Open Access Article
Open Access ArticleFunneling aromatic products of chemically depolymerized lignin into 2-pyrone-4-6-dicarboxylic acid with Novosphingobium aromaticivorans†
Jose M.
Perez
 abc,
Wayne S.
Kontur
bc,
Manar
Alherech
bd,
Jason
Coplien
bc,
Steven D.
Karlen
abc,
Wayne S.
Kontur
bc,
Manar
Alherech
bd,
Jason
Coplien
bc,
Steven D.
Karlen
 bce,
Shannon S.
Stahl
bce,
Shannon S.
Stahl
 bd,
Timothy J.
Donohue
bd,
Timothy J.
Donohue
 bcf and
Daniel R.
Noguera
bcf and
Daniel R.
Noguera
 *abc
*abc
aDepartment of Civil and Environmental Engineering, University of Wisconsin-Madison, Madison, WI, USA. E-mail: dnoguera@wisc.edu
bDOE Great Lakes Bioenergy Research Center, Madison, WI, USA
cWisconsin Energy Institute, University of Wisconsin-Madison, Madison, WI, USA
dDepartment of Chemistry, University of Wisconsin-Madison, Madison, WI, USA
eDepartment of Biochemistry, University of Wisconsin-Madison, Madison, WI, USA
fDepartment of Bacteriology, University of Wisconsin-Madison, Madison, WI, USA
First published on 27th February 2019
Abstract
Lignin is an aromatic heteropolymer found in plant biomass. Depolymerization of lignin, either through biological or chemical means, invariably produces heterogenous mixtures of low molecular weight aromatic compounds. Microbes that can metabolize lignin-derived aromatics have evolved pathways that funnel these heterogeneous mixtures into a few common intermediates before opening the aromatic ring. In this work, we engineered Novosphingobium aromaticivorans DSM12444, via targeted gene deletions, to use its native funneling pathways to simultaneously convert plant-derived aromatic compounds containing syringyl (S), guaiacyl (G), and p-hydroxyphenyl (H) aromatic units into 2-pyrone-4,6-dicarboxylic acid (PDC), a potential polyester precursor. In batch cultures containing defined media, the engineered strain converted several of these depolymerization products, including S-diketone and G-diketone (non-natural compounds specifically produced by chemical depolymerization), into PDC with yields ranging from 22% to 100%. In batch cultures containing a heterogeneous mixture of aromatic monomers derived from chemical depolymerization of poplar lignin, 59% of the measured aromatic compounds were converted to PDC. Overall, our results show that N. aromaticivorans has ideal characteristics for its use as a microbial platform for funneling heterogeneous mixtures of lignin depolymerization products into PDC or other commodity chemicals.
Introduction
The impact of fossil carbon utilization on the global environment has encouraged the search for sustainable strategies to convert renewable resources into fuels and chemicals. Biorefining, the industrial activity of deriving fuels and chemicals from plant biomass in a sustainable and economically viable manner, is essential to reduce the proportion of fossil fuels that power the global economy. Plant biomass, the most abundant renewable organic resource on Earth, is primarily composed of sugars and phenolic compounds.3,4 While there are already established approaches to derive fuels from the sugar components of plant biomass,5 effective methods for biomass deconstruction to recover and valorize the phenolic components are only starting to emerge.6,7 One source of phenolic compounds is lignin, an alkyl-aromatic heteropolymer that is interlinked with cellulose and hemicellulose in plant cell walls and accounts for up to 30% of the total lignocellulosic biomass weight.8 There are other sources of phenolics in plant biomass, such as arabinofuranosides in grasses9,10 or lignin bound p-hydroxybenzoate in some hardwoods.11 We are interested in evaluating the potential for bio-based production of valuable chemicals from the phenolic components of plant biomass.The most abundant biomass-derived phenolics can be classified based on the number of methoxy groups attached to the main phenyl structure; these are syringyl (S; two methoxy groups), guaiacyl (G; one methoxy group), and p-hydroxyphenyl (H; no methoxy groups) units.12 Several approaches have been recently described for biomass deconstruction and lignin depolymerization that result in recovery of S, G, and H aromatic units.6 However, the heterogeneity of the resulting mixtures presents a major challenge for conversion into commodity chemicals because of the low quantity of valuable marketable compounds in deconstructed lignin samples and the technical limitations for their separation or purification from other components.7
We are exploring microbial strategies for the conversion of deconstructed lignin into commodity chemicals since microorganisms have evolved strategies to gain energy from the degradation of a large variety of aromatics compounds.13,14 Such strategies could be harnessed for the valorization of aromatic mixtures if the metabolic pathways are routed towards production of desirable chemical products.15 In general, microbial transformation of aromatic compounds occurs by a combination of upper metabolic pathways, which convert multiple compounds into key aromatic intermediates13 in what has been called “biological funneling”,16 and a central aromatic pathway that breaks the aromaticity and renders metabolic products that enter central carbon metabolism.13,14 Biological funneling has been recently described for the conversion of plant-derived phenolics to aromatic compounds such as vanillin17 and benzoic acid,18 and to non-aromatic compounds, such as cis,cis-muconate,19 β-keto adipate,20 muconolactone,20 2-pyrone-4,6-dicarboxylic acid (PDC),21,22 pyridine-2,4-dicarboxylic acid,23 and polyhydroxyalkanoates.16 Some of these approaches require extensive metabolic re-routing and introduction of foreign pathways,19,22 while others rely on a small number of mutations that redirect aromatic metabolism to the product of interest.17,18
Here we report on the impact of gene deletions in the central aromatic catabolic pathways of Novosphingobium aromaticivorans DSM12444, an organism known to degrade aromatic compounds24 and to break down interlinkages in lignin,25 that allow it to funnel a large diversity of plant-derived phenolics into PDC, a potential bioplastic and epoxy adhesives precursor.26 A complete genome sequence is available for this α-proteobacterium (GenBank NC_007794.1), and the organism is amenable to genetic and genomic techniques needed to test the role of individual genes in aromatic metabolism, and model, engineer, or improve its pathways.25 Specifically, we show that by using a defined set of mutations, N. aromaticivorans can be engineered to simultaneously produce PDC from all three major types of plant-derived phenolic compounds (S, G, and H). In addition, we find that this organism can metabolize aromatics simultaneously with the use of other organic carbon sources (such as glucose), a feature that allows mutant strains to excrete compounds derived from the incomplete metabolism of the aromatics. This work represents a valuable advance in using bacteria to funnel aromatic compounds into defined single commodities and shows that N. aromaticivorans could be an ideal microbial chassis for valorization of lignin and other plant-derived aromatics.
Results
Model of aromatic metabolism by N. aromaticivorans DSM12444 and justification of experimental approach
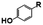
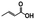
N. aromaticivorans DSM12444, a bacterium isolated from a polyaromatic hydrocarbon-contaminated sediment in the deep subsurface, aerobically utilizes a variety of aromatic compounds as sole carbon and energy sources for growth.24 Based on its genome content, a recent analysis of N. aromaticivorans aromatic metabolism using a transposon library,2 and the known metabolism of lignin-derived aromatics in the related α-proteobacterium Sphingobium sp. SYK-6,1 we propose a model for the degradation pathways of plant-derived aromatic compounds in this organism (Fig. 1). Consistent with the predicted pathways in N. aromaticivorans and Sphingobium sp. SYK-6, we propose that G and H aromatic units are degraded via protocatechuic acid (Fig. 1), with ring opening by LigAB, a 4,5 extradiol dioxygenase that yields 4-carboxy-2-hydroxy-cis,cis-muconate-6-semialdehyde (CHMS). CHMS is then oxidized to PDC by the dehydrogenase LigC. LigI is predicted to hydrolyze PDC to produce 4-oxalomesaconate (OMA),27 which is further transformed to the central carbon metabolites pyruvate and oxaloacetate (Fig. 1).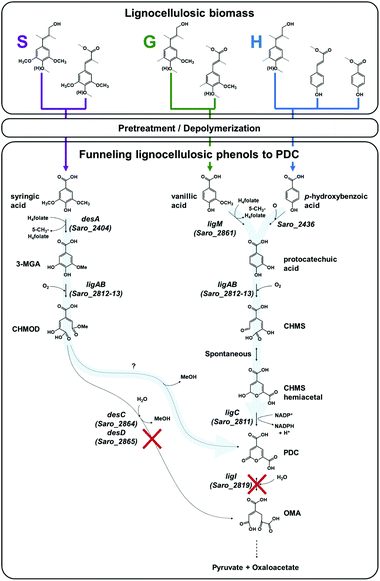 | ||
Fig. 1 Predicted pathways of S unit (syringic acid), G unit (vanillic acid), and H unit (p-hydroxybenzoic acid) metabolism in N. aromaticivorans DSM12444, based on work in Sphingobium sp. SYK-6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and N. aromaticivorans DSM12444.2 In this model, deletions of the genes ligI (Saro_2819), desC (Saro_2864), and desD (Saro_2865) are hypothesized to enable the funneling (represented by light blue arrows) of S, G, and H lignocellulosic biomass-derived aromatic compounds into 2-pyrone-4,6-dicarboxylic acid (PDC). Abbreviations: 3-methylgallate, 3-MGA; 4-carboxy-2-hydroxy-6-methoxy-6-oxohexa-2,4-dienoate, CHMOD; 4-carboxy-2-hydroxy-cis,cis-muconate-6-semialdehyde, CHMS; 4-oxalomesaconate, OMA. 1 and N. aromaticivorans DSM12444.2 In this model, deletions of the genes ligI (Saro_2819), desC (Saro_2864), and desD (Saro_2865) are hypothesized to enable the funneling (represented by light blue arrows) of S, G, and H lignocellulosic biomass-derived aromatic compounds into 2-pyrone-4,6-dicarboxylic acid (PDC). Abbreviations: 3-methylgallate, 3-MGA; 4-carboxy-2-hydroxy-6-methoxy-6-oxohexa-2,4-dienoate, CHMOD; 4-carboxy-2-hydroxy-cis,cis-muconate-6-semialdehyde, CHMS; 4-oxalomesaconate, OMA. | ||
Dimethoxylated aromatics (S aromatics) are predicted to be degraded via a separate pathway, with demethylation of syringic acid to 3-methylgallate (3-MGA) carried out by the O-demethylase DesA (Fig. 1). In N. aromaticivorans, LigAB has been proposed to catalyze ring opening to produce a mixture of stereoisomers of 4-carboxy-2-hydroxy-6-methoxy-6-oxohexa-2,4-dienoate (CHMOD); a cis–trans isomerase, DesD, isomerizes one of the stereoisomers, and the methylesterase DesC completes demethylation of CHMOD to OMA.2 Two other routes of 3-MGA degradation are proposed in Sphingobium sp. SYK-6, one requiring ring opening by the 3,4-dioxygenase DesZ and cyclization to PDC and another one requiring O-demethylation to gallate by LigM followed by ring opening by the dioxygenase DesB.1 While LigM is present in N. aromaticivorans, homologues of DesZ and DesB are not encoded in its genome.2 In addition, the LigAB of Sphingobium sp. SYK-6 has been shown to produce a combination of CHMOD and PDC when 3-MGA is the substrate,28 and there are reports of slow abiotic transformation of CHMOD to PDC.29 Therefore, in our model (Fig. 1), we hypothesize that the main enzymatic route of 3-MGA degradation in N. aromaticivorans is via CHMOD to OMA, but that PDC may also be a product of enzymatic or abiotic CHMOD transformation.
We used the above model to hypothesize which disruptions in the aromatic degradation pathways in N. aromaticivorans would lead to accumulation of specific pathway intermediates. We chose to focus on creating mutations that could lead to accumulation of PDC (Fig. 1), which is of particular interest since this dicarboxylic acid has been shown to be a suitable precursor for polyesters.30 We hypothesized that a disruption of the proposed G and H degradation pathway via the deletion of the ligI gene (Fig. 1) would prevent PDC degradation and lead to its accumulation in cultures fed G and H aromatics as substrates. Furthermore, we predicted that this metabolic disruption would result in strains with limited ability to grow on G and H aromatics, since most of the carbon in these compounds would remain in the PDC molecule. If this latter prediction is correct, then the addition of another substrate would be needed to support growth of cells on G or H aromatics lacking a functional ligI gene. In addition, given the possibility of PDC production from CHMOD (Fig. 1), we also hypothesized that deleting the desCD genes would result in accumulation of upstream intermediates and redirection of metabolism via PDC (Fig. 1).
Below we describe how we tested these hypotheses and how the defined mutations lead to PDC accumulation from (i) G and H units, (ii) S, G, and H units, and (iii) aromatics that are present in depolymerized lignin.
Construction of a N. aromaticivorans mutant that accumulates PDC from G and H aromatics
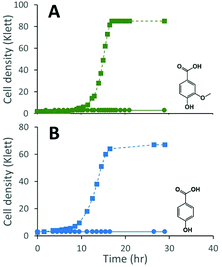
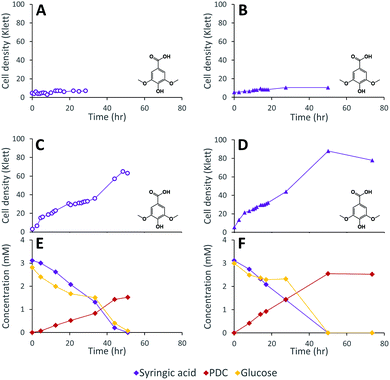
We constructed strain 12444ΔligI by deleting the ligI gene and cultured it initially in minimal media containing glucose since this gene was not predicted to be necessary for glucose metabolism. To test the role of this gene in metabolism of aromatic compounds, we attempted to grow strain 12444ΔligI on minimal media containing 3 mM vanillic acid or 3 mM p-hydroxybenzoic acid as representative of G and H aromatics, respectively. As expected, strain 12444ΔligI was unable to grow on either of these substrates as sole carbon sources (Fig. 2). When glucose was provided in addition to vanillic acid or p-hydroxybenzoic acid, strain 12444ΔligI was able to grow (Fig. 3 panels A and B), and, in both cases, glucose and the aromatic substrate were removed from the media, a small amount of protocatechuic acid transiently accumulated, and PDC accumulated as the final product of the transformations (Fig. 3 panels C and D). The PDC yield from vanillic acid and p-hydroxybenzoic acid by strain 12444ΔligI in these cultures were 81% (±17%) and 73% (±1.7%), respectively (Table 1).In theory, other G and H aromatics metabolized by N. aromaticivorans would also produce PDC when fed to strain 12444ΔligI (Fig. 1). We tested this prediction with the G aromatics vanillin and ferulic acid and the H aromatics p-hydroxybenzaldehyde and p-coumaric acid (Fig. S1† and Table 1). Cultures grown on minimum media with 3 mM vanillin plus 3 mM glucose showed transient accumulation of vanillic acid (Fig. S1A†), then a nearly stoichiometric accumulation of PDC. In the cultures grown with glucose and p-hydroxybenzaldehyde (Fig. S1B†), a transient accumulation of extracellular p-hydroxybenzoic acid and protocatechuic acid was observed, then accumulation of PDC with a 79% (±2%) yield (Table 1). Cultures grown on ferulic acid plus glucose showed a transient accumulation of vanillic acid and protocatechuic acid (Fig. S1C†), then accumulation of PDC with a 76% (±10%) yield (Table 1). Similarly, the cultures grown with p-coumaric acid and glucose transiently accumulated extracellular p-hydroxybenzoic and protocatechuic acids (Fig. S1D†), then accumulated PDC with an efficiency of 84% (±5.4%) (Table 1).
These results are consistent with transformation of G and H aromatics via the predicted pathway of Fig. 1. The observed PDC yields (Table 1) suggest that PDC is the main intermediate that accumulates, and that disruption of the ligI gene is sufficient to prevent PDC catabolism.
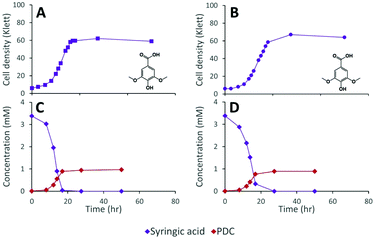
The inability of 12444ΔligI to metabolize PDC is not predicted to affect the degradation of aromatics containing S units, since the metabolism of these compounds would follow the 3-MGA, CHMOD, OMA pathway (Fig. 1). In agreement with this hypothesis, when strain 12444ΔligI was fed 3 mM syringic acid as the sole carbon source, growth of this mutant reached final cell densities similar to those of parent strain 12444Δ1879 and this aromatic was metabolized to a similar extent in both strains (Fig. 4). This observation confirms that LigI is not necessary for syringic acid degradation. However, these experiments also showed that PDC accumulates in the growth media in both cases, representing 28% (0.97 mM) and 26% (0.89 mM) of the initial concentration of syringic acid for strains 12444Δ1879 and 12444ΔligI, respectively.
Construction of an N. aromaticivorans mutant that accumulates PDC from S aromatics
Dimethoxylated phenolics, such as syringic acid, are predicted to be degraded by N. aromaticivorans via the 3-MGA, CHMOD, OMA pathway (Fig. 1). Based on this prediction, we hypothesize that deleting the desCD genes would disrupt the degradation of S aromatics (Fig. 1), leading to the accumulation of the intermediate CHMOD. However, this mutation may not be sufficient to prevent growth of N. aromaticivorans on S aromatics because CHMOD may undergo abiotic or enzymatic transformation to PDC,29 which could then be hydrolysed by LigI. Thus, to test these hypotheses, we constructed strain 12444ΔdesCD by deleting the desCD genes from strain 12444Δ1879.Growth was not observed when strain 12444ΔdesCD was cultured in minimal media with 3 mM syringic acid as the sole carbon source (Fig. 5A), indicating that either desC, desD or both genes are essential for growth on syringic acid, in agreement with observations reported previously.2 To test the 12444ΔdesCD strain for a defect in S aromatic metabolism when growth was occurring, we inoculated the strain into media containing both 3 mM glucose and 3 mM syringic acid (Fig. 5C). The 12444ΔdesCD strain grew, with consumption of both syringic acid and glucose, and with increased PDC accumulation compared to strain 12444Δ1879, converting 49% (±0.9%) of the syringic acid into PDC (versus 28% for 12444Δ1879; Fig. 4C). This suggests that increased cyclization of CHMOD to PDC took place, although this observation is not sufficient to determine whether the reaction is abiotic or enzymatic. Growth of 12444ΔdesCD on vanillic acid as the only carbon source demonstrated that the disruption in desCD does not affect the catabolism of G units and does not lead to detectable PDC accumulation (Fig. S2†).
Construction of an N. aromaticivorans mutant that accumulates PDC from S, G, and H aromatics
Based on the observations with strains 12444ΔligI and 12444ΔdesCD, we hypothesized that a mutant missing ligI and desCD would be able to produce a higher yield of PDC from S aromatics. We generated this strain (12444ΔligIΔdesCD) and found that when it was cultured in minimal media with 3 mM syringic acid as the sole carbon source, it did not grow, as expected from previously presented data (Fig. 5B). When glucose was added to the growth media, strain 12444ΔligIΔdesCD grew (Fig. 5D), glucose and syringic acid were removed from the media, and PDC accumulated (Fig. 5F). Indeed, the PDC yield of 12444ΔligIΔdesCD (66% ± 13%), was higher than that of 12444ΔdesCD (49% ± 0.9%) (Table 1).PDC production from syringaldehyde by strain 12444ΔligIΔdesCD was also tested. When this strain was grown on 1 mM syringaldehyde plus 3 mM glucose (Fig. S3E†), syringaldehyde disappeared from the growth media, syringic acid was transiently detected, and PDC accumulated with a 90% (±7%) yield (Table 1).
The fate of unconverted aromatic carbon
Since PDC yields were typically less than 100%, it is possible that some aromatic compounds are degraded via alternative routes not blocked by the ΔligI and ΔdesCD mutations, and therefore, a fraction of aromatics may be still used as carbon and energy sources for growth in strain 12444ΔligIΔdesCD. To evaluate this hypothesis, we compared cell yields in 12444ΔligIΔdesCD cultures grown on either 3 mM glucose or 3 mM glucose plus 3 mM protocatechuic acid. The cultures grown on glucose reached a final density of165 (±1) Klett units and no glucose or PDC was detected in the culture media (Table 2). The cultures receiving glucose plus protocatechuic acid reached a final cell density of 202 (±2) Klett units (Table 2). In these cultures, all glucose was consumed and 0.2 mM (±0.03) protocatechuic acid remained in the growth media (Table 2). The calculated yield of PDC based on the consumed protocatechuic acid was 85% (±1%) (Table 2). Since in both conditions the same amount of glucose was provided, the higher cell density observed in the cultures containing glucose plus protocatechuic acid can be explained by the use of a fraction of protocatechuic as a carbon and energy source for cell growth, presumably via a less efficient alternative pathway. The absence of PDC in the cultures containing only glucose shows that strain 12444ΔligIΔdesCD does not produce PDC from glucose.| Glucose | Glucose + protocatechuic acid | |
|---|---|---|
| Maximum cell density (Klett) | 165.3 (±0.58) | 201.7 (±2.08) |
| Metabolities concentration immediately after inoculation | ||
| Glucose (mM) | 3.1 (±0.02) | 3.1 (±0.04) |
| Protocatechuic acid (mM) | 0.0 | 2.9 (±0.02) |
| PDC (mM) | 0.0 | 0.0 |
| Metabolities concentration at stationary phase | ||
| Glucose (mM) | 0.0 | 0.0 |
| Protocatechuic acid (mM) | 0.0 | 0.2 (±0.03) |
| PDC (mM) | 0.0 | 2.3 (±0.04) |
| PDC yield (%) | 0.0 | 85 (±1.10) |
Production of PDC from chemically depolymerized lignin
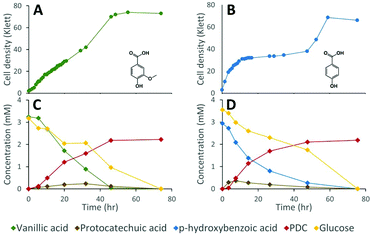
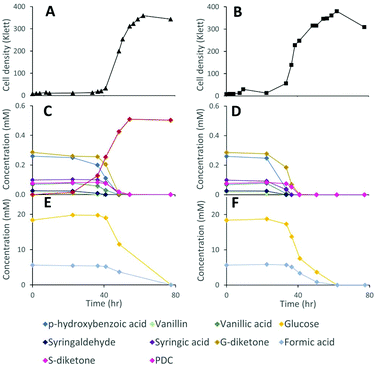
Lignocellulosic biomass pretreatment and chemical depolymerization of lignin typically result in heterogeneous mixtures of aromatics with variable molar yields of monomers recovered.6,7 Based on the above results, a strain lacking both LigI and DesCD activity might also be able to simultaneously convert all three classes (S, G, and H) of plant-derived aromatics into PDC. To test the ability of strain 12444ΔligIΔdesCD to produce PDC simultaneously from multiple S, G, and H aromatic compounds, we cultured it in glucose-containing media supplemented with the products of depolymerized poplar lignin,4 which contained a mixture of S, G, and H aromatic compounds (Fig. 6). For comparison, strain 12444Δ1879 was cultured in the same media. In addition, a flask containing the same media without cells was incubated as an abiotic control. A large proportion of the aromatic compounds present in this type of depolymerized lignin are S and G type diketones4 and no information has been previously reported about the ability of bacteria to degrade them. Thus, in the experiments below we also tested for metabolism of the S and G diketones and their potential conversion into PDC.In the abiotic control, none of the aromatic compounds were transformed after 77.5 h of incubation (Fig. S3D, S6B and S7†). In the inoculated cultures, both strains grew, and, in both cases, all the major aromatic compounds (G-diketone, S-diketone, p-hydroxybenzoic acid, vanillin, vanillic acid, syringaldehyde, and syringic acid) disappeared from the growth media (Fig. 6 and Fig. S3† panels B and C). PDC only accumulated in the 12444ΔligIΔdesCD cultures, reaching a concentration of 0.49 mM (±0.02), which corresponds to a molar yield of 59% (±1.9%) assuming that all of the above aromatics were used as a source of this compound (Fig. 6).
Gel permeation chromatography (GPC) was performed to determine the presence of, and evaluate changes in, oligomeric lignin fragments found in these depolymerized lignin samples (Fig. S6 and S7†). This analysis showed presence of compounds with a wide range of molecular weights (Mw), which we grouped in 2 ranges (see Materials and methods). Based on the analysis of standards, compounds eluting between 17.0 and 22.7 min corresponded to oligomeric lignin fragments, while compounds eluting after 22.7 min are dimeric and monomeric compounds. An abiotic control showed that during 78 hours of incubation there was an observable increase in low Mw oligomers, likely from reactive monomer condensation, that resulted in an average Mw decrease from 857 to 722 Da (Fig. S7†). Both microbial cultures showed a decrease in the dimeric and monomeric compounds (signals eluting after 22.7 min) compared to the abiotic control sample. As with the sample before incubation, both microbial cultures showed the decrease in oligomer Mw attributed to reactive monomer condensation, but not as much as in the abiotic control (Fig. S7†). Accumulation of PDC in experiments with 12444ΔligIΔdesCD was observable by a peak at 23.55 min (Fig. S6†), corresponding to that of the PDC standard, which was not observed in the abiotic control or the experiment with the parent strain 12444Δ1879.
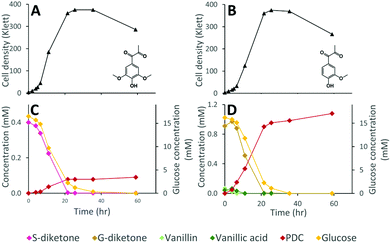
While the above data suggest that 12444ΔligIΔdesCD is able to convert the G, S, and H units found in depolymerized lignin into PDC, the lack of stoichiometric conversion into PDC makes it difficult to assess how well each substrate is metabolized and converted into this product. To specifically test PDC production from the S and G aromatic diketones, we grew cultures of N. aromaticivorans strain 12444ΔligIΔdesCD on minimum media supplemented with chemically synthesized S-diketone plus glucose or G-diketone plus glucose (see ESI† for aromatic diketone synthesis procedures). In the cultures containing S-diketone, 12444ΔligIΔdesCD grew, glucose and the aromatic diketone disappeared from the growth media, and PDC accumulated with a yield of 22.0% (±0.7%) (Table 1, Fig. 7, panels A and C). On the other hand, in the cultures supplemented with G-diketone (which contained small amounts of vanillic acid and vanillin as impurities from the synthesis method) both glucose and the aromatic substrates disappeared and PDC accumulated (Fig. 7, panels B and D), with a nearly stoichiometric yield (107% ± 1.6%, Table 1) for G-diketone (assuming a 100% yield from the vanillic acid and vanillin impurities). From this, we conclude that strain 12444ΔligIΔdesCD metabolizes these S and G diketones, using pathways that are also involved in degradation of the S, G and H aromatics normally found in lignin, and converts them into PDC, albeit at different efficiencies.
Production of PDC from vanillic acid and vanillin in a fed-batch reactor
To study the feasibility of PDC production by strain 12444ΔligIΔdesCD at titers higher than those observed in batch cultures, we cultured the mutant strain in a pH-controlled fed-batch reactor in which a concentrated solution containing vanillic acid, vanillin, and glucose was intermittently fed. In this experiment, a maximum concentration of 26.7 mM (4.9 g L−1) of PDC was reached after 48 hours of incubation (Fig. S8†), which represents a more than 8 times higher concentration than observed in the batch experiments reported here. As the reaction progressed, an accumulation of glucose, vanillic, and protocatechuic acid was observed. Further experiments are necessary to optimize bioreactor conditions to increase PDC titer and minimize substrate and intermediate metabolite accumulation.Discussion
The economic and environmental viability of producing fuels and chemicals from lignocellulose is tightly connected to the efficiency of its utilization. New methods are needed to efficiently utilize the recalcitrant aromatic fractions, such as lignin.31 Multiple chemical approaches have shown promising results for breaking down the complex lignin polymer into small molecule aromatic units.6,7 However, the heterogeneous nature of the depolymerization products obtained pose challenges for further upgrading to valuable products.32 One successful strategy to address the chemical heterogeneity is to funnel the mixture of compounds through convergent aromatic biodegradation pathways into one valuable product by interruption and/or redirection of the metabolic flow to a pathway intermediate.19,22,23 These studies suggest that a mixed approach that integrates chemical and biological tools has the potential to be an effective strategy to maximize the yield of desired products from lignin transformation. Some of the major challenges in biological funneling are the transformation of unnatural products resulting from chemical depolymerization for which microbial metabolic capabilities are unknown, the maximization of target product yield while minimizing the accumulation of undesired intermediates or end products, and the identification of industrially useful target molecules that could most readily be produced from lignin components via known metabolic pathways.15The present study addresses each of these issues using mutant strains of N. aromaticivorans DSM12444, a microbe naturally capable of degrading S, G, and H type aromatic compounds, as a well as lignin derived aromatic dimers.25,33 We chose N. aromaticivorans DSM12444 due to its known or predicted ability to grow in the presence of multiple aromatic compounds, its suitability for genetic analysis and modification, its ability to co-metabolize aromatics in the presence of other organic compounds (such as sugars, which are another plentiful product of plant biomass degradation), and the potential to produce single valuable products using defined mutants.
The efficiency of carbon recovery in valuable compounds depends on factors such as the target product, the minimization of undesired metabolic byproducts, and number or amount of substrates being metabolized by the bacterium. Products derived from metabolic intermediates in the upper aromatic catabolic pathways of bacteria like N. aromaticivorans DSM12444 should yield higher carbon recovery than products derived from lower pathways, where more carbon may have already been lost during degradation. We selected PDC as the target product for this study because, in addition to its proven potential as a polyester precursor,26 it is the earliest compound in which the degradation pathways for S, G, and H aromatic compounds were predicted to converge in defined N. aromaticivorans mutants (Fig. 1).
The observation of PDC accumulation when strain 12444Δ1879 was grown on syringic acid (28%; Fig. 4C) was surprising, since we had predicted that the majority of the syringic acid would follow the 3-MGA, CHMOD, OMA pathway (Fig. 1) when the pathway was not altered by mutation. Furthermore, we had predicted that any PDC formed during syringic acid degradation in this strain would be oxidized by LigI to OMA (Fig. 1). The sequential increase in PDC yield in strains 12444ΔdesCD (49%; Fig. 5E) and 12444ΔligIΔdesCD (66%; Fig. 5F) confirms the participation of DesC, DesD, and LigI in the degradation of S type aromatics in N. aromaticivorans and suggests that a large fraction of the syringic acid is naturally channeled through PDC. Since PDC does not accumulate in 12444Δ1879 cultures grown on the products from chemically depolymerized lignin (Fig. 6D) we offer two alternative hypotheses that would need to be tested in the future. First, it is possible that G or H substrates regulate expression of LigI in N. aromaticivorans. Thus, LigI would be poorly or not expressed when S type aromatics are the sole carbon source, allowing for some PDC accumulation by strain 12444Δ1879 grown on syringic acid. On the other hand, LigI would be expressed when 12444Δ1879 is grown on the mixtures of S, G, and H aromatics present in depolymerized lignin, preventing PDC accumulation. Alternatively, since it is not known whether CHMOD transformation to PDC is abiotic or enzymatic, it may be possible that CHMOD is secreted into the growth media where it undergoes spontaneous cyclization, resulting in extracellular PDC accumulation. Higher PDC yields by 12444ΔdesCD and 12444ΔligIΔdesCD could then be explained by increased CHMOD secretion when the aromatic degradation pathways are blocked.
We observe nearly stoichiometric conversion of vanillin and G-diketone into PDC, without extracellular accumulation of other aromatics. However, conversion of p-coumaric acid, p-hydroxybenzaldehyde, p-hydroxybenzoic acid, ferulic acid, vanillic acid, syringaldehyde, syringic acid, and S-diketone to PDC was found to have somewhat lower efficiencies (Table 1). The non-stoichiometric conversion of these aromatic compounds into PDC by N. aromaticivorans is not due to accumulation of intermediate metabolites such as syringic acid, vanillic acid, p-hydroxybenzoic acid and protocatechuic acid, since they only accumulated transiently. Instead, the lower conversion efficiencies could potentially be explained by the presence of alternative, less efficient, and poorly studied pathways for the degradation of those compounds. For instance, the N. aromaticivorans genome contains multiple genes annotated as aromatic ring cleavage dioxygenases for which specificity has not yet been established.34 The presence of a catechol degradation pathway in N. aromaticivorans that uses 2,3-cleavage of the aromatic ring has been suggested as a possible alternative pathway for protocatechuic acid degradation.2 Such alternative non-specific reaction of a catechol dioxygenase could explain the observed lower efficiencies in the transformation of some G and H aromatics to PDC. This hypothesis is supported by the increased cell density observed in cultures of strain 12444ΔligIΔdesCD grown in media containing glucose plus protocatechuic acid compared to cultures only fed glucose (Table 2). Another enzyme with low substrate specificity appears to be the O-demethylase LigM, included in our model as catalyzing the demethylation of vanillic acid (Fig. 1). In Sphingobium sp. SYK-6, LigM is also predicted to catalyze O-demethylation of 3-MGA to gallate,1 which is then proposed to be oxidized to OMA by either LigAB, a dioxygenase with broad specificity (Fig. 1), or DesB, an enzyme not present in N. aromaticivorans. Although this route for degradation of S aromatics is not predicted to be important in N. aromaticivorans,2 LigM activity with 3-MGA and LigAB activity with gallate could contribute to lowering the efficiency of PDC formation from S aromatics by bypassing the blockage in S aromatic degradation intended with the desCD mutation. Thus, future identification and analysis of additional pathways involved in aromatic metabolism by N. aromaticivorans DSM12444 could provide useful information for further increasing the yield of PDC or other target chemicals by preventing aromatic substrates from being degraded by alternative routes.
Fed-batch experiments in a pH-controlled bioreactor showed an increase of up to 8.7 times in PDC titers with respect to titers obtained in batch experiments. These results show a promising potential for production of PDC from aromatic compounds. However, in this experiment, a progressive accumulation of aromatic substrates and glucose was observed. Additional research will be necessary to optimize culture conditions.
The efficiency of lignin conversion to a desired product is also impacted by the nature of the aromatic compounds that result from chemical lignin depolymerization, which may be different from natural products of environmental lignin depolymerization. Therefore, the existence of microbial pathways to metabolize these products could be crucial to increase product recovery. For example, formic-acid-induced depolymerization of oxidized lignin produces a high proportion of aromatic diketones,4 compounds that have also been reported to be present in lignocellulose dilute acid hydrolysates.35 Biological sources of these or structurally related compounds have not been reported, so it was previously unknown whether N. aromaticivorans DSM12444 could metabolize these products or convert them into PDC or other valuable materials. In this study, we found that N. aromaticivorans can convert both S- and G-type diketones into PDC, indicating that they are also degraded via the predicted aromatic degradation pathways (Fig. 1). However, the upper pathway enzymes that transform the diketones to known intermediates in the aromatic degradation pathways remain unknown.
Finally, chemically depolymerized lignin yields a variety of higher molecular weight lignin derived products in addition to monomeric units.4 Sphingomonad bacteria, such as N. aromaticivorans DSM12444, are known or predicted to be capable of breaking most of the linkages found between aromatic subunits in natural lignin in defined ways that yield predictable mono-aromatic products that can be further metabolized.1,36N. aromaticivorans, specifically, is known to be capable of degrading model aromatic dimers containing β-aryl–ether bonds25 and its genome contains homologs of genes that code for the degradation of other aromatic dimers in Sphingobium sp. SYK-6.1 This is an unexplored, but potentially important aspect of employing N. aromaticivorans as a platform microbe for valorization of mixtures of low molecular weight aromatic compounds generated from chemical depolymerization of lignin.
Materials and methods
Bacterial strains, growth media and culturing conditions
A variant of N. aromaticivorans DSM12444 (strain 12444Δ1879) that lacks the gene Saro_1879 (putative sacB; SARO_RS09410 in the recently reannotated genome in NCBI)25 was used as a parent strain to create the deletion mutant strains 12444ΔligI (lacks gene Saro_2819; SARO_RS14300), 12444ΔdesCD (lacks the genes Saro_2864 and Saro_2865; SARO_RS14525 and SARO_RS14530), and 12444ΔligIΔdesCD (lacks genes Saro_2819, Saro_2864, and Saro_2865). All genetic modifications used a variant of the plasmid pk18mobsacB,37 which contains sacB and a kanamycin resistance gene. A detailed procedure for constructing strains with gene deletions is contained in the ESI.† All bacterial strains and plasmids used in this study are listed in Table S1.† Primers used in the construction of the mutant strains are listed in Table S2.†Escherichia coli cultures were grown in LB media containing 50 μg mL−1 kanamycin at 37 °C. N. aromaticivorans cultures were grown in SISnc-V0 media supplemented with the indicated carbon source at 30 °C. SISnc-V0 media is a modification of Sistrom's minimal media38 in which succinate, L-glutamate, L-aspartate, and vitamins were omitted. For routine culture and storage, the growth media was supplemented with 1 g L−1 glucose. For gene modifications, the growth media was supplemented with 1 g L−1 glucose and 50 μg mL−1 kanamycin, or 1 g L−1 glucose and 10% sucrose.
N. aromaticivorans growth experiments
Cell cultures were grown overnight in SISnc-V0 media supplemented with 1 g L−1 glucose, then diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 with fresh SISnc-V0 containing 1 g L−1 glucose and incubated for one hour. Then, 2 ml of the growing culture was spun for 5 min at 5000 rpm, and the cell pellets were resuspended into fresh SISnc-V0 media containing no added carbon source. The resuspended cells were diluted 1
1 with fresh SISnc-V0 containing 1 g L−1 glucose and incubated for one hour. Then, 2 ml of the growing culture was spun for 5 min at 5000 rpm, and the cell pellets were resuspended into fresh SISnc-V0 media containing no added carbon source. The resuspended cells were diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 into SISnc-V0 media supplemented with the indicated carbon source, then shaken at 200 rpm and 30 °C. Cell growth was monitored by measuring cell density using a Klett-Summerson photoelectric colorimeter with a red filter. For N. aromaticivorans, 1 Klett unit (KU) is equal to ∼8 × 106 cfu ml−1.25 Culture samples (1 mL) were collected at various time points, spun for 5 min at 5000 rpm and 4 °C, and the supernatants were filtered through 0.22 μM nylon syringe tip filters (Fisher Scientific), then stored at −20 °C. Each culture was grown at least three times and the data shown corresponds to the results obtained from a representative culture. Conversion efficiency of aromatics to product was calculated by dividing the total amount of product by the total amount of aromatic substrates consumed. Conversion efficiencies reported correspond to the average and standard deviation of the efficiencies calculated for all replicates.
100 into SISnc-V0 media supplemented with the indicated carbon source, then shaken at 200 rpm and 30 °C. Cell growth was monitored by measuring cell density using a Klett-Summerson photoelectric colorimeter with a red filter. For N. aromaticivorans, 1 Klett unit (KU) is equal to ∼8 × 106 cfu ml−1.25 Culture samples (1 mL) were collected at various time points, spun for 5 min at 5000 rpm and 4 °C, and the supernatants were filtered through 0.22 μM nylon syringe tip filters (Fisher Scientific), then stored at −20 °C. Each culture was grown at least three times and the data shown corresponds to the results obtained from a representative culture. Conversion efficiency of aromatics to product was calculated by dividing the total amount of product by the total amount of aromatic substrates consumed. Conversion efficiencies reported correspond to the average and standard deviation of the efficiencies calculated for all replicates.
Production of PDC in a fed-batch bioreactor
A 250 ml bioreactor (Infors, model Multifors 2) containing 130 ml minimum media with 12 mM glucose was inoculated with 2 ml of N. aromaticivorans strain 12444ΔligIΔdesCD culture that had been pre-grown overnight with glucose. After 7.5 h of batch incubation, the bioreactor was intermittently fed media containing 226 mM vanillic acid, 34 mM vanillin, 550 mM glucose, 15 g L−1 ammonium sulfate, and 5% (v/v) DMSO. Culture pH was controlled by the addition of 1 M KOH when needed, to maintain pH 7. Temperature was maintained at 30 °C and the stirrer speed between 250 and 320 rpm. Air was used to deliver oxygen at a flow rate of 1 L min−1. During 50 hours of operation, a total of 29 ml of feed solution was added.Analysis of extracellular metabolites
Metabolite identification was performed by gas chromatography-mass spectrometry (GC-MS) of filtered culture supernatants. Sample aliquots (150 μL) were combined with 70 μL of 1 mM m-coumaric acid in water (internal standard), acidified with HCl to pH < 2, and ethyl acetate extracted (3 × 500 μL). The three ethyl acetate extractions were combined, dried under a stream of N2 at 40 °C, and derivatized by the addition of 150 μL of pyridine and 150 μL of N,O-bis(trimethylsilyl)trifluoro-acetamide with trimethylchlorosilane (99![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, w/w, Sigma) and incubated at 70 °C for 45 min. The derivatized samples were analyzed on an Agilent GC-MS (GC model 7890A, MS Model 5975C) equipped with a (5% phenyl)-methylpolysiloxane capillary column (Agilent model HP-5MS). The injection port temperature was held at 280 °C and the oven temperature program was held at 80 °C for 1 min, then ramped at 10 °C min−1 to 220 °C, held for 2 min, ramped at 20 °C min−1 to 310 °C, and held for 6 min. The MS used an electron impact (EI) ion source (70 eV) and a single quadrupole mass selection scanning at 2.5 Hz, from 50 to 650 m/z. The data was analyzed with Agilent MassHunter software suite, using m-coumaric acid as internal standard.
1, w/w, Sigma) and incubated at 70 °C for 45 min. The derivatized samples were analyzed on an Agilent GC-MS (GC model 7890A, MS Model 5975C) equipped with a (5% phenyl)-methylpolysiloxane capillary column (Agilent model HP-5MS). The injection port temperature was held at 280 °C and the oven temperature program was held at 80 °C for 1 min, then ramped at 10 °C min−1 to 220 °C, held for 2 min, ramped at 20 °C min−1 to 310 °C, and held for 6 min. The MS used an electron impact (EI) ion source (70 eV) and a single quadrupole mass selection scanning at 2.5 Hz, from 50 to 650 m/z. The data was analyzed with Agilent MassHunter software suite, using m-coumaric acid as internal standard.
Quantitative analysis of glucose and formic acid were performed on an Agilent 1260 infinity HPLC equipped with a refractive index detector (HPLC-RID) (Agilent Technologies, Inc., Palo Alto, CA) and an Aminex HPX-87H with Cation-H guard column (BioRad, Inc. Hercules, CA). The mobile phase was 0.02 N sulfuric acid at a flow rate of 0.5 ml min−1.
Quantitative analysis of aromatic compounds and PDC were performed on a Shimadzu triple quadrupole liquid chromatography mass spectrometer (LC-MS) (Nexera XR HPLC-8045 MS/MS). The mobile phase was a binary gradient consisting of solvent A (water) and solvent B (0.1% formic acid in a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of acetonitrile and methanol, v/v). The stationary phase was a Phemonenex Kinetex F5 column (2.6 μm pore size, 2.1 mm ID, 150 mm length, P/N: H18-105937). All compounds were detected by multiple-reaction-monitoring (MRM) and quantified using the strongest MRM transition (Table S3†).
1 mixture of acetonitrile and methanol, v/v). The stationary phase was a Phemonenex Kinetex F5 column (2.6 μm pore size, 2.1 mm ID, 150 mm length, P/N: H18-105937). All compounds were detected by multiple-reaction-monitoring (MRM) and quantified using the strongest MRM transition (Table S3†).
1H-NMR analysis
Nuclear magnetic resonance (NMR) spectroscopy was performed on a Bruker Biospin (Billerica, MA) Avance 500 MHz spectrometer equipped with a 5 mm quadruple-resonance 1H/31P/13C/15N QCI gradient cryoprobe with inverse geometry (proton coils closest to the sample). Samples were prepared as ∼1 mg in 600 μL acetone-d6.Gel permeation chromatography (GPC) analysis
Analytical GPC was performed on a Shimadzu LC20 with a photodiode array detector (SPD-M20A). Separation was performed using a PSS PolarSil linear S column (7.8 mm × 30 cm, 5 μm) at 35 °C. The mobile phase was 5.2 mM sodium phosphate buffer at pH 8, pumped at 0.5 mL min−1, 60 min run time. The molecular weight distribution was calibrated at λ = 254 nm using PDC (184 g mol−1, 23.55 min) and poly(styrene sulfonate) sodium salts, Mp (retention time): 976 kDa (13.20 min), 258 kDa (13.55 min), 65.4 kDa (14.78 min), 47 kDa (16.07 min), 9.74 kDa (17.96 min), 4.21 kDa (19.433 min), and 2.18 kDa (20.35 min) from the PSS-psskit (Polymer Standards Service-USA, Inc, Amherst, MA, USA). Monomer standards were also ran to establish the lower threshold of the column and confirmed that some of them interact with the stationary phase in the alkaline-water mobile phase, these were: rosmarinic acid (360 g mol−1, 21.49 min), ferulic acid (194 g mol−1, 26.63 min), p-coumaric acid (164 g mol−1, 24.96 min), vanillic acid (168 g mol−1, 24.22 min), p-hydroxybenzoic acid (138 g mol−1, 24.87 min), and guaiacol (124 g mol−1, 39.82 min). Compounds eluting from 17.0–22.7 min correspond to oligomeric lignin, while compounds eluting after 22.7 min, correspond to dimeric and monomeric compounds. It should be noted that no Mw values were calculated for peaks detected after 22.7 min, as they were outside the calibration range of the GPC column. In the control samples there were strong monomer signals eluting after 26.0 min, especially a pair of signals at ∼30 min with an absorption band at 375 nm. Most of these monomer signals were not present, or were much weaker, in the inoculated samples after 78 hours of incubation.Preparation of media containing depolymerized lignin products
Lignin was isolated by acid precipitation from pretreatment liquor of poplar biomass that had been pretreated by the copper alkaline hydrogen peroxide method (AHP-Cu).39–41 The lignin was depolymerized using an adaptation of the oxidative methods described previously.4 Depolymerization products were recovered by ethyl acetate extraction, followed by solvent evaporation. This material was re-dissolved in water while adjusting the pH to 7.0 to favor solubilization of aromatic compounds. Consistent with reported products of oxidative depolymerization,4 quantitative HPLC-MS analysis showed concentrations of 1 mM G-diketone, 0.35 mM S-diketone, 0.37 mM syringic acid, 0.12 mM syringaldehyde, 0.44 mM vanillic acid, 0.1 mM vanillin, and 0.93 mM p-hydroxybenzoic acid in the final aqueous solution. For experiments with N. aromaticivorans, aliquots of this solution (25 mL) were mixed with concentrated (5×) SISnc-V0 media containing 1 g L−1 glucose (20 mL) and water (55 mL).Chemicals
Syringic acid, syringaldehyde, ferulic acid, vanillic acid, vanillin, p-coumaric acid, p-hydroxybenzoic acid, p-hydroxybenzaldehyde, and protocatechuate were purchased from Sigma-Aldrich (St Louis, MO). G- and S-diketones were synthesized according to the methods described in the ESI.† PDC was produced by culturing N. aromaticivorans 12444ΔligI in 1 L of SISnc-V0 media supplemented with 3 mM vanillic acid and 0.5 g L−1 (2.8 mM) glucose, and purified following a simplified version of published methods,42 obtaining a >97% pure chemical standard for GC-MS and LC-MS quantifications. Specific details of these procedures are detailed in the ESI.† The identity of PDC was confirmed by comparing the GC-MS spectrum of TMS derivatives (Fig. S4†) and the 1H-NMR spectrum (Fig. S5†) with those reported previously.43Conclusions
A promising path to produce valuable products from the abundant and renewable raw material lignin is to integrate chemical and biological strategies to chemically depolymerize lignin into heterogeneous mixtures of compounds that are then funneled into a single valuable product using microbial catalysts. An ideal microbial catalyst would be capable of simultaneously converting aromatic compounds containing S, G, and H structures, including non-natural compounds generated by chemical depolymerization, into a single compound with high efficiency.Here, we focused on the microbial production of PDC from aromatic products known to be generated by chemical methods of lignin depolymerization. PDC has been shown to have potential as a precursor for polyesters and there is growing interest in using microbes to generate it from lignin.21,22 However, the range of lignin-derived aromatic substrates that could be converted into PDC was limited.21,22 This study expanded the range, as we demonstrated how we could take advantage of N. aromaticivorans’ natural ability to degrade plant-derived aromatics to create mutant strains that simultaneously funnel a wider range of lignocellulose-derived aromatic compounds (including S, G, and H units, and the non-natural S- and G-diketones) into PDC. It is also important that N. aromaticivorans naturally produces PDC via its native metabolic pathways, and therefore, creating PDC-producing strains did not require extensive genetic engineering and optimization steps. Future improvement in PDC yields would require identification of alternative pathways that may be contributing to aromatic degradation in this organism. Ultimately, the information and strategies developed here and in future optimizations of PDC production by N. aromaticivorans DSM12444 could potentially be used to develop this and other microbes into platforms for producing a wide range of additional valuable compounds from lignin.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by U.S. Department of Energy (DOE) Great Lakes Bioenergy Research Center grants (DOE Office of Science BER DE-FC02-07ER64494 and DE-SC0018409). Additional funding from the Chilean National Commission for Scientific and Technological Research (CONICYT) as a fellowship to Jose M. Perez is also acknowledged. We thank Eric Hegg for providing isolated poplar lignin from AHP-Cu pretreatment and Mick McGee for HPLC-RID analyses.References
- N. Kamimura, K. Takahashi, K. Mori, T. Araki, M. Fujita, Y. Higuchi and E. Masai, Environ. Microbiol. Rep., 2017, 9, 679–705 CrossRef CAS PubMed
.
- J. H. Cecil, D. C. Garcia, R. J. Giannone and J. K. Michener, Appl. Environ. Microbiol., 2018, 84, 1–13 CrossRef PubMed
.
- J. Vogel, Curr. Opin. Plant Biol., 2008, 11, 301–307 CrossRef CAS PubMed
.
- A. Rahimi, A. Ulbrich, J. J. Coon and S. S. Stahl, Nature, 2014, 515, 249–252 CrossRef CAS PubMed
.
- H. Jørgensen, J. B. Kristensen and C. Felby, Biofuels, Bioprod. Biorefin., 2007, 1(2), 119–134 CrossRef
.
- Z. Sun, B. Fridrich, A. de Santi, S. Elangovan and K. Barta, Chem. Rev., 2018, 118, 614–678 CrossRef CAS PubMed
.
- W. Schutyser, T. Renders, S. Van den Bosch, S. F. Koelewijn, G. T. Beckham and B. F. Sels, Chem. Soc. Rev., 2018, 47, 852–908 RSC
.
-
J. Ralph, G. Brunow and W. Boerjan, eLS, 2007 Search PubMed
.
- P. J. Harris and R. D. Hartley, Nature, 1976, 259, 508 CrossRef CAS
.
- P. J. Harris and R. D. Hartley, Biochem. Syst. Ecol., 1980, 8, 153–160 CrossRef CAS
.
- D. C. C. Smith, Nature, 1955, 176, 267 CrossRef CAS
.
- R. Vanholme, K. Morreel, C. Darrah, P. Oyarce, J. H. Grabber, J. Ralph and W. Boerjan, New Phytol., 2012, 196, 978–1000 CrossRef CAS PubMed
.
- G. Fuchs, M. Boll and J. Heider, Nat. Rev. Microbiol., 2011, 9, 803–816 CrossRef CAS PubMed
.
- T. D. Bugg, M. Ahmad, E. M. Hardiman and R. Rahmanpour, Nat. Prod. Rep., 2011, 28, 1883–1896 RSC
.
- G. T. Beckham, C. W. Johnson, E. M. Karp, D. Salvachua and D. R. Vardon, Curr. Opin. Biotechnol., 2016, 42, 40–53 CrossRef CAS PubMed
.
- J. G. Linger, D. R. Vardon, M. T. Guarnieri, E. M. Karp, G. B. Hunsinger, M. A. Franden, C. W. Johnson, G. Chupka, T. J. Strathmann, P. T. Pienkos and G. T. Beckham, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 12013–12018 CrossRef CAS PubMed
.
- P. D. Sainsbury, E. M. Hardiman, M. Ahmad, H. Otani, N. Seghezzi, L. D. Eltis and T. D. H. Bugg, ACS Chem. Biol., 2013, 8, 2151–2156 CrossRef CAS PubMed
.
- S. Austin, W. S. Kontur, A. Ulbrich, J. Z. Oshlag, W. Zhang, A. Higbee, Y. Zhang, J. J. Coon, D. B. Hodge, T. J. Donohue and D. R. Noguera, Environ. Sci. Technol., 2015, 49, 8914 CrossRef CAS PubMed
.
- D. Vardon, M. A. Franden, C. Johnson, E. Karp, M. Guarnieri, J. Linger, M. Salm, T. Strathmann, G. Beckham and G. Ferguson, Energy Environ. Sci., 2015, 8, 617–628 RSC
.
- Y. Okamura-Abe, T. Abe, K. Nishimura, Y. Kawata, K. Sato-Izawa, Y. Otsuka, M. Nakamura, S. Kajita, E. Masai, T. Sonoki and Y. Katayama, J. Biosci. Bioeng., 2016, 121, 652–658 CrossRef CAS PubMed
.
- Y. Otsuka, M. Nakamura, K. Shigehara, K. Sugimura, E. Masai, S. Ohara and Y. Katayama, Appl. Microbiol. Biotechnol., 2006, 71, 608–614 CrossRef CAS PubMed
.
- Y. Qian, Y. Otsuka, T. Sonoki, B. Mukhopadhyay, M. Nakamura, J. Jellison and B. Goodell, BioResources, 2016, 11, 6097–6109 CAS
.
- Z. Mycroft, M. Gomis, P. Mines, P. Law and T. D. H. Bugg, Green Chem., 2015, 17, 4974–4979 RSC
.
- J. K. Fredrickson, D. L. Balkwill, G. R. Drake, M. F. Romine, D. B. Ringelberg and D. C. White, Appl. Environ. Microbiol., 1995, 61, 1917 CAS
.
- W. S. Kontur, C. A. Bingman, C. N. Olmsted, D. R. Wassarman, A. Ulbrich, D. L. Gall, R. W. Smith, L. M. Yusko, B. G. Fox, D. R. Noguera, J. J. Coon and T. J. Donohue, J. Biol. Chem., 2018, 293, 4955–4968 CrossRef CAS PubMed
.
- K. Shikinaka, Y. Otsuka, M. Nakamura, E. Masai and Y. Katayama, J. Oleo Sci., 2018, 67, 1059–1070 CrossRef CAS PubMed
.
- E. Masai, S. Shinohara, H. Hara, S. Nishikawa, Y. Katayama and M. Fukuda, J. Bacteriol., 1999, 181, 55–62 CAS
.
- D. Kasai, E. Masai, K. Miyauchi, Y. Katayama and M. Fukuda, J. Bacteriol., 2004, 186, 4951–4959 CrossRef CAS PubMed
.
- I. S. Sze and S. Dagley, J. Bacteriol., 1987, 169, 3833–3835 CrossRef CAS PubMed
.
- T. Michinobu, M. Hishida, M. Sato, Y. Katayama, E. Masai, M. Nakamura, Y. Otsuka, S. Ohara and K. Shigehara, Polym. J., 2008, 40, 68–75 CrossRef CAS
.
- A. J. Ragauskas, G. T. Beckham, M. J. Biddy, R. Chandra, F. Chen, M. F. Davis, B. H. Davison, R. A. Dixon, P. Gilna, M. Keller, P. Langan, A. K. Naskar, J. N. Saddler, T. J. Tschaplinski, G. A. Tuskan and C. E. Wyman, Science, 2014, 344, 1246843 CrossRef PubMed
.
- J. Zakzeski, P. C. A. Bruijnincx, A. L. Jongerius and B. M. Weckhuysen, Chem. Rev., 2010, 110, 3552–3599 CrossRef CAS PubMed
.
- D. L. Gall, J. Ralph, T. J. Donohue and D. R. Noguera, Curr. Opin. Biotechnol., 2017, 45, 120–126 CrossRef CAS PubMed
.
- V. D'Argenio, E. Notomista, M. Petrillo, P. Cantiello, V. Cafaro, V. Izzo, B. Naso, L. Cozzuto, L. Durante, L. Troncone, G. Paolella, F. Salvatore and A. Di Donato, Acta Vet. Scand., 2014, 15, 384 Search PubMed
.
- V. D. Mitchell, C. M. Taylor and S. Bauer, BioEnergy Res., 2014, 7, 654–669 CrossRef CAS
.
- E. Masai, Y. Katayama and M. Fukuda, Biosci., Biotechnol., Biochem., 2007, 71, 1–15 CrossRef CAS PubMed
.
- A. Schäfer, A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach and A. Pühler, Gene, 1994, 145, 69–73 CrossRef
.
- W. R. Sistrom, J. Gen. Microbiol., 1962, 28, 607–616 CrossRef CAS PubMed
.
- A. Bhalla, N. Bansal, R. Stoklosa, M. Fountain, J. Ralph, D. Hodge and E. Hegg, Biotechnol. Biofuels, 2016, 9, 34 CrossRef PubMed
.
- A. Das, A. Rahimi, A. Ulbrich, M. Alherech, A. H. Motagamwala, A. Bhalla, L. da Costa Sousa, V. Balan, J. A. Dumesic, E. L. Hegg, B. E. Dale, J. Ralph, J. J. Coon and S. S. Stahl, ACS Sustainable Chem. Eng., 2018, 6, 3367–3374 CrossRef CAS
.
- Z. Li, C. H. Chen, T. Liu, V. Mathrubootham, E. L. Hegg and D. B. Hodge, Biotechnol. Bioeng., 2013, 110, 1078–1086 CrossRef CAS PubMed
.
- T. Michinobu, M. Bito, Y. Yamada, Y. Katayama, K. Noguchi, E. Masai, M. Nakamura, S. Ohara and K. Shigehara, Bull. Chem. Soc. Jpn., 2007, 80, 2436–2442 CrossRef CAS
.
- D. Kasai, E. Masai, Y. Katayama and M. Fukuda, FEMS Microbiol. Lett., 2007, 274, 323–328 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available: Supplemental figures, tables, and methods. See DOI: 10.1039/c8gc03504k |
| This journal is © The Royal Society of Chemistry 2019 |