Red-emitting pyrene–benzothiazolium: unexpected selectivity to lysosomes for real-time cell imaging without alkalinizing effect†
Chathura S.
Abeywickrama
a,
Kaveesha J.
Wijesinghe
b,
Robert V.
Stahelin
 c and
Yi
Pang
c and
Yi
Pang
 *ad
*ad
aDepartment of Chemistry, University of Akron, Akron, Ohio 44325, USA. E-mail: yp5@uakron.edu
bDepartment of Chemistry and Biochemistry, University of Notre Dame, Notre Dame, Indiana, USA
cDepartment of Medicinal Chemistry and Molecular Pharmacology, Purdue University, West Lafayette, Indiana, 47907, USA
dMaurice Morton Institute of Polymer Science, University of Akron, Akron, Ohio 44325, USA
First published on 4th March 2019
Abstract
A series of pyrene–benzothiazolium probes were synthesized. By replacing the pyridinium with a benzothiazolium unit, the selectivity of pyrene-derivatives is found to switch from nuclear to cellular lysosomes. New probes do not require proton participation and exhibit high biocompatibility and long-term imaging ability.
Lysosomes are membrane-encapsulated organelles that contain degradative enzymes to digest macromolecules within the cell.1,2 Since the discovery of lysosomes by Duve in 1955,3 the organelles are known to contain a variety of digestive enzymes called “hydrolases”, which exhibit optimal activity in a narrow acidic pH environment within the lysosomal lumen (pH 4.5–6).4 Abnormal lysosome activities or lysosome malfunction has been attributed to many disease conditions including neurodegenerative disorders, lysosome storage diseases, cancer and inflammations.4–13
Imaging lysosomes in live or fixed cell samples has recently received much attention due to their involvement in cell processing activities and cancer.12,14–17 Most probe design incorporates a basic amino group for lysosome selectivity, which include commercial LysoTracker® Green DND-26 (λex ∼ 504 nm, λem ∼ 510 nm) and LysoTracker® Red DND-99 (λex ∼ 577 nm, λem ∼ 590 nm).18 Since the existing fluorescent probes require an acidic media to operate, their accumulation in lysosomes will cause the pH increase (the “alkalinizing effect”) that perturbs the normal cell activity.19–22 Although new lysosome probes have been developed recently with spirolactams23 and peptide24 based molecules, protonation of an amine group remains an essential step, in order to guide the probe molecules into the acidic lysosome lumen. There are also interests in developing new probes that exhibit emission in the near infrared (NIR) region.24–26 However, no lysosome probes are found to operate without resorting to protonation in the lumen, which is known to alter the intracellular pH to affect normal cell activities.
As a planar molecular skeleton with extensive conjugation, pyrene has been used for developing fluorescent sensors,27–32 including those for intracellular membranes30 and mitochondria.31 Our recent study shows that probe 1 is a bright red-emitting dye for cell nucleus staining.32 The nucleus selectivity of 1 is likely achieved via binding to DNA minor grove (Fig. 1a). During the interaction, the ammonium cation is assumed to fit into the shallow space of the DNA minor grove for effective polar interaction with the DNA backbone. In an effort to examine the substituent impact on the cell staining, herein we report the synthesis of 2, where the substituent R on the benzothiazolium could provide sufficient steric hindrance to prevent the ammonium cation to fully enter the DNA minor grove, thereby decreasing its interaction with the cell nucleus. Surprisingly, compound 2 exhibited remarkable selectivity for intracellular lysosomes, in sharp contrast to 1. Since 2 has no group for protonation, its ability to generate bright red fluorescence, upon binding to lysosomes, provides a promising strategy to achieve long term tracking of lysosomes by eliminating harmful “alkalinizing effects”.
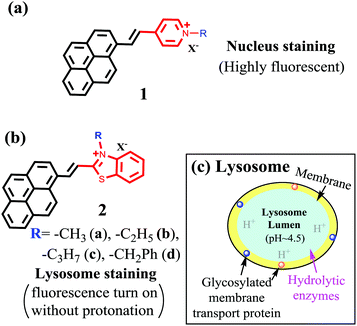 | ||
| Fig. 1 Structure of pyrene-based probes 1 (a) and (b) 2 and their selectivity switch from nucleus to lysosomes. And schematic illustration of lysosome structure (c). | ||
Synthesis: The probe 2 with different substituents was synthesized in good yields (ESI,† Fig. S1).26,32 Chemical structures of probe 2 were characterized by 1H NMR and 13C NMR spectroscopy and high-resolution mass spectrometry (ESI,† Fig. S2 and S3).
Optical properties: In dichloromethane (DCM), probes 2 exhibited λabs ≈ 530–540 nm (Table 1 and ESI,† Fig. S4), which, is significantly red-shifted from the monomeric pyrene (λabs ≈ 352 nm). The observed large bathochromic shift could be attributed to the strong intra-molecular charge transfer (ICT) from pyrene to benzothiazolium moiety in 2, which was verified by spectroscopic study at low temperature (in liquid nitrogen at −188 °C) (ESI,† Fig. S5). The probe 2 also showed bright red emission with λem ∼ 630–640 nm (ϕfl ≈ 0.22–0.48 in DMSO), along with a large Stokes’ shift (Δλ ≈ 140 nm, 4170 cm−1). Interestingly, fluorescent quantum yield of 2 was significantly lower in an aqueous environment (ϕfl ≈ 0.05) than that in a non-aqueous media (Table 1), making it possible for cell staining under “wash-free” conditions.
| Solvent | Property | Probe | |||
|---|---|---|---|---|---|
| 2a | 2b | 2c | 2d | ||
| DCM | λ abs | 530 nm | 531 nm | 532 nm | 542 nm |
| λ em | 633 nm | 632 nm | 634 nm | 641 nm | |
| (QY) | (0.15) | (0.16) | (0.12) | (0.17) | |
| DMSO | λ abs | 479 nm | 483 nm | 481 nm | 492 nm |
| λ em | 638 nm | 636 nm | 638 nm | 645 nm | |
| (QY) | (0.22) | (0.27) | (0.31) | (0.48) | |
| EtOH | λ abs | 491 nm | 495 nm | 494 nm | 507 nm |
| λ em | 627 nm | 628 nm | 628 nm | 637 nm | |
| (QY) | (0.13) | (0.15) | (0.19) | (0.21) | |
| Water | λ abs | 468 nm | 470 nm | 471 nm | 481 nm |
| λ em | 632 nm | 631 nm | 630 nm | 640 nm | |
| (QY) | (0.054) | (0.051) | (0.063) | (0.061) | |
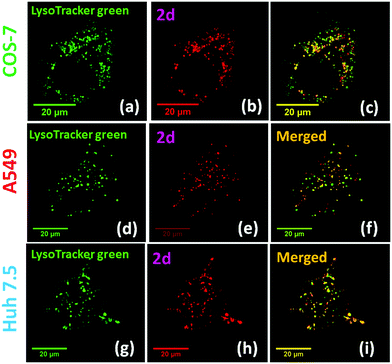
Biological cell studies: Bright red emission was observed when using 2 (500 nM) to stain different cell lines (COS-7, HEK293, A549 and Huh 7.5) (ESI,† Fig. S7). Clear cell imaging was observable after 30 minutes of staining without any post-staining washing. The non-uniform pattern observed from the cell imaging (Fig. 2) suggested that the dyes might be selectively bound to certain intracellular organelles. Possibility of binding to mitochondria was ruled out by co-staining 2 with commercial MitoTracker® Green FM on COS-7 cells (ESI,† Fig. S8). The intracellular location of 2 was eventually confirmed by co-staining with LysoTracker® Green DND-26, which showed excellent co-localization (Fig. 2 and ESI,† Fig. S16). The calculated Mander's correlation coefficients for probes 2a–2d were >0.9 in all cell lines, indicating the exceptional lysosome specificity (ESI,† Fig. S9). Consistent results were also observed in the co-localization with LysoTracker® Green DND-26 in HEK 293 cells (ESI,† Fig. S10) and in cancer cell lines (A549 and Huh 7.5), showing the probe's ability to visualize lysosome in live cells. The observed lysosome selectivity from 2 was in sharp contrast to the nucleus selectivity reported from 1,32 showing a large impact of the associated cyanine segment on the organelle selectivity.
The LC50 values for probe 2 were calculated to be in the 25–30 μM range by using CellTiter-Glow® luminescencent cell viability assay in COS-7 cells (ESI,† Fig. S22). The result further confirmed that these probes are suitable for biological cell studies since the working concentration of the probes are far below their calculated LC50 value. Bright fluorescent confocal microscopy images could be obtained when probe 2 concentration was as low as 100 nM (ESI,† Fig. S15).
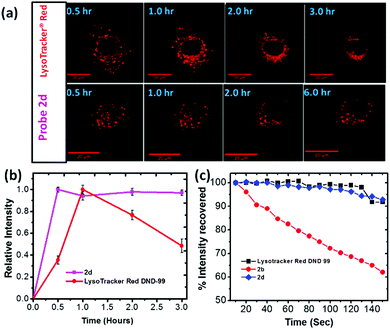
Long term imaging: Lack of cellular pH perturbation, in addition to its low toxicity, raises possibility of using 2 for long term lysosome imaging. Thus, A549 cells were treated with 2d (500 nM) over a period of 6 hours, which gave stable fluorescent confocal microscopy images (Fig. 3a and ESI,† Fig. S18). In sharp contrast, cells stained with commercial LysoTracker® Red DND-99 showed significant morphological changes and notable cell shrinkage after 1.5 hours (Fig. 4a and ESI,† Fig. S19). The fluorescence signals of 2d and LysoTracker® Red was further analyzed by plotting their average fluorescence intensity vs. staining time (Fig. 3b). No apparent intensity change were observed from 2b in A549 cells over a period of 6 hours. However, LysoTracker® exhibited a significant decrease in relative intensity with time, which could be attributed to the fluorescence quenching due to pH elevation within lysosomes (i.e. alkalinizing effect). In another experiment, COS-7 cells were incubated with probe 2d for 30 min and 24 h, and clear fluorescence signal was still observable from 2d even after 24 h in comparison with initial confocal microscopy images (ESI,† Fig. S21). The ability of 2 in giving stable fluorescence signals in intracellular environment (e.g. on lysosomes, Fig. 3b) thus opened the possibility for long term imaging, due to elimination of pH sensitivity.
Photostability: Photostability was evaluated by studying the samples of A549 cells that were stained with 2b, 2d or LysoTracker® Red for 30 minutes separately. The resulting samples were continuously irradiated with a 561 nm laser on the microscope (ESI,† Fig. S23 and S24). The fluorescence intensity, averaged by measurement on samples (n > 30), was plotted as a function of irradiation time (Fig. 3c). Probe 2d exhibited excellent stability in comparison with the commercial LysoTacker® Red, without showing significant photobleaching during the experiment period (Fig. 3c). The result also suggested that the substituent on the benzothiazolium segment had a large impact on the photostability of 2. A possible reason for poor photostability of 2b is attributed to possible photodegradation of the excited probe in the presence of molecular oxygen where singlet oxygen (1O2) or superoxide anion (O2−) upon interactions of the excited probe 2 with molecular oxygen (see ESI,† Fig. S28).33,34 Therefore, relative electron density on cyanine N atom can affect their photostability (2d exhibited a greater photostability as a result of having electron rich benzyl substituent).34
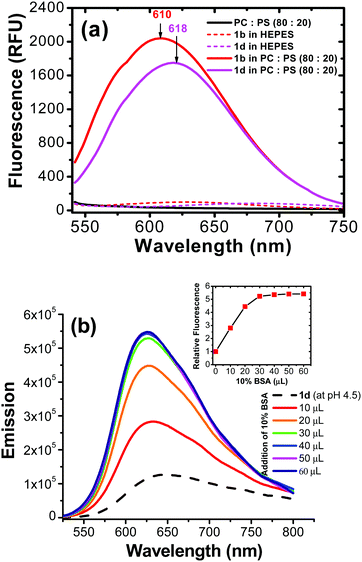
Possible probe interaction: Since the probe structure does not include a basic amino group, the intriguing question is how the probe 2 was accumulated in cellular lysosomes. Since both pyrene and benzothiazolium segments are quite inert to protonation, the acidic environment in the organelle might not promote accumulation of 2 in the lysosome lumen. The hydrophobic nature of pyrene segment thus led us to assume that probe 2 might enter hydrophobic regions (i.e., lysosomal membrane or lysosomal proteins) rather than the aqueous lysosomal lumen. One possibility is that 2 could be associated with the lysosomal membrane (hydrophobic region). In order to evaluate the hypothesis, we carried out a study involving in vitro generated lipid vesicles that can mimic an artificial phospholipid bilayer structure similar to cellular membranes. In the control experiment, probe 2 did not show any appreciable fluorescent signal in aqueous HEPES buffer (pH = 7.4). Interestingly, the pyrene-based molecules (2b and 2d) exhibited a large fluorescent turn on in the presence of liposome vesicles (Fig. 4a). This observation indicated that the possibility to associate with hydrophobic membrane components of the lysosome, generating fluorescence turn on signal. The assumption is consistent with the literature report where pyrene-based probes are able to stain biological membranes due to their hydrophobic nature.30
Fluorescence of 2 in different pH aqueous solutions revealed that emission was only slightly affected (about ±10%) in a wide pH range (pH = 3.2–11.4) (ESI,† Fig. S6). Since the fluorescence of 2 did not change significantly from neutral (pH ≈ 7.2) to acidic (pH ≈ 4.5 within lysosome lumen), in intracellular it is possible that the dye is binding to a hydrophobic region of lysosomes. In another experiment, an acidic aqueous solution of (pH = 4.5) probe 2d was titrated with 10% bovine serum albumin (BSA). Interestingly, addition of BSA led to a significant fluorescent enhancement (≈600%) (Fig. 4b). It should be pointed out that BSA binding did not cause fluorescence enhancement in commercial LysoTracker® probes, since the PET effect from an amine group would be stronger in BSA than in acidic aqueous.35 Therefore, interaction mechanism of the probe 2 in cellular lysosomes should be significantly different from that of acidotropic commercial LysoTracker® probes.
In conclusion, new fluorescent probes series have been successfully developed by coupling pyrene and benzothiazolium segments. In addition to bright red emission with large Stokes’ shift (Δλ > 130 nm), the pyrene-based probes 2 exhibited the following attractive properties that are distinctive from the existing lysosome probes: (1) low cytotoxicity (LC50 > 25 μM); (2) large fluorescence turn on in hydrophobic environments, such as hydrophobic lysosomal lipid membrane, to enable wash-free staining; and (3) stable emission throughout a wide pH range (1.0–12.0), as the probe structure is insensitive to acidity. Although their structures do not contain an amino group, the new probes (2a–2d) exhibit remarkable selectivity for visualizing lysosomes in both healthy and tumor cells. By eliminating the “alkalinizing effect”, the new probes have significant advantages over the existing commercial LysoTracker® probes and can be used for long term tracking of lysosome activity. This study points to the possibility of developing lysosome probes by targeting the lysosome membranes, rather than the acidic lysosome lumens where the enzymatic activities occur. Since probes 2 do not exhibit “alkalinizing effect” and exert minimal perturbation on the digestive activities of enzymes, they could be valuable tools for monitoring lysosome activities in biological research.
Y. P. acknowledges support from the University of Akron via Coleman endowment, and partial support from NIH (Grant no. 1R15GM126438-01A1). K. J. W. was supported by a NIH CBBI fellowship (T32GM075762). The imaging studies were supported by the Indiana University School of Medicine-South Bend Imaging and Flow Cytometry core (to R. V. S.).
Conflicts of interest
There are no conflicts to declare.References
- N. B. Yapici, Y. Bi, P. Li, X. Chen, X. Yan, S. R. Mandalapu, M. Faucett, S. Jockusch, J. Ju and K. M. Gibson, Sci. Rep., 2015, 5, 8576 CAS.
- X. Chen, Y. Bi, T. Wang, P. Li, X. Yan, S. Hou, C. E. Bammert, J. Ju, K. M. Gibson, W. J. Pavan and L. Bi, Sci. Rep., 2015, 5, 9004 CrossRef CAS PubMed.
- C. de Duve and R. Wattiaux, Annu. Rev. Physiol., 1966, 28, 435–492 CrossRef CAS PubMed.
- T. Kirkegaard and M. Jäättelä, Biochim. Biophys. Acta, Mol. Cell Res., 1793, 2009, 746–754 Search PubMed.
- H. Maes and P. Agostinis, Mitochondrion, 2014, 58–68 CrossRef CAS PubMed.
- P. Honscheid, K. Datta and M. H. Muders, Int. J. Radiat. Biol., 2014, 90, 628–635 CrossRef PubMed.
- P. Jiang and N. Mizushima, Cell Res., 2014, 24, 69 CrossRef CAS PubMed.
- X. He, J. Li, S. An and C. Jiang, Ther. Delivery, 2013, 4, 1499–1510 CAS.
- J. Reyjal, K. Cormier and S. Turcotte, Advances in Experimental Medicine and Biology, 2014, vol. 772, pp. 167–188 Search PubMed.
- F. M. Platt, Nat. Rev. Drug Discovery, 2004, 5, 642–649 Search PubMed.
- R. Ashoor, R. Yafawi, B. Jessen and S. Lu, PLoS One, 2013, 11, 82481 CrossRef PubMed.
- M. Jaattela, Oncogene, 2004, 23, 2746–2756 CrossRef PubMed.
- P. Boya and G. Kroemer, Oncogene, 2008, 27, 6434–6451 CrossRef CAS PubMed.
- T. Reinheckel, J. Deussing, W. Roth and C. Peters, Biol. Chem., 2001, 382, 735–741 CAS.
- K. N. Balaji, N. Schaschke, W. Machleidt, M. Catalfamo and P. A. Henkart, J. Exp. Med., 2002, 196, 493–503 CrossRef CAS PubMed.
- U. Felbor, B. Kessler, W. Mothes, H. H. Goebel, H. L. Ploegh, R. T. Bronson and B. R. Olsen, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 7883–7888 CrossRef CAS PubMed.
- M. E. Guicciardi, M. Leist and G. J. Gores, Oncogene, 2004, 23, 2881–2890 CrossRef CAS PubMed.
- R. P. Haugland, The handbook: a guide to fluorescent probes and labeling technologies, molecular probes, 2005 Search PubMed.
- G. Y. Wiederschain, Biochemistry, 2011, 76, 1276 CAS.
- X. Zhang, C. Wang, Z. Han and Y. Xiao, ACS Appl. Mater. Interfaces, 2014, 6, 21669–21676 CrossRef CAS PubMed.
- H. Zhu, J. Fan, Q. Xu, H. Li, J. Wang, P. Gao and X. Peng, Chem. Commun., 2012, 48, 11766–11768 RSC.
- H. M. Kim, M. J. An, J. H. Hong, B. H. Jeong, O. Kwon, J. Y. Hyon, S. C. Hong, K. J. Lee and B. R. Cho, Angew. Chem., Int. Ed., 2008, 47, 2231–2234 CrossRef CAS PubMed.
- B. Wang, S. Yu, X. Chai, T. Li, Q. Wu and T. Wang, Chem. – Eur. J., 2016, 22, 5649–5656 CrossRef CAS PubMed.
- Y. Han, M. Li, F. Qiu, M. Zhang and Y.-H. Zhang, Nat. Commun., 2017, 8, 1307 CrossRef PubMed.
- X. Zhang, C. Wang, Z. Han and Y. Xiao, ACS Appl. Mater. Interfaces, 2014, 6, 21669–21676 CrossRef CAS PubMed.
- D. Dahal, L. McDonald, X. Bi, C. Abeywickrama, F. Gombedza, M. Konopka, S. Paruchuri and Y. Pang, Chem. Commun., 2017, 53, 3697–3700 RSC.
- Y. Niko, Y. Cho, S. Kawauchi and G. Konishi, RSC Adv., 2014, 4, 36480 RSC.
- Y. Niko, S. Kawauchi and G. I. Konishi, Chem. – Eur. J., 2013, 19, 9760–9765 CrossRef CAS PubMed.
- Y. Niko, S. Kawauchi, S. Otsu, K. Tokumaru and G. I. Konishi, J. Org. Chem., 2013, 78, 3196–3207 CrossRef CAS PubMed.
- Y. Niko, P. Didier, Y. Mely, G. I. Konishi and A. S. Klymchenko, Sci. Rep., 2016, 6, 18870 CrossRef CAS PubMed.
- Y. Niko, H. Moritomo, H. Sugihara, Y. Suzuki, J. Kawamata and G. Konishi, J. Mater. Chem. B, 2015, 3, 184–190 RSC.
- C. S. Abeywickrama, K. J. Wijesinghe, R. V. Stahelin and Y. Pang, Chem. Commun., 2017, 53, 5886–5889 RSC.
- C. Chen, B. Zhou, D. Lu and G. Xu, J. Photochem. Photobiol., A, 1995, 89, 25–29 CrossRef CAS.
- X. Chen, X. Peng, A. Cui, B. Wang, L. Wang and R. Zhang, J. Photochem. Photobiol., A, 2006, 181, 79–85 CrossRef CAS.
- K. A. Bertman, C. S. Abeywickrama, H. J. Baumann, N. Alexander, L. McDonald, L. P. Shriver, M. Konopka and Y. Pang, J. Mater. Chem. B, 2018, 6, 5050–5058 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9cc01068h |
| This journal is © The Royal Society of Chemistry 2019 |

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
: