood and is reviewed elsewhere,1,33 we focus on certain alternative uses of optical microscopy before addressing other characterisation methods.Chemical tracers and time-lapse imaging
The main limitation of optical microscopy is that it is not directly applicable to chemical detection. Fluorescence microscopy can be transformed into a chemical imaging tool by taking advantage of the spectral properties of some fluorophores.76 One microfluidic-compatible method loads freely diffusing fluorescent pH indicators into the biofilm from the liquid phase.77–79 However, methods that avoid direct interaction between fluorescent probe molecules and the biofilm are preferable for reasons of biocompatibility and photostability. Recently, a pH-imaging microfluidic flow cell was reported that used a glass sealing layer with a layer of nanoparticles capable of metal-enhanced florescence using a pH sensitive fluorophore. Quantitative pH images of intact lactic acid biofilms (Streptococcus salivarius) were acquired with a fluorescence microscope while manipulating the hydrodynamic and chemical conditions for dental applications.80 The continued development of such nanoparticles offers promising avenues for chemical imaging of other parameters, including ionic strength, electrolyte concentration and even potentially molecular activity markers within the biofilms.81,82Optical microscopy can also be used to follow dynamic processes via time-lapse imaging with motion-tracking analyses. Such studies are readily conducted using microfluidics thanks to the ability to modulate or maintain the experimental conditions over both the short- and long-term. Typically, physiochemical processes that involve events on the order of seconds or minutes, include initial bacterial adhesion,83 response to applied forces61 and changing flow84,85 or chemical conditions.79 Slower processes are usually biological in nature, such as self-aggregation of pre-biofilm sessile cells,86,87 growth kinetics,45 changes to biofilm morphology and bacterial phenotype,88–90 gene dynamics91 and quorum sensing. Many of these phenomena require sensitivity over a wide range of time scales to characterise the entire process. For example, Fig. 3a and b show biofilm stream lengths during different stages of Pseudomonas fluorescens development from formation to failure, with characteristic time scales ranging from less than 0.1 s to hours.92 Another example is the use of a semi-automated tracking algorithm to monitor changes in viscosity of Pseudomonas sp. biofilms over a period of days under different nutrient concentrations93 or ionic strengths.94 In this work, the velocities of moving biofilm segments (e.g., Fig. 3c) were fed into a semi-empirical numerical model to obtain the time-varying viscosity. Interestingly, after a certain time (which was correlated with the ionic strength), an abrupt thickening process was observed, resulting in increases in viscosity by more than an order of magnitude over a duration of less than ten hours (Fig. 3d).94 This observation enabled the identification of the rapid viscosity transition as the likely cause of streamer formation via a sudden partial detachment mechanism observed in another microfluidic experiment.95
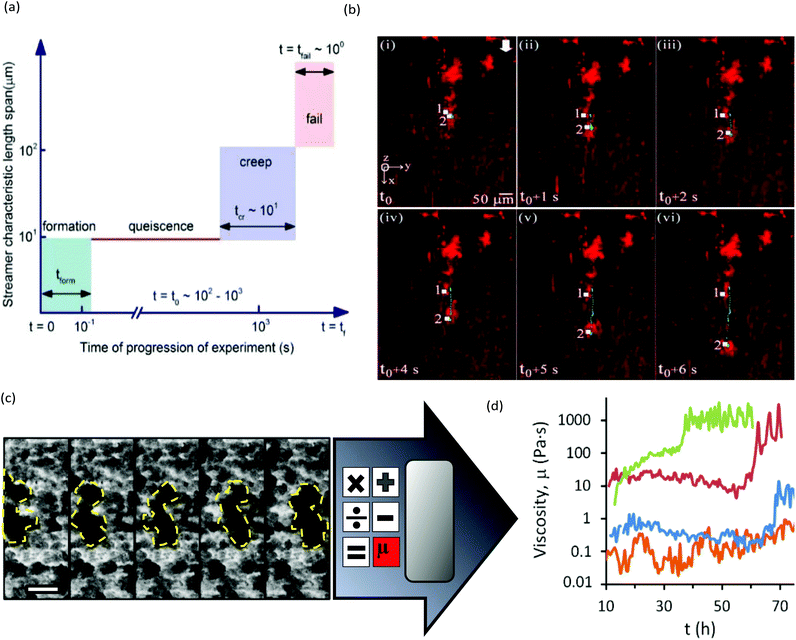 | ||
| Fig. 3 (a) Schematic of space–time scales of different regimes associated with biofilm streamer formation. (b) An example of streamer creeping motion occurring in the time segment shown in (a) immediately before failure.91 (c) Moving biofilm segment at intervals of 1 h. (d) Resulting biofilm viscosity (μbiofilm) after application of a semi-empirical model for [NaCl] at 0 wt% (orange), 0.05 wt% (blue), 0.1 wt% (red) and 0.2 wt% (green). | ||
Despite the use of chemical probes or motion tracking that expand the scope of optical microscopy using, fundamental constraints still remain. These include image resolution, dimensionality (two) and artefacts from biofilm segments outside of the focal plane. In the following section, we discuss high-resolution imaging techniques.
High-resolution microscopy
We define high-resolution microscopy as imaging techniques providing either higher spatial resolution or dimensionality than transmission or fluorescence optical microscopy. Nanoscale resolution measurements on biofilm segments and individual biofilm components can be achieved using electron and scanning probe microscopy. The most prevalent methods include scanning electron microscopy (SEM) and atomic force microscopy (AFM). Due to the nature of these approaches and the sample preparation required, they typically cannot be conducted in closed microchannels. One notable study overcame this limitation for AFM by including specialised access ports through which AFM tips could be placed in contact with hydrated biofilms during growth.96 This method could also be used in the future to introduce other scanning probes into microchannels, such as the powerful emerging technique of scanning electrochemical microscopy.97 As well, new developments in self-contained microfluidic vacuum devices may open the way for the application of SEM for in situ measurements.98 Because nanoscale resolution is not required for most biofilm studies, no strong push has been made to overcome the barriers outlined above.The arrival of widely available confocal laser scanning microscopy (CLSM) has been credited as an important factor in the re-intensification of biofilm research in the 1990s. In contrast to regular fluorescence microscopy, CLSM excites fluorophores using a scanning laser in the x–y plane while z-axis resolution is achieved with a combination of stage positioning and confocal optics. Together with image reconstruction software, the acquisition of so-called image stacks enables measurements of biofilm heterogeneity in all three dimensions, yielding more representative and information-intensive images.99 Similar to other optical microscopy techniques, CLSM is well-suited to most microfluidic environments and can accommodate chemically sensitive fluorescent probes and motion-tracking analysis, as discussed above. Although CLSM is fundamentally diffraction-limited it is included among the high-resolution techniques because the focused laser source and the confocal aperture enable practical enhancements to resolution and image dimensionality compared to most optical microscopy methods. As voxels are scanned one-by-one, the addition of a grating and array detector also enables emission-side spectral imaging as a standard option for most CLSM instruments.
A useful application of CLSM is the study of streamers, which have notably distinctive three-dimensional patterns and can form efficiently due to laminar secondary flow streams near microstructures2 or at channel turning points.68 For example, with the use of bacteria carrying different fluorophores, CLSM enabled the study of the trapping kinetics of planktonic bacteria via pre-established streamers in a serpentine microchannel (Fig. 4a). Another application combined time-lapse CLSM imaging at the attachment surface with electrochemical and vibrational spectroscopy to confirm a restructuring process in Pseudomonas fluorescens biofilms which was responsible for the formation of mushroom structures at the early growth stages.100 Other reports include the use of CLSM to assess the effect of shear forces on adhesion, structure, and planktonic cell proliferation,101 the effect of Reynolds number on bacteria distribution within biofilms47 and high-throughput assessment of antimicrobials.102 CLSM is also beneficial for examination of multi-species biofilms due to its high spatial resolution and ability to identify different bacterial lines expressing different fluorescent proteins. This aspect is regularly exploited in microfluidic studies to promote multispecies biofilm studies under controlled growth conditions for quorum sensing103 and multispecies synergistic interactions.104 For example, using a microfluidic flow velocity gradient device, it was shown that the competition between bacteria in mixed species biofilms composed of Pseudomonas aeruginosa and Flavobacterium sp. was highly dependent on the local flow velocity (Fig. 4b).106
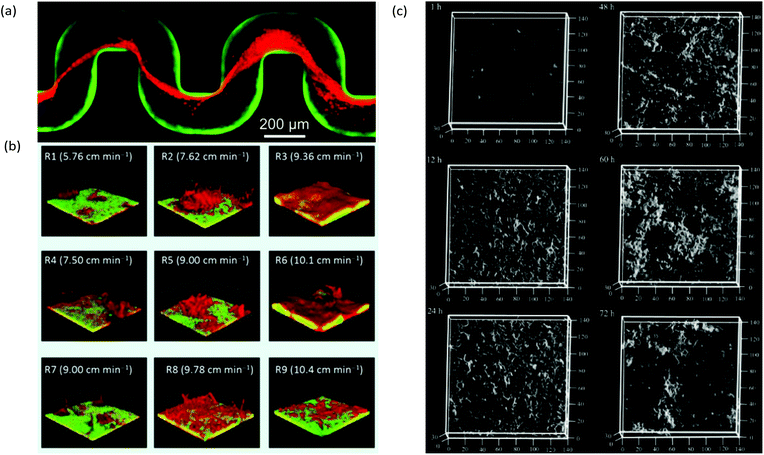 | ||
| Fig. 4 (a) Captured Pseudomonas aeruginosa bacteria (red) by pre-formed streamers in serpentine channels from a biofilm of the same type from previous inoculation and growth (green).68 (b) Dual-species 7-day-old biofilms of Pseudomonas aeruginosa (green) and Flavobacterium sp. (red) under a range of local flow rates showing the representative biofilm morphology at each of the nine regions in a microfluidic flow velocity gradient system.105 (c) Confocal reflection microscopy images of S. mutans NBRC13955 biofilm growth in a microfluidic device at different growth times.107 | ||
Drawbacks to CLSM include cost, portability and potential for background signals due to autofluorescence from some microfluidic device materials under laser irradiation. In addition, competition between the high resolution and low working distance in most objective lenses can limit the depth of observation, resulting in certain regions of the biofilm becoming inaccessible unless the spatial resolution is reduced. Additionally, attenuation of laser excitation or fluorescence emission within the biofilm interior can cause artificially low intensities relative to the local number of fluorescent species. Therefore, photon intensity is usually treated in a qualitative manner, despite the potential for quantitative information. Multiphoton systems operating with longer-wavelength lasers can reduce this problem but at an increased cost. Another drawback is difficulty in observing the EPS without the use of specialised staining methods,106 which can affect metabolic activity. This drawback has been addressed by confocal reflection microscopy, which uses the same optical setup as CLSM but does not require the presence of fluorophores for detection. This method has been demonstrated for biofilm studies in microchannels that generated high-quality topographical images of biofilms at during growth (Fig. 4c).107 Finally, microscopy generally cannot perform chemical characterisation directly on the unmodified biofilm. Solutions to this missing element are discussed in the following sections on in situ electrochemical, spectroscopic and other emerging analytical techniques.
Electrochemical measurements
Electrode integration remains at the heart of the movement towards highly integrated, system-level microfluidic platforms, with examples in biosensing, point-of-care diagnostics, sample manipulation, high current applications, imaging, and other areas.108–112 These technical successes are attributed to the diverse backgrounds of microfluidic community researchers, and a wide range of suitable materials and microfabrication techniques for creation and integration of miniaturised electrodes.113–115 Therefore, the stage has been set for innovative electrochemical studies of biofilms in microchannels. In general, these studies focus on electroactive biofilms, which are targeted as functional materials intended for used in bioelectrochemical systems (BES) for sustainable energy, bioremediation and also for new “living sensors”. In addition, certain electrochemical techniques such as electrochemical impedance spectroscopy (EIS) are applicable to non-faradaic biofilms. In the following sections, we discuss two- and three-electrode microfluidic biofilm growth cells separately because of the differences in the applicable electrochemical techniques for each case.Two-electrode systems
Typical two-electrode BES include microbial fuel cells (MFC), which produce usable electric current via the oxidation of dissolved organic molecules. This process is accomplished by an anode-adhered electroactive biofilm in which extracellular electron transfer is enhanced, thereby efficiently completing the Krebs cycle. Typical electroactive biofilms include those from Geobacter and Shewanella genera. MFCs are considered a sustainable energy technology because bioremediation is achieved while energy is produced.MFCs were first reported over 90 years ago116 and are currently one of the most significant biofilm applications in microfluidics. Laminar flow isolation of anolyte and catholyte solutions is possible in microfluidic MFCs (Fig. 5a). This reduces the internal resistance of the device and decreases the cost and maintenance compared to microfluidic MFCs that use proton exchange membranes (Fig. 5b).117,118 Characterisation of two-electrode microfluidic MFCs is easily accomplished by measuring the voltage across an external resister (Rext) between the anode and cathode (e.g., Fig. 5a). The voltage measurements can be converted to current using the familiar Ohm's law, V = IRext. The polarization curve shows the voltage drop vs. current for different Rext. The power output curve vs. current are calculated from the same data used to generate polarization curves with P = V2/Rext. Fig. 5c shows typical power and polarization curves plotted in the same figure from a 1.5 μL MFC containing Shewanella oneidensis strain MR-1.119 This single figure can reveal the maximum power output plus information on internal resistances, including quantification of different modes of energy losses. Such measurements have been used to demonstrate flow-induced performance improvements in microfluidic MFCs (Fig. 5d).120–124 Because the biofilms are constantly in contact with fresh nutrient media, constant growth conditions are maintained, unlike in batch reactors where continuous consumption of nutrients results in varying outputs and requires periodic replacement of the liquid phase.
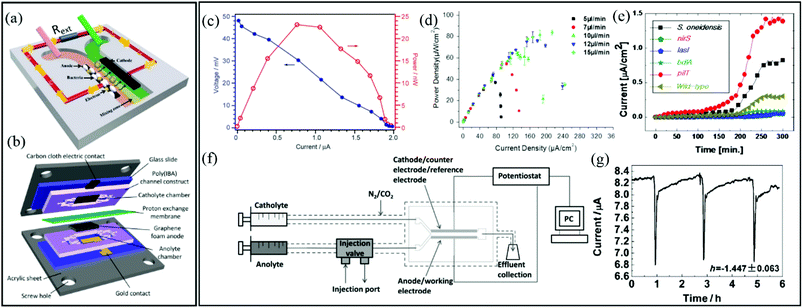 | ||
| Fig. 5 Two-electrode electrochemical microflow cells including (a) a membraneless microfluidic microbial fuel cell, with external resistor, Rext, connecting the anode and cathode117 and (b) a membrane-based microfluidic microbial fuel cell. (c) Polarization curve (blue) and power output curve (red) for a microfluidic MFC.119 (d) Power density curves for a microfluidic MFC at different flow rates.122 (e) Current density versus time growth curves of six different electroactive biofilms during a parallel growth experiment.126 (f) Two-electrode bioelectrochemical device with special injection valve used to record the effect of different chemical compounds on the output current from an anode-adhered Geobacter biofilm. (g) Perturbations to current output from device shown in (f) during 2 min applications of sodium fumarate.50 | ||
Microfluidic MFCs can also been used as analytical platforms in which outputs from electroactive biofilms are screened for electrochemical activity.125,126 For example, in one study, the small scale was exploited to parallelise multiple microchannels and enabled rapid assays of electrochemical activity from six different electroactive biofilms, two from wild type (S. oneidensis and P. aeruginosa) and four from genetically engineered (P. aeruginosa) bacteria (Fig. 5e).126 This design increased throughput and also eliminated problems that can arise in sequential experiments due to batch-to-batch differences in nutrient solutions, temperature profiles, pumping inconsistencies, etc. In another class of applications, mature electroactive biofilms exposed to different chemical or biological analytes can convert the MFCs into living biosensors. In addition to high-throughput screening, microfluidics promotes rapid response times and excellent control over concentrations and analyte application times. Therefore, overexposure of the biofilm to certain analytes can be avoided, preventing changes in baseline outputs under control conditions, thereby improving accuracy.118,127 For example, using a two-electrode microfluidic device with a special injection port on the anode side (Fig. 5f), real-time quantitative analysis of the electrochemical activity of a Geobacter sulfurreducens biofilm was performed during application of various chemical stimuli and toxins.50a Accurate pulsation in current was induced by a periodic 2 min exposure to sodium fumarate. The short and accurate exposure time led to complete reversibility after approximately 3 h (Fig. 5g). Recently a useful advancement in microfluidic MFCs was reported whereby PDMS devices were protected with parylene C coatings. This reduced oxygen permeability, supporting bench-top experiments involving anaerobic electroactive biofilms such as Geobacter.50b
Three-electrode systems
In three-electrode systems, the current flowing between the working and counter electrodes is usually measured while the working electrode potential is set relative to a reference electrode. This approach increases the number of applicable electrochemical techniques, using application of static potential (e.g., chronoamperometry, CA), swept potential (e.g., cyclic voltammetry, CV) and variable frequency AC voltages (e.g., EIS). These systems offer the potential for sophisticated investigation of charge transfer and reaction kinetics as well as isolation and identification of the different electrical resistances present in the system. In microfluidic channels, a precise laminar flow is directed across electrode-adhered biofilms to control mass transport. Thus microfluidic three-electrode devices offer a cost-effective alternative to hydrodynamic voltammetry with conventional rotating disk electrodes (RDE)128–130 while reducing the experimental footprint and improving control over the liquid flow against the biofilm. It is important to acknowledge that accurate results require integrating a stable reference electrode into microfluidic channels. A recent publication shows a microfluidic three-electrode electrochemical cell with an Ag/AgCl reference electrode in a three-electrode configuration.73 A Y-junction was used to produce side-by-side co-flowing streams, enabling isolation of the analyte stream against the working and counter electrodes and a KCl stream in contact with the calibrated reference electrode (Fig. 6a). The co-flow setup facilitated constant application of the electrolyte solution against the reference electrode while enabling ion exchange with the analyte solution without requiring a complex setup. Using this system, biofilms from Geobacter sulfurreducens bacteria were treated as a transducer after a linear current response to concentration of an electron accepter produced a calibration curve. Application of a known concentrations of electron acceptor molecules was also then used to monitor the short- and long-term system response to addition of solutions containing toxic molecules with good repeatability (Fig. 6b). The same device enabled in situ measurements via cyclic voltammetry, showing increases in limiting current and cytochrome c signals during the initial growth stages (Fig. 6c and d, respectively).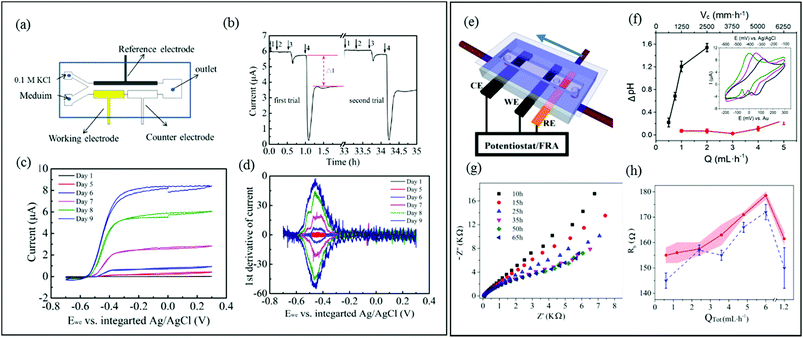 | ||
| Fig. 6 (a) Schematic of a three-electrode microfluidic device with co-flow configuration and (b) response from a working electrode-adhered Geobacter sulfurreducens biofilm showing the effect and recovery from different concentrations of a toxic compound. (c) Cyclic voltammetry results for the same Geobacter biofilm. (d) First derivative results from (c) showing the redox potential for the Geobacter cytochrome c groups over 9 days.73 (e) Schematic of a three-electrode microfluidic device with sequential electrode arrangement and the upstream placement of a Au pseudo reference electrode. Blue arrow indicates direction of flow. (f) Results from an electrochemical study of de-acidification of Geobacter sulfurreducens under flow of a standard acetate nutrient solution under turnover (red) and nutrient limited concentrations (black). Inset shows the shifting CV curves that were used to monitor flow-dependant changes to biofilm pH.131a (g) Nyquist plots obtained from EIS in the first 65 hours of growth using similar flow cell shown in (e). Changes to biofilm resistance (h) with total flow rate (QTot) modulation before (red bands) and after (blue dash lines) shear removal of biofilm upper layers.100 | ||
A different three-electrode microfluidic setup used a sequential electrode arrangement, with a gold pseudo reference electrode placed the furthest upstream (Fig. 6e).100,131a The constant flow of fresh nutrient solution ensured that constant physiochemical conditions, in particular pH, were applied to the reference electrode, resulting in a stable reference potential throughout the experiment. By calibrating the pseudo reference potential to the potential of a known reference electrode, the results can be accurately compared with other results in the literature. This simplified the setup, avoiding the need for complex device design or a co-flowing electrolyte streams. This design has been used to monitor the effect of flow conditions of a typical nutrient solution on the de-acidification of a Geobacter sulfurreducens biofilm (Fig. 6f).131a Electrochemical impedance spectroscopy is another powerful electrochemical technique (typically used in a three-electrode configuration) that can be applied to both electroactive and non-electroactive biofilms. In this approach, a low amplitude AC voltage perturbation is applied to an electrode-adhered biofilm, and the resulting AC current is recorded at different frequencies. The corresponding Nyquist or Bode plots can be modelled as an equivalent electrical circuit composed of electrical elements such as resistors, capacitors, etc.131b The electrical properties of biofilms subjected to the applied AC voltage at different frequencies can be evaluated in terms of the individual electrical elements in the equivalent model circuit, which correspond to real biophysical elements in the biofilm. Using a nearly identical microfluidic three-electrode flow cell as shown in Fig. 6e, a microfluidic EIS study was conducted to generate Nyquist plots (Fig. 6g) which capacitance and resistance of a Pseudomonas biofilm were monitored to reveal subtle changes in the biofilm biomass and structure at the electrode-biofilm interface, which were not apparent from parallel microscope measurements.100Fig. 6h shows changes to biofilm resistance with flow rate, which were interpreted as resulting from structural changes in the biofilm at electrode surface.
Spectroscopy and chemical imaging
On-chip spectroscopic studies of biofilms continue to be a niche area. However, we believe that this area will be the next to experience rapid growth. Spectral microscopy can be used to attain a global understanding of structure–function relationships by performing both morphological and chemical characterisations simultaneously.14 The importance of spectroscopic tools for biofilms is already recognised due to their ability to monitor chemical signatures on native components, including the EPS, cells, and their metabolites without significant sample preparation of addition of foreign probe molecules.130–133 In this section, we focus on vibrational spectroscopy because technical advances in integration with microfluidic systems is maturing, already leading to a number of relevant biofilm studies. With initial examples having already been accomplished at a technical level, the way is paved for implementation on biofilms.134–136 To accelerate movement in this direction, we outline the basis of operation for each technique and highlight selected achievements and challenges in application to biofilms studied in microchannels.In vibrational spectroscopy, energy exchange typically occurs between an electromagnetic field and different molecular vibrational modes of the analyte. Two popular approaches are infrared and Raman spectroscopy. Both of these techniques are useful in identifying biofilm-bound polysaccharides, proteins, DNA and RNA molecules, organic pigments (carotenoids), and lipids based on the position of the absorbance bands (Table 1).13 Measurements of band intensity can quantify the amount of each analyte present. In addition, comparison of different band intensities can yield deeper information. For example, bioactivity is an important parameter that can be quantified by considering the polysaccharide-to-protein absorbance ratios or the relative intensities of CH, CH2 and CH3 bands.137,138 Subtle changes in band position or width can reveal additional information on the biofilm and its environment but might be difficult to monitor in the crowded mid-infrared spectral window where most analysis occurs. Computer-aided analysis, including machine learning and principal component analysis, have been used in biofilm studies to find subtle correlations in complex data sets.139,140 Furthermore, as noted below, some spectral sensing can investigate the attachment surface specifically.
| Raman band (cm−1) | IR band (cm−1) | Assignment | ||||
|---|---|---|---|---|---|---|
| Polysaccharide | Proteins | DNA/RNA | Carotenoids | Lipids | ||
| 3232, 2940 | 3650–3250, 3100–2800 | O–H and N–H, free OH | O–H and N–H, free OH, C–H | — | — | O–H, C–H |
| 1750–1540 | 1760–1600 | C–C, C–O | Amide I | — | — | C–C, C–O |
| 1617–1600 | 1600–1585 | Aromatic ring | C–C Tyr, Trp | — | — | — |
| 1607 | 1500–1400 | — | C–C Try, Phe | — | — | — |
| 1580–1575 | 1550 | — | Amide II, Trp | G, A ring | — | — |
| 1460–1314 | 1450–1398 | CH2, COO−, CH | CH2, CH | — | — | CH2 |
| 1304–1200 | 1450–1250 | — | Amide III | G ring | C–H, CH2 | |
| 1160–1125, 1095–1000, 899–855 | 1152–900 | C–C, C–O, C–O–C, C–O–H, C–H | C–N, C–C | PO2− | C–CH3 | C–C, C–N, PO2− |
| 750 | — | Cytoc | T ring | — | — | |
Raman spectroscopy
Raman spectroscopy is based on photon scattering. For molecules with a high polarizability, scattering can perturb their electron configuration, and approximately 1 in 106 scattered photons undergoes a shift in energy due to induced molecular vibrations. Of note, the low polarizability of water means that water signals are largely absent from Raman spectra, which is a strong advantage over infrared (IR) spectroscopy for in situ measurements of hydrated biofilms without significant interferences. Another advantage is that visible-wavelength laser sources can easily access the transparent microfluidic channels for high spatial resolution studies, including spectral microscopy,141 even at the single-cell level and in three-dimensions.142,143However, additional attention should be focused on the strong autofluorescence from some microfluidic fabrication materials, specifically parylene and certain thermoplastics.144–147 The major drawback of Raman spectroscopy is its inherently low signal. Even biofilms accumulated at the attachment surfaces are difficult to measure due to their relatively low density and small volumes compared with that of the microfluidic device material and the liquid contained within the channel. Nevertheless, device interference can be avoided with judicious selection of isolated biofilm absorbance peaks and advanced analysis techniques. For example, without any special precautions, measurements of Pseudomonas aeruginosa biofilms in standard PSMS/glass microchannels showed statistically significant data clusters in principal component analysis that were consistent with the early-, mid- and late-stage growth phases (Fig. 7a).148 Signal enhancement via surface enhanced Raman spectroscopy (SERS) can increase the Raman signal of the biofilm signal over the background. And as SERS is a surface sensitive technique, it is well-suited for surface adhered biofilms. A method for formation of a nanostructured SERS surface in typical PDMS/glass devices was demonstrated for early stage biofilm studies in microchannels with the use of electroless deposition followed by an air-plasma treatment to achieve nanostructuration.149 In addition to increasing the sensitivity by a factor of 2.5 × 104, this technique also shielded the underlying PDMS from the probe light, thus eliminating it as a source of spectral contamination (Fig. 7b). However, as illumination and detection are usually from the sample side, the biofilm can block the light from reaching the SERS surface. This problem was avoided in another study, where chemical analysis was conducted on the effluent following its interaction with a Pseudomonas aeruginosa biofilm using a separate downstream SERS chamber. This eliminated biofilm contact with the SERS surface and simplified both the experiment and the analysis of the results (Fig. 7c).150 Another Raman spectroscopy method involves studies of biofilms containing cytochrome c heme groups, which undergo resonance enhancement with light excitation at 532 nm, a standard laser wavelength on most Raman systems. For example, this feature has been exploited for studies of biofilms formed from the Geobacter genus.150 Cytochrome c is also an indicator of bacterial phenotype151 and can be used as a marker of cellular apoptosis.152 Although not yet demonstrated for biofilm studies, hollow-core crystal fibres with sub-millimetre inner diameters could be used as micro-scale sensing channels for in situ analytical studies.153–156
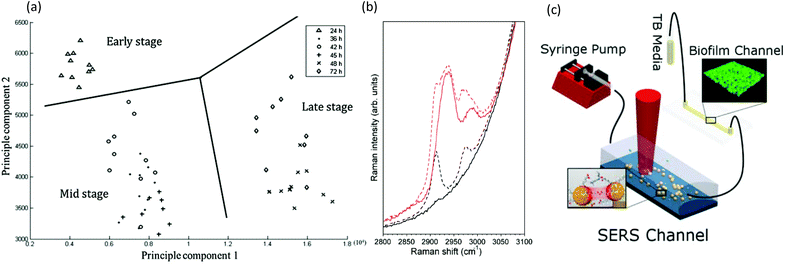 | ||
| Fig. 7 Raman spectroscopy in microchannels. (a) Representative two-dimensional principle component analysis of P. aeruginosa biofilms for identification of clustering at different developmental stages.148 (b) Spectra of 500 mM sodium citrate solution (red) and water (black) as measured using confocal Raman spectroscopy. Solid lines showing spectra acquired in channels with surfaces coated by an opaque silver layer, thereby eliminating the background signal from the PDMS wall, and with broken lines showing comparison with spectra acquired in the non-silver coated channel.149 (c) Schematic of the setup for measuring low-concentration bacterial metabolites in a SERS channel down-stream of a growing Pseudomonas aeruginosa biofilm.150 | ||
Infrared spectroscopy
Similar to Raman methods, Fourier transform infrared (FTIR) spectroscopy is sensitive to chemical signatures from biofilm EPS, microbes, and their metabolites.157,158 In this method, molecular vibrations are induced via direct photon absorption by polar molecules, leading to much higher sensitivities than in Raman spectroscopy. The main drawback in applying infrared spectroscopy to hydrated biofilms is that the signals of interest can be obscured by strong water absorption in the mid-IR region. Therefore, IR spectroscopy is often applied on dehydrated samples. Further complications arise in sealed microchannels due to absorption by the device material. To solve these problems, intense synchrotron radiation (SR-FTIR) has been demonstrated for in situ spectroscopy on hydrated living biosystems.159 The results were improved using an open microchannel design that eliminated background signals from the device and enabled controlled evaporation to limit water to the minimum possible amount while maintaining biofilm hydration (Fig. 8a).160 Another benefit was that the highly focused synchrotron beam could be rapidly rastered across the sample to produce real-time chemical maps corresponding to distribution of different EPS chemical groups (Fig. 8b). Nevertheless, due to the specialised setup, SR-FTIR is not generalisable.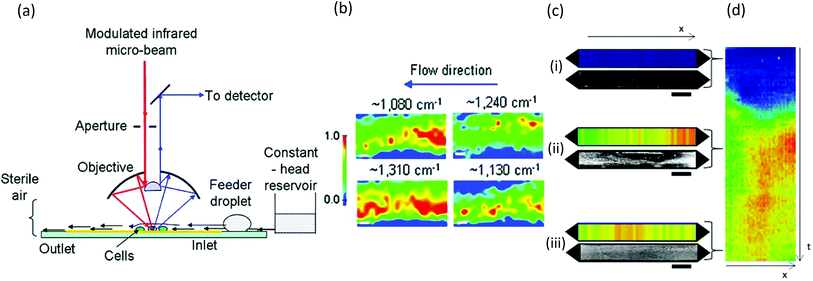 | ||
| Fig. 8 FTIR studies of biofilms in microchannels. (a) Schematic of synchrotron FTIR applied to Escherichia coli biofilms adhered to a reflective surface in an open microchannel and (b) resulting chemical maps generated from absorption bands at 1080 cm−1 (C–O–C and C–O–P vibrations in polysaccharides and PO2− vibrations in nucleic acids), 1130 cm−1 (C–O vibrations in carbohydrate glycocalyx), 1240 cm−1 (vibrations from PO2− in DNA/RNA), and 1310 cm−1 (amide III peak in proteins).160 (c) Dual imaging of growing Pseudomonas sp. biofilm growth on top of an embedded germanium multibounce ATR element in a microfluidic device consisting of linear chemical maps from absorption band at 1540 cm−1 (amide II signal from proteins) and dark-field optical microscopy after 5 h (i), 30 h (ii) and 60 h (iii). (d) Space–time image showing the variation of amide II absorbance along the channel length (horisontal) at different times (vertical). | ||
Another more accessible approach is the use of attenuated total reflection (ATR), which differentiates itself as a surface-sensitive technique, achieving a path length of less than 3 μm in hydrated samples due to the limited penetration depth of the evanescent field from the surface into the sample. Integration into microchannels has been well developed following the initial reports nearly 10 years ago,135,161 and as a surface-sensitive technique, ATR is well suited to the study of biofilms. Recently, a new approach was demonstrated in which the excitation beam was steered to different locations on a multi-bounce ATR crystal to monitor different parallel microchannels or to monitor different locations along the length of one microchannel to create one-dimensional images.162 Parallel optical imaging on the same Pseudomonas sp. biofilm was achieved by coupling with a microscope in dark-field mode (Fig. 8c). Continuous measurements in time generated position-time plots (Fig. 8d) showing trends throughout the experiment.
Other techniques and emerging approaches
Lastly, we review biofilm characterisation modalities that have a high probability of use in microfluidic studies, although they might have only recently been demonstrated or not yet even proven on-chip.Advanced microscopy
In addition to the standard techniques discussed in the first part of this paper, new high performance imaging methods hold much potential for future biofilm studies in microflow cells. For example, studies using white light interferometry and holography have been demonstrated in on- and off-chip applications and could be easily be extended to on-chip biofilm studies. Continued development is likely due to an impressive axial resolution on the nanometre scale.163–166 In addition, other “super resolution” microscopy techniques have been applied to biofilm research167 and might be applied to future on-chip studies as their integration with microfluidics matures. Particle tracking methods are also promising. One method, particle image velocimetry (PIV), is a popular technique used in the study of flow profiles. Micro-PIV is often applied to verify predictions from numerical simulations in microchannels, and is especially useful for experimental measurement of flow profiles in complex flow environments (e.g. microchannels containing biofilms) that are not easily modelled. Applications to simple agitated biofilm growth cells have been demonstrated,168 but not yet for microfluidic biofilm studies, though this is likely due to its popularity in other areas of microfluidics. Another tracking method, known as micro-rheology, monitors the Brownian motion of microparticles in different target materials as a passive monitor of the local mechanical properties. It was first applied to biofilms 10 years ago with various follow-up studies since then,169,170 including a recent on-chip study that should stimulate further work.171,172 In that study, a parallel channel microfluidic device collected particle diffusion coefficient measurements and identification of 5 different Sinorhizobium meliloti bacterial strains during the first 4 days of growth. Nanoparticle tracking analysis (NTA) is a relatively new technique that monitors the motion of nanoparticles via the light scattered by the particles in dark-field mode. The smaller size of particles in NTA compared with those used in micro-rheology can improve sensitivity, but this technique is neither well tested on biofilms in regular growth cells nor on those in microchannels.Strictly surface imaging techniques that have been used previously for biofilm research include surface plasmon resonance imaging (SPRi)173 and total internal reflection (TIR) imaging.174 Both are able to visualise sub-monolayer alterations at a sensing surface. In the case of SPRi, these are based on quantitative changes to optical density, whereas for TIR, quantitative information about cell size and distance for the surface can be obtained. While the extreme surface sensitivity can obtain high contrast images from cells and other adsorbates just a few nanometers from the surface, the lateral field of view is on the centimetre-scale, which is sufficiently large to observe long-range behaviour and heterogeneity at the same time.
Electrochemistry
Electrochemical measurements from two-electrode devices are well-established, but with the achievement of new microfluidic three-electrode configurations discussed above, it is anticipated that hydrodynamic voltammetry biofilm studies will proliferate. As well, microfluidic devices fabricated using flexible, low-cost techniques such as and material printers, printed circuit boards have enabled high electrode-density arrays and easy integration into microchannels opening the for biological applications,175 electrochemical imaging112 and other novel applications.108 Future application may include scanning electrochemical microscopy (SECM), a very versatile tool for precision three-dimensional electrochemical imaging which combines the high spatial resolution of scanning probes, such as AFM with electrochemical sensing of voltammetry techniques.97,176,177 SECM has already been demonstrated for off-chip biofilm studies,178,179 has great potential by imaging and has the potential to be used on-chip by exploiting previous advances in on-chip AFM.96Spectroscopy
Among the most promising emerging spectral techniques is nuclear magnetic resonance (NMR) and magnetic resonance imaging (MRI) applied on-chip. Much interest was generated from the announcement of on-chip NMR by “remote detection” in 2005,180,181 but we are not aware of any studies using this technique for biofilm studies. Another promising approach is MRI, which enables visualization of biofilm spatial and chemical heterogeneity via a range of in situ measurements, often within the same platform.182 These measurements include high-resolution two- and three-dimensional mapping of structures, chemical groups, liquid velocities, and diffusion coefficients as well as localised measurements by spectroscopy, relaxometry, mechanical properties and porosity.14 A recent report thoroughly demonstrated MRI capabilities on Shewanella oneidensis biofilms within a microfluidic flow cell, opening the door to a new area of microfluidic bioanalytical science.183Mass spectrometry
Mass spectrometry is growing in popularity in terms of its application to biofilm studies, identification of bacteria, biochemical groups and drug molecules.184–187 Some examples are available in the literature demonstrating the use of mass spectrometry to study biofilms in microchannels. For example time-of-flight secondary ion mass spectrometry (ToF-SIMS) has been used for chemical imaging of fragments from different fatty acid molecules from biofilm EPS and bacteria.188,189 This approach involved the use of a special microfluidic device which enabled controlled ion beam milling of a thin (ca. 100 nm) silicon nitride biofilm support layer, opening 2–3 μm diameter observation windows. While the technique causes the irreversible local alteration of the device and the biofilm, many such windows can be milled without compromising the overall integrity of the SiN layer, enabling acquisition of replicate measurements. It should be noted that in addition to ToF-SIMS similar self-contained microfluidic vacuum devices have been used for other for imaging modalities for aqueous surfaces in microchannels such as electron microscopy, NMR, CLSM and electrochemistry,183,190–194 opening the way for deeper in situ biofilm studies in the future.Respirometry
Respirometry is another emerging approach enabling the real-time monitoring of biofilm metabolic activity via detection of molecular metabolites. In such work, a biofilm is grown on a gas permeable surface, such as the inner side of a Teflon tube, while a gas analyser collects and senses changes to CO2 levels passing through to a controlled gas sheath on the other side. Researchers have used this to monitor activity of various Pseudomonas biofilms as a function of time,13a properties of the growth solution and biofilm architecture,194 as well as in multi-species biofilms.195,196 To date, these studies have been conducted in small inner diameter tubes, technically classified as millifluidic, but could be compatible with traditional microfluidic devices from PDMS, which are also known to be permeable to CO2 and other small gas molecules. Another approach to respirometry has recently been demonstrated using a calorimetry, which can measure heat released by metabolically active microorganisms. This technique has been demonstrated with on-chip measurements of biofilm activity during exposure to antibiotics, with specific benefits including fast thermal equilibrium times and low medium consumption.13bConclusions and outlooks
Bacterial biofilms are living materials with a strong propensity to change their properties as a response to environmental cues. Therefore, studying them analytically represents a great challenge. This review has advanced the argument that extracting the best, most quantitative results from powerful analytical tools and quantitative microscopy requires a parallel focus on the experimental control parameters. As such, microfluidics can be an indispensable component for modern bioanalytical platforms. In this work, we exhaustively reviewed the literature for microfluidic studies of bacterial biofilms with a focus on the historical and future trajectory of different approaches. We specifically focused on the most important on-chip characterisation techniques, including microscopy, electrochemical measurements, spectroscopy and chemical imaging. We also provided an outlook on emerging techniques that have the potential to make important impacts, including mass spectrometry and respirometry. The goal of integrating modern analytical chemistry tools in microchannels to study complex biological systems is a driver for continued technical development in microfluidics. Therefore, we can consider the state of the art in microfluidic biofilm research as a good gauge in the overall progress in microfluidic analytical platforms.Next we provide a summary comparison between the different analytical modalities discussed in this work and their ability to perform measurements of biofilm structural, chemical, mechanical, surface and metabolic properties. Table 2 organises details from the previous sections regarding the ability to meet these requirements using microscopy (all modes), electrochemistry and spectroscopy, as well as major emerging modalities, mass spectrometry and respirometry.
| Structural | Chemical | Mechanical | Surface | Metabolic activity | |
|---|---|---|---|---|---|
| Microscopy | Yes | With additional probes | Via time-lapse and micro-rheology | Via SPRi, TIR | Via Monod kinetics during growth |
| Electrochemistry | Via ESI | Yes | No | Yes | Yes |
| Spectroscopy | Via spectral imaging | Yes | Via NMR | Via ATR-FTIR, SERS | Yes |
| Mass spectrometry | Via mapping techniques | Yes | No | Via some variants, e.g. SIMS | Yes |
| Respirometry | No | Largely limited to CO2 | No | No | Yes |
Structural measurements are naturally provided by microscopy techniques, which can be quantitative with computer aided image analysis. Spatially resolved spectroscopy and some mass spectrometry techniques such as ToF-SIMS can deepen the spatial analysis by providing chemical structural maps. Some electrochemical techniques, namely EIS, can provide indirect information on biofilm structural properties based on electrical equivalence elements. Spectroscopy and mass spectrometry are the most naturally suited for direct measurements of a wide range of chemical groups simultaneously. Electrochemical measurements can open the way for targeted studies of redox active biochemical groups, such as cytochrome c proteins, via potential scan techniques. On the other hand, chemical analysis by respirometry and microscopy, are limited in their scope, being isolated to very specific biofilm metabolites (usually CO2) or based on sensitivity of optically active probe molecules or materials (e.g. pH or ionic strength). Measurements of mechanical properties are generally considered to be difficult in a microchannel due to the limitations in physically accessing the biofilm within the enclosed channel. Nevertheless, time-lapse optical microscopy combined with control over shear flow fields can provide an excellent work-around. Micro-rheology also brings a new, passive, approach to measuring a host of mechanical properties in situ. Surface measurements can be useful given the dominance of questions regarding biofilm attachment. These measurements can be readily conducted by integrating a sensing surface as one of the microchannel walls. For example, these include electrochemical measurements at an electrode surface, advance imaging techniques such as SPRi and TIR and spectroscopic measurements using an ATR element or SERS surfaces. Some mass spectroscopy, such as ToF-SIMS, can probe the attachment surface during its erosion by an ion beam. Finally, metabolic measurements are ideally made using respirometry techniques. However, almost every other analytical modality can provide insights as well. The versatility of spectroscopy and mass spectrometry enables the characterisation of metabolites, whose rate of production and accumulated concentrations within the biofilm are regularly used as proxies for metabolic activity. On the other hand, microscopy can usually only report on metabolic activity during initial growth via Monod kinetics.
In the next 3–5 years, we predict that general trends in lab-on-a-chip development will continue to be transferred to new biofilm research methodology. This includes the tendency towards more options for in situ multi-modal characterisation and high throughput analysis. Specific analytical advances for biofilm studies will likely also include a focus on chemical imaging and surface-sensitive approaches in order to match characterisation applicability to biofilm heterogeneity and surface dwelling nature, respectively. At the same time, a greater focus should be applied to parallel assaying capabilities to ensure that results are statistically significant and transferable to other studies. In the longer term, all of these advances should be integrated into a system-level configuration. This should include a streamlined user interface to achieve control over flow conditions and data acquisition, so that users can focus on scientific enquiry, without being burdened by technical details.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
J. G. is the recipient of an Early Researcher Award and an AUDACE grant (high risk, high reward) to study microbiological systems using microfluidics from the Fonds de recherche du Québec—nature et technologies (FRQNT). This research was supported by funding through the Natural Sciences and Engineering Research Council, Canada. Thanks to Molly Gregas for help with technical editing.Notes and references
- J. Kim, H.-D. Park and S. Chung, Molecules, 2012, 17, 9818–9834 CrossRef CAS PubMed.
- A. Karimi, D. Karig, A. Kumar and A. Ardekani, Lab Chip, 2015, 15, 23–42 RSC.
- M. Alles and A. Rosenhahn, Biofouling, 2015, 31, 469–480 CrossRef CAS PubMed.
- J. Dai, M. Hamon and S. Jambovane, Bioengineering, 2016, 3, 25 CrossRef PubMed.
- S. Choi, Biosens. Bioelectron., 2015, 69, 8–25 CrossRef CAS PubMed.
- S. Goel, in Microbial Fuel Cell, Springer, 2018, 229–247 Search PubMed.
- H. Ren, H. S. Lee and L. J. Chae, Microfluid. Nanofluid., 2012, 13, 353 CrossRef CAS.
- G. Sanket, Appl. Mater. Today, 2018, 11, 270–279 CrossRef.
- E. Samiei, M. Tabrizian and M. Hoorfar, Lab Chip, 2016, 16, 2376–2396 RSC.
- Y. Yang, E. Noviana, M. P. Nguyen, B. J. Geiss, D. S. Dandy and C. S. Henry, Anal. Chem., 2016, 89, 71–91 CrossRef PubMed.
- (a) R. Bakke, R. Kommedal and S. Kalvenes, J. Microbiol. Methods, 2001, 44, 13–26 CrossRef CAS PubMed; (b) C. J. Rupp, C. A. Fux and P. Stoodley, Appl. Environ. Microbiol., 2005, 71, 2175–2178 CrossRef CAS PubMed; (c) J. Mathias and P. Stoodley, Biofouling, 2009, 25, 695–703 CrossRef CAS PubMed.
- P. Zhang, Y.-P. Chen, J.-H. Qiu, Y.-Z. Dai and B. Feng, Trends Biotechnol., 2018 DOI:10.1016/j.tibtech.2018.07.006 , in press.
- (a) O. Kroukamp and G. M. Wolfaardt, Appl. Environ. Microbiol., 2009, 75, 4391–4397 CrossRef CAS PubMed; (b) F. Marianaa, F. Buchholza, J. Lerchnerb, T. R. Neuc, H. Harmsa and T. Maskowa, Int. J. Med. Microbiol., 2013, 303, 158–165 CrossRef PubMed.
- F. Paquet-Mercier, M. Safdar, M. Parvinzadeh and J. Greener, Microscopy: Advances in Scientific Research and Education, 2014, pp. 638–649 Search PubMed.
- K. Wandelt, Encyclopedia of Interfacial Chemistry: Surface Science and Electrochemistry, Elsevier, 2018 Search PubMed.
- R. M. Donlan, Emerging Infect. Dis., 2002, 8, 881 CrossRef PubMed.
- P. Thomen, J. Robert, A. Monmeyran, A.-F. Bitbol, C. Douarche and N. Henry, PLoS One, 2017, 12, e0175197 CrossRef PubMed.
- A. Azúa-Bustos, C. González-Silva, R. A. Mancilla, L. Salas, R. E. Palma, J. J. Wynne, C. P. McKay and R. Vicuña, Microb. Ecol., 2009, 58, 485–497 CrossRef PubMed.
- L. L. Jahnke, W. Eder, R. Huber, J. M. Hope, K.-U. Hinrichs, J. M. Hayes, D. J. Des Marais, S. L. Cady and R. E. Summons, Appl. Environ. Microbiol., 2001, 67, 5179–5189 CrossRef CAS PubMed.
- A.-L. Reysenbach, M. Ehringer and K. Hershberger, Extremophiles, 2000, 4, 61–67 CAS.
- A. Orell, S. Fröls and S.-V. Albers, Annu. Rev. Microbiol., 2013, 67, 337–354 CrossRef CAS PubMed.
- H.-C. Flemming, J. Wingender, U. Szewzyk, P. Steinberg, S. A. Rice and S. Kjelleberg, Nat. Rev. Microbiol., 2016, 14, 563 CrossRef CAS PubMed.
- S. C. Dexter and S.-H. Lin, Int. Biodeterior. Biodegrad., 1992, 29, 231–249 CrossRef CAS.
- C. G. Kumar and S. Anand, Int. J. Food Microbiol., 1998, 42, 9–27 CrossRef CAS PubMed.
- V. Zijnge, M. B. M. van Leeuwen, J. E. Degener, F. Abbas, T. Thurnheer, R. Gmür and H. J. Harmsen, PLoS One, 2010, 5, e9321 CrossRef PubMed.
- A. I. Terr, Med. Mycol., 2009, 47, S217–S222 CrossRef CAS PubMed.
- L. Antunes, M. Caetano and R. B. R. Ferreira, Rev. Med. Microbiol., 2011, 22, 12–16 CrossRef.
- N. Høiby, O. Ciofu and T. Bjarnsholt, Future Microbiol., 2010, 5, 1663–1674 CrossRef PubMed.
- L. Hall-Stoodley, J. W. Costerton and P. Stoodley, Nat. Rev. Microbiol., 2004, 2, 95 CrossRef CAS PubMed.
- J. W. Costerton, K. Cheng, G. G. Geesey, T. I. Ladd, J. C. Nickel, M. Dasgupta and T. J. Marrie, Annu. Rev. Microbiol., 1987, 41, 435–464 CrossRef CAS PubMed.
- B. Halan, K. Buehler and A. Schmid, Trends Biotechnol., 2012, 30, 453–465 CrossRef CAS PubMed.
- X. Z. Li, B. Hauer and B. Rosche, J. Microbiol. Biotechnol., 2013, 23, 195–204 CrossRef CAS PubMed.
- M. J. Franklin, C. Chang, T. Akiyama and B. Bothner, Microbiol. Spectrum, 2015, 3, 1–23 CAS.
- S. Haruta, T. Yoshida, Y. Aoi, K. Kaneko and H. Futamata, Microbes Environ., 2013, 28, 285–294 CrossRef PubMed.
- J. Azeredo, N. F. Azevedo, R. Briandet, N. Cerca, T. Coenye, A. R. Costa, M. Desvaux, G. Di Bonaventura, M. Hébraud, Z. Jaglic, M. Kačániová, S. Knøchel, A. Lourenço, F. Mergulhão, R. L. Meyer, G. Nychas, M. Simões, O. Tresse and C. Sternberg, Crit. Rev. Microbiol., 2017, 43, 313–351 CrossRef CAS PubMed.
- H. Ceri, M. Olson, C. Stremick, R. Read, D. Morck and A. Buret, J. Clin. Microbiol., 1999, 37, 1771–1776 CAS.
- J. Kim, H.-S. Kim, S. Han, J.-Y. Lee, J.-E. Oh, S. Chung and H.-D. Park, Lab Chip, 2013, 13, 1846–1849 RSC.
- Y. Liu and J.-H. Tay, Water Res., 2002, 36, 1653–1665 CrossRef CAS PubMed.
- P. Stoodley, J. Wefel, A. Gieseke and C. von Ohle, J. Am. Dent. Assoc., JADA, 2008, 139, 1182–1190 CrossRef CAS PubMed.
- A. Brito and L. Melo, Water Res., 1999, 33, 3673–3678 CrossRef CAS.
- D. De Beer, P. Stoodley and Z. Lewandowski, Water Res., 1996, 30, 2761–2765 CrossRef CAS.
- V. T. Nguyen, E. Morgenroth and H. Eberl, Water Sci. Technol., 2005, 52, 167–172 CrossRef.
- H. Eberl, C. Picioreanu, J. Heijnen and M. Van Loosdrecht, Chem. Eng. Sci., 2000, 55, 6209–6222 CrossRef CAS.
- E. Lauga, W. R. DiLuzio, G. M. Whitesides and H. A. Stone, Biophys. J., 2006, 90, 400–412 CrossRef CAS PubMed.
- N. B. Aznaveh, M. Safdar, G. Wolfaardt and J. Greener, Lab Chip, 2014, 14, 2666–2672 RSC.
- M. J. Vieira, L. F. Melo and M. M. Pinheiro, Biofouling, 1993, 7, 67–80 CrossRef CAS.
- M. O. Pereira, M. Kuehn, S. Wuertz, T. Neu and L. F. Melo, Biotechnol. Bioeng., 2002, 78, 164–171 CrossRef CAS PubMed.
- W. Zhang, T. S. Sileika, C. Chen, Y. Liu, J. Lee and A. I. Packman, Biotechnol. Bioeng., 2011, 108, 2571–2582 CrossRef CAS PubMed.
- R. Rusconi, S. Lecuyer, L. Guglielmini and H. A. Stone, J. R. Soc., Interface, 2010, 7, 1293–1299 CrossRef PubMed.
- (a) Z. Li, A. Venkataraman, M. A. Rosenbaum and L. T. Angenent, ChemSusChem, 2012, 5, 1119–1123 CrossRef CAS PubMed; (b) J. Y. Yoon, Y. Ahn and U. Schröder, J. Power Sources, 2018, 398, 209–214 CrossRef CAS.
- K. Lee, C. Kim, Y. Kim, B. Ahn, J. Bang, J. Kim, R. Panchapakesan, Y.-K. Yoon, J. Y. Kang and K. W. Oh, Microfluid. Nanofluid., 2011, 11, 75–86 CrossRef CAS.
- D. Voicu, C. Scholl, W. Li, D. Jagadeesan, I. Nasimova, J. Greener and E. Kumacheva, Macromolecules, 2012, 45, 4469–4475 CrossRef CAS.
- M. A. Unger, H.-P. Chou, T. Thorsen, A. Scherer and S. R. Quake, Science, 2000, 288, 113–116 CrossRef CAS PubMed.
- G. Wegner, N. Allard, A. Al Shboul, M. Auger, A.-M. Beaulieu, D. Bélanger, P. Bénard, I. Bilem, M. Byad and F. Byette, Functional Materials: For Energy, Sustainable Development and Biomedical Sciences, Walter de Gruyter GmbH & Co KG, 2014 Search PubMed.
- J. Atencia and D. J. Beebe, Nature, 2004, 437, 648 CrossRef PubMed.
- T. M. Squires and S. R. Quake, Rev. Mod. Phys., 2005, 77, 977 CrossRef CAS.
- R. Rusconi, S. Lecuyer, N. Autrusson, L. Guglielmini and H. A. Stone, Biophys. J., 2011, 100, 1392–1399 CrossRef CAS PubMed.
- M. Hassanpourfard, R. Ghosh, T. Thundat and A. Kumar, Lab Chip, 2016, 16, 4091–4096 RSC.
- J. Bruchmann, K. Sachsenheimer, B. E. Rapp and T. Schwartz, PLoS One, 2015, 10, e0117300 CrossRef PubMed.
- S. Yazdi and A. M. Ardekani, Biomicrofluidics, 2012, 6, 044114 CrossRef PubMed.
- M. Meyer, S. Subramanian, Y. Kim, H. Ben-Yoav, M. Gnerlich, K. Gerasopoulos, W. Bentley and R. Ghodssi, J. Micromech. Microeng., 2015, 25, 095003 CrossRef.
- D. N. Hohne, J. G. Younger and M. J. Solomon, Langmuir, 2009, 25, 7743–7751 CrossRef CAS PubMed.
- J. Kim, M. Hegde, S. H. Kim, T. K. Wood and A. Jayaraman, Lab Chip, 2012, 12, 1157–1163 RSC.
- S. H. Hong, M. Hegde, J. Kim, X. Wang, A. Jayaraman and T. K. Wood, Nat. Commun., 2012, 3, 613 CrossRef PubMed.
- H. Jang, R. Rusconi and R. Stocker, NPJ Biofilms Microbiomes., 2017, 3, 6 CrossRef PubMed.
- S. Khodaparast, M. K. Kim, J. E. Silpe and H. A. Stone, Environ. Sci. Technol., 2017, 51, 1340–1347 CrossRef CAS PubMed.
- F. Asayesh, M. P. Zarabadi and J. Greener, Biomicrofluidics, 2017, 11, 064109 CrossRef PubMed.
- K. Drescher, Y. Shen, B. L. Bassler and H. A. Stone, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 4345–4350 CrossRef CAS PubMed.
- T. H. R. Niepa, L. Hou, H. Jiang, M. Goulian, H. Koo, K. J. Stebe and D. Lee, Sci. Rep., 2016, 6, 30578 CrossRef CAS PubMed.
- C. B. Chang, J. N. Wilking, S. H. Kim, H. C. Shum and D. A. Weitz, Small, 2015, 11, 3954–3961 CrossRef CAS PubMed.
- Z. Zhang, P. Boccazzi, H.-G. Choi, G. Perozziello, A. J. Sinskey and K. F. Jensen, Lab Chip, 2006, 6, 906–913 RSC.
- F. Asayesh, M. P. Zarabadi, N. B. Aznaveh and J. Greener, Anal. Methods, 2018, 10, 4579–4587 RSC.
- F. Li, Z. Zheng, B. Yang, X. Zhang, Z. Li and L. Lei, Electrochim. Acta, 2016, 199, 45–50 CrossRef CAS.
- Z. Lewandowski and H. Beyenal, Fundamentals of biofilm research, CRC press, 2013 Search PubMed.
- Z. Lewandowski, D. Webb, M. Hamilton and G. Harkin, Water Sci. Technol., 1999, 39, 71–76 CrossRef.
- D. Wencel, T. Abel and C. McDonagh, Anal. Chem., 2013, 86, 15–29 CrossRef PubMed.
- R. C. Hunter and T. J. Bevridge, Appl. Environ. Microbiol., 2005, 71, 2501–2510 CrossRef CAS PubMed.
- A. L. Rios, J. Wierzchos, L. G. Sancho and C. Ascaso, Environ. Microbiol., 2003, 5, 231–237 CrossRef PubMed.
- A. E. Franks, K. P. Nevin, H. Jia, M. Izallalen, T. L. Woodard and D. R. Lovley, Energy Environ. Sci., 2009, 2, 113–119 RSC.
- M. P. Gashti, J. Asselin, J. Barbeau, D. Boudreau and J. Greener, Lab Chip, 2016, 16, 1412–1419 RSC.
- J. Asselin, M.-P. Lambert, N. Fontaine and D. Boudreau, ChemComm, 2017, 53, 755–758 RSC.
- D. Brouard, O. Ratelle, J. Perreault, D. Boudreau and M. St-Louis, Vox Sang., 2015, 108, 197–204 CrossRef CAS PubMed.
- N. Debnath, M. Hassanpourfard, R. Ghosh, T. Thundat, M. Sadrzadeh and A. Kumar, Soft Matter, 2017, 13, 8698–8705 RSC.
- C. J. Rupp, C. A. Fux and P. Stoodley, Appl. Environ. Microbiol., 2005, 71, 2175–2178 CrossRef CAS PubMed.
- T. Shaw, M. Winston, C. J. Rupp, I. Klapper and P. Stoodley, Phys. Rev. Lett., 2004, 93, 098102 CrossRef CAS PubMed.
- L. De La Fuente, T. J. Burr and H. C. Hoch, Appl. Environ. Microbiol., 2008, 74, 5579–5582 CrossRef CAS PubMed.
- K. A. Nolte, J. Schwarze and A. Rosenhahn, Biofouling, 2017, 33, 531–543 CrossRef CAS PubMed.
- A. Valiei, A. Kumar, P. P. Mukherjee, Y. Liu and T. Thundat, Lab Chip, 2012, 12, 5133–5137 RSC.
- A. Meyer, J. A. Megerle, C. Kuttler, J. Müller, C. Aguilar, L. Eberl, B. A. Hense and J. O. Rädler, Phys. Biol., 2012, 9, 026007 CrossRef PubMed.
- M. K. Kim, F. Ingremeau, A. Zhao, B. L. Bassler and H. A. Stone, Nat. Microbiol., 2016, 1, 15005 CrossRef CAS PubMed.
- J. C. Locke and M. B. Elowitz, Nat. Rev. Microbiol., 2009, 7, 383 CrossRef CAS PubMed.
- I. Biswas, R. Ghosh, M. Sadrzadeh and A. Kumar, Sci. Rep., 2016, 6, 32204 CrossRef CAS PubMed.
- J. Greener, M. Parvinzadeh Gashti, A. Eslami, M. Zarabadi and S. Taghavi, Biomicrofluidics, 2016, 10, 064107 CrossRef CAS PubMed.
- F. Paquet-Mercier, M. P. Gashti, J. Bellavance, S. M. Taghavi and J. Greener, Lab Chip, 2016, 16, 4710–4717 RSC.
- M. Parvinzadeh Gashti, J. Bellavance, O. Kroukamp, G. Wolfaardt, S. M. Taghavi and J. Greener, Biomicrofluidics, 2015, 9, 041101 CrossRef PubMed.
- A. P. Mosier, A. E. Kaloyeros and N. C. Cady, J. Microbiol. Methods, 2012, 91, 198–204 CrossRef PubMed.
- J. L. Connell, J. Kim, J. B. Shear, A. J. Bard and M. Whiteley, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 18255–18260 CrossRef CAS PubMed.
- L. Yang, X.-Y. Yu, Z. Zhu, M. J. Iedema and J. P. Cowin, Lab Chip, 2011, 11, 2481–2484 RSC.
- S. Schlafer and R. L. Meyer, J. Microbiol. Methods, 2017, 138, 50–59 CrossRef PubMed.
- M. P. Zarabadi, F. o. Paquet-Mercier, S. J. Charette and J. Greener, Langmuir, 2017, 33, 2041–2049 CrossRef CAS PubMed.
- E. Bester, G. M. Wolfaardt, N. B. Aznaveh and J. Greener, Int. J. Mol. Sci., 2013, 14, 21965–21982 CrossRef PubMed.
- W. C. Nance, S. E. Dowd, D. Samarian, J. Chludzinski, J. Delli, J. Battista and A. H. Rickard, J. Antimicrob. Chemother., 2013, 68, 2550–2560 CrossRef CAS PubMed.
- W. Timp, U. Mirsaidov, P. Matsudaira and G. Timp, Lab Chip, 2009, 9, 925–934 RSC.
- A. Culotti and A. I. Packman, PLoS One, 2014, 9, e107186 CrossRef PubMed.
- W. Zhang, T. Sileika and A. I. Packman, FEMS Microbiol. Ecol., 2013, 84, 344–354 CrossRef CAS PubMed.
- T. R. Neu and J. R. Lawrence, in Methods Enzymol, Elsevier, 1999, vol. 310, pp. 145–152 Search PubMed.
- Y. Yawata, K. Toda, E. Setoyama, J. Fukuda, H. Suzuki, H. Uchiyama and N. Nomura, J. Biosci. Bioeng., 2010, 110, 377–380 CrossRef CAS PubMed.
- D. G. Rackus, M. H. Shamsi and A. R. Wheeler, Chem. Soc. Rev., 2015, 44, 5320–5340 RSC.
- T. Z. Jubery, S. K. Srivastava and P. Dutta, Electrophoresis, 2014, 35, 691–713 CrossRef CAS PubMed.
- M. Abdelgawad and A. R. Wheeler, Adv. Mater., 2009, 21, 920–925 CrossRef CAS.
- E. V. Fanavoll, D. A. Harrington, S. Sunde, G. Singh and F. Seland, Electrochim. Acta, 2017, 225, 69–77 CrossRef CAS.
- A. Kara, A. Reitz, J. Mathault, S. Mehou-Loko, M. A. Amirdehi, A. Miled and J. Greener, Lab Chip, 2016, 16, 1081–1087 RSC.
- M. Wei, F. Zhang, W. Wang, P. Alexandridis, C. Zhou and G. Wu, J. Power Sources, 2017, 354, 134–147 CrossRef CAS.
- M. Hourmand, A. A. Sarhan and M. Sayuti, Int. J. Adv. Des. Manuf. Technol., 2017, 91, 1023–1056 CrossRef.
- R. K. L. Tan, S. P. Reeves, N. Hashemi, D. G. Thomas, E. Kavak, R. Montazami and N. N. Hashemi, J. Mater. Chem. A, 2017, 5, 17777–17803 RSC.
- B. Cohen, J. Bacteriol., 1931, 21, 18–19 CAS.
- Y. Yang, D. Ye, J. Li, X. Zhu, Q. Liao and B. Zhang, J. Power Sources, 2016, 324, 113–125 CrossRef CAS.
- R. Gupta, W. Bekele and A. Ghatak, Bioresour. Technol., 2013, 147, 654–657 CrossRef CAS PubMed.
- F. Qian, M. Baum, Q. Gu and D. E. Morse, Lab Chip, 2009, 9, 3076–3081 RSC.
- D. Vigolo, T. T. Al-Housseiny, Y. Shen, F. O. Akinlawon, S. T. Al-Housseiny, R. K. Hobson, A. Sahu, K. I. Bedkowski, T. J. DiChristina and H. A. Stone, Phys. Chem. Chem. Phys., 2014, 16, 12535–12543 RSC.
- H. Ren, S. Pyo, J.-I. Lee, T.-J. Park, F. S. Gittleson, F. C. Leung, J. Kim, A. D. Taylor, H.-S. Lee and J. Chae, J. Power Sources, 2015, 273, 823–830 CrossRef CAS.
- H. Ren, C. I. Torres, P. Parameswaran, B. E. Rittmann and J. Chae, Biosens. Bioelectron., 2014, 61, 587–592 CrossRef CAS PubMed.
- S. Choi and J. Chae, Sens. Actuators, A, 2012, 177, 10–15 CrossRef CAS.
- J. Y. Yoon, Y. Ahn and U. Schröder, J. Power Sources, 2018, 398, 209–214 CrossRef CAS.
- A. Fraiwan, D. F. Call and S. Choi, J. Renewable Sustainable Energy, 2014, 6, 023125 CrossRef.
- S. Mukherjee, S. Su, W. Panmanee, R. T. Irvin, D. J. Hassett and S. Choi, Sens. Actuators, A, 2013, 201, 532–537 CrossRef CAS.
- W. Yang, X. Wei and S. Choi, IEEE Sens. J., 2016, 16, 8672–8677 CAS.
- S. Boulêtreau, J.-Y. Charcosset, J. Gamby, E. Lyautey, S. Mastrorillo, F. Azémar, F. Moulin, B. Tribollet and F. Garabetian, Water Res., 2011, 45, 1347–1357 CrossRef PubMed.
- J. T. Babauta and H. Beyenal, Biotechnol. Bioeng., 2014, 111, 285–294 CrossRef CAS PubMed.
- D. M. Barry and P. B. McGrath, Materials, 2016, 9, 568 CrossRef PubMed.
- (a) M. P. Zarabadi, S. J. Charette and J. Greener, 2018, DOI:10.1002/celc.201800968, in press;; (b) H. Ben-Yoav, A. Freeman, M. Sternheim and Y. Shacham-Diamand, Electrochim. Acta, 2011, 56, 7780–7786 CrossRef CAS.
- M. Pan, L. Zhu, L. Chen, Y. Qiu and J. Wang, BioResources, 2016, 11, 8092–8115 CAS.
- B. Zhang and R. Powers, Future Med. Chem., 2012, 4, 1273–1306 CrossRef CAS PubMed.
- J. Greener, B. Abbasi and E. Kumacheva, Lab Chip, 2010, 10, 1561–1566 RSC.
- S. G. Kazarian, Anal. Bioanal. Chem., 2007, 388, 529–532 CrossRef CAS PubMed.
- Q. Zhou and T. Kim, Sens. Actuators, B, 2016, 227, 504–514 CrossRef CAS.
- D. Serra, A. Bosch, D. M. Russo, M. E. Rodríguez, Á. Zorreguieta, J. Schmitt, D. Naumann and O. Yantorno, Anal. Bioanal. Chem., 2007, 387, 1759–1767 CrossRef CAS PubMed.
- J. Pink, T. Smith-Palmer, T. Beveridge and D. Pink, Biofilms, 2004, 1, 157–163 CrossRef.
- B. D. Beier, R. G. Quivey and A. J. Berger, AMB Express, 2012, 2, 35 CrossRef PubMed.
- K. Loutherback, L. Chen and H.-Y. N. Holman, Anal. Chem., 2015, 87, 4601–4606 CrossRef CAS PubMed.
- A. F. Chrimes, K. Khoshmanesh, P. R. Stoddart, A. Mitchell and K. Kalantar-zadeh, Chem. Soc. Rev., 2013, 42, 5880–5906 RSC.
- C. D. Syme, N. M. Sirimuthu, S. L. Faley and J. M. Cooper, Chem. Commun., 2010, 46, 7921–7923 RSC.
- J. P. Harrison and D. Berry, Front. Microbiol., 2017, 8, 675 Search PubMed.
- K. R. Hawkins and P. Yager, Lab Chip, 2003, 3, 248–252 RSC.
- E. W. Young, E. Berthier and D. J. Beebe, Anal. Chem., 2012, 85, 44–49 CrossRef PubMed.
- B. Lu, S. Zheng, B. Q. Quach and Y.-C. Tai, Lab Chip, 2010, 10, 1826–1834 RSC.
- A. Piruska, I. Nikcevic, S. H. Lee, C. Ahn, W. R. Heineman, P. A. Limbach and C. J. Seliskar, Lab Chip, 2005, 5, 1348–1354 RSC.
- J. Feng, C. De La Fuente-Núñez, M. J. Trimble, J. Xu, R. E. Hancock and X. Lu, Chem. Commun., 2015, 51, 8966–8969 RSC.
- F. Paquet-Mercier, N. B. Aznaveh, M. Safdar and J. Greener, Sensors, 2013, 13, 14714–14727 CrossRef PubMed.
- C. Q. Nguyen, W. J. Thrift, A. Bhattacharjee, S. Ranjbar, T. Gallagher, M. Darvishzadeh-Varcheie, R. N. Sanderson, F. Capolino, K. Whiteson and P. Baldi, ACS Appl. Mater. Interfaces, 2018, 10, 12364–12373 CrossRef CAS PubMed.
- N. Lebedev, S. M. Strycharz-Glaven and L. M. Tender, Front. Energy Res., 2014, 2, 34 Search PubMed.
- M. Whiteley, M. G. Bangera, R. E. Bumgarner, M. R. Parsek, G. M. Teitzel, S. Lory and E. Greenberg, Nature, 2001, 413, 860 CrossRef CAS PubMed.
- M. Okada, N. I. Smith, A. F. Palonpon, H. Endo, S. Kawata, M. Sodeoka and K. Fujita, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 28–32 CrossRef CAS PubMed.
- A. Khetani, J. Riordon, V. Tiwari, A. Momenpour, M. Godin and H. Anis, Opt. Express, 2013, 21, 12340–12350 CrossRef PubMed.
- F. Eftekhari, A. Lee, E. Kumacheva and A. S. Helmy, Opt. Lett., 2012, 37, 680–682 CrossRef CAS PubMed.
- A. Khetani, V. S. Tiwari, A. Harb and H. Anis, Opt. Express, 2011, 19, 15244–15254 CrossRef CAS PubMed.
- Y.-P. Chen, P. Zhang, J.-S. Guo, F. Fang, X. Gao and C. Li, Chemosphere, 2013, 92, 633–638 CrossRef CAS PubMed.
- J. Schmitt and H.-C. Flemming, Int. Biodeterior. Biodegrad., 1998, 41, 1–11 CrossRef CAS.
- K. Loutherback, G. Birarda, L. Chen and H.-Y. N. Holman, Protein Pept. Lett., 2016, 23, 273–282 CrossRef CAS PubMed.
- H.-Y. N. Holman, R. Miles, Z. Hao, E. Wozei, L. M. Anderson and H. Yang, Anal. Chem., 2009, 81, 8564–8570 CrossRef CAS PubMed.
- K. A. Chan, S. Gulati, J. B. Edel, A. J. de Mello and S. G. Kazarian, Lab Chip, 2009, 9, 2909–2913 RSC.
- M. Pousti and J. Greener, Surf. Sci., 2018, 676, 56–60 CrossRef CAS.
- M. Brann, J. D. Suter, R. S. Addleman and C. Larimer, Biomicrofluidics, 2017, 11, 044113 CrossRef PubMed.
- M. Salta, L. Capretto, D. Carugo, J. A. Wharton and K. R. Stokes, Biomicrofluidics, 2013, 7, 064118 CrossRef PubMed.
- V. Bianco, M. Paturzo, O. Gennari, A. Finizio and P. Ferraro, Opt. Express, 2013, 21, 23985–23996 CrossRef CAS PubMed.
- F. C. Cheong, S. Duarte, S.-H. Lee and D. G. Grier, Rheol. Acta, 2009, 48, 109–115 CrossRef CAS.
- V. Berk, J. C. Fong, G. T. Dempsey, O. N. Develioglu, X. Zhuang, J. Liphardt, F. H. Yildiz and S. Chu, Science, 2012, 337, 236–239 CrossRef CAS PubMed.
- M. M. Salek, P. Sattari and R. J. Martinuzzi, Ann. Biomed. Eng., 2012, 40, 707–728 CrossRef PubMed.
- S. Rogers, C. Van Der Walle and T. Waigh, Langmuir, 2008, 24, 13549–13555 CrossRef CAS PubMed.
- S. C. Chew, B. Kundukad, T. Seviour, J. R. Van der Maarel, L. Yang, S. A. Rice, P. Doyle and S. Kjelleberg, mBio, 2014, 5, e01536–e01514 CrossRef PubMed.
- C. Cheng, Y. Dong, M. Dorian, F. Kamili and E. Seitaridou, Open J. Biophys., 2017, 7, 157 CrossRef.
- L. Eland, A. Wipat, S. Lee, S. Park and L. Wu, in Methods in Microbiology, Elsevier, 2016, pp. 69–111 Search PubMed.
- S. Paul, P. Vadgama and A. Ray, IET Nanobiotechnol., 2009, 3, 71–80 CrossRef CAS PubMed.
- T. Velinov, Y. Asenovska, D. Marinkova, L. Yotova, S. Stoitsova, M. Bivolarska and L. Stavitskaya, Colloids Surf., B, 2011, 88, 407–412 CrossRef CAS PubMed.
- C. Dixon, J. Lamanna and A. R. Wheeler, Adv. Funct. Mater., 2017, 27, 1604824 CrossRef.
- Scanning Electrochemical Microscopy, ed. A. J. Baird and M. V. Mirkin, CRC Press, London, UK, 2nd edn, 2012 Search PubMed.
- A. J. Wain, A. J. Pollard and C. Richter, Anal. Chem., 2014, 86, 5143–5149 CrossRef CAS PubMed.
- X. Liu, M. M. Ramsey, X. Chen, D. Koley, M. Whiteley and A. J. Bard, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 2668–2673 CrossRef CAS PubMed.
- D. Koley, M. M. Ramsey, A. J. Bard and M. Whiteley, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 19996–20001 CrossRef CAS PubMed.
- E. E. McDonnell, S. Han, C. Hilty, K. L. Pierce and A. Pines, Anal. Chem., 2005, 77, 8109–8114 CrossRef CAS PubMed.
- C. Hilty, E. E. McDonnell, J. Granwehr, K. L. Pierce, S. I. Han and A. Pines, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 14960–14963 CrossRef CAS PubMed.
- B. Manz, F. Volke, D. Goll and H. Horn, Biotechnol. Bioeng., 2003, 84, 424–432 CrossRef CAS PubMed.
- R. S. Renslow, M. J. Marshall, A. E. Tucker, W. B. Chrislera and X.-Y. Yu, Analyst, 2017, 142, 2363–2371 RSC.
- A. Gaudreau, J. Labrie, C. Goetz, S. Dufour and M. Jacques, J. Microbiol. Methods, 2018, 145, 79–81 CrossRef CAS PubMed.
- T. Si, B. Li, K. Zhang, Y. Xu, H. Zhao and J. V. Sweedler, J. Proteome Res., 2016, 15, 1955–1962 CrossRef CAS PubMed.
- X. Hua, C. Szymanski, Z. Wang, Y. Zhou, X. Ma, J. Yu, J. Evans, G. Orr, S. Liu and Z. Zhu, Integr. Biol., 2016, 8, 635–644 RSC.
- W. Wei, Y. Zhang, R. Komorek, A. Plymale, R. Yu, B. Wang, Z. Zhu, F. Liu and X.-Y. Yu, Biointerphases, 2017, 12, 05G601 CrossRef PubMed.
- X. Hua, X.-Y. Yu, Z. Wang, L. Yang, B. Liu, Z. Zhu, A. E. Tucker, W. B. Chrisler, E. A. Hill and T. Thevuthasan, Analyst, 2014, 139, 1609–1613 RSC.
- X. Hua, M. J. Marshall, Y. Xiong, X. Ma, Y. Zhou, A. E. Tucker, Z. Zhu, S. Liu and X.-Y. Yu, Biomicrofluidics, 2015, 9, 031101 CrossRef PubMed.
- L. Yang, X.-Y. Yu, Z. Zhu, T. Thevuthasan and J. P. Cowin, J. Vac. Sci. Technol., A, 2011, 29, 061101 CrossRef.
- B. Liu, X.-Y. Yu, Z. Zhu, X. Hua, L. Yang and Z. Wang, Lab Chip, 2014, 14, 855–859 RSC.
- L. Yang, X.-Y. Yu, Z. Zhu, M. J. Iedema and J. P. Cowin, Lab Chip, 2011, 11, 2481–2484 RSC.
- X.-Y. Yu, B. Liu and L. Yang, Microfluid. Nanofluid., 2013, 15, 725–744 CrossRef CAS.
- B. G. Chung, J. W. Park, J. S. Hu, C. Huang, E. S. Monuki and N. L. Jeon, BMC Biotechnol., 2007, 7, 60 CrossRef PubMed.
- E. Bester, O. Kroukamp, M. Hausner, E. A. Edwards and G. M. Wolfaardt, J. Appl. Microbiol., 2017, 110, 387–398 CrossRef PubMed.
- L. M. D. Jackson, O. Kroukamp and G. M. Wolfaardt, Front. Microbiol., 2015, 6, 953 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8an01526k |
| This journal is © The Royal Society of Chemistry 2019 |