Open Access Article
Open Access ArticleLiquid biopsy using the nanotube-CTC-chip: capture of invasive CTCs with high purity using preferential adherence in breast cancer patients†
Masoud S.
Loeian
 a,
Sadegh
Mehdi Aghaei
a,
Sadegh
Mehdi Aghaei
 a,
Farzaneh
Farhadi
a,
Veeresh
Rai
a,
Hong Wei
Yang
a,
Farzaneh
Farhadi
a,
Veeresh
Rai
a,
Hong Wei
Yang
 b,
Mark D.
Johnson
b,
Farrukh
Aqil
b,
Mark D.
Johnson
b,
Farrukh
Aqil
 c,
Mounika
Mandadi
c,
Shesh N.
Rai
c,
Mounika
Mandadi
c,
Shesh N.
Rai
 c and
Balaji
Panchapakesan
c and
Balaji
Panchapakesan
 *a
*a
aSmall Systems Laboratory, Department of Mechanical Engineering, Worcester Polytechnic Institute, Worcester, MA 01609, USA. E-mail: bpanchapakesan@wpi.edu
bDepartment of Neurological Surgery, UMass Memorial Healthcare, University of Massachusetts Medical School, Worcester, MA 01655, USA
cJames Graham Brown Cancer Center, University of Louisville School of Medicine, The University of Louisville, Louisville, KY 40292, USA
First published on 3rd May 2019
Abstract
In this paper, we report the development of the nanotube-CTC-chip for isolation of tumor-derived epithelial cells (circulating tumor cells, CTCs) from peripheral blood, with high purity, by exploiting the physical mechanisms of preferential adherence of CTCs on a nanotube surface. The nanotube-CTC-chip is a new 76-element microarray technology that combines carbon nanotube surfaces with microarray batch manufacturing techniques for the capture and isolation of tumor-derived epithelial cells. Using a combination of red blood cell (RBC) lysis and preferential adherence, we demonstrate the capture and enrichment of CTCs with a 5-log reduction of contaminating WBCs. EpCAM negative MDA-MB-231/luciferase-2A-green fluorescent protein (GFP) cells were spiked in the blood of wild mice and enriched using an RBC lysis protocol. The enriched samples were then processed using the nanotube-CTC-chip for preferential CTC adherence on the nanosurface and counting the GFP cells yielded anywhere from 89% to 100% capture from the droplets. Electron microscopy (EM) studies showed focal adhesion with filaments from the cell body to the nanotube surface. We compared the nanotube preferential adherence to collagen adhesion matrix (CAM) scaffolding, reported as a viable strategy for CTC capture in patients. The CAM scaffolding on the device surface yielded 50% adherence with 100% tracking of cancer cells (adhered vs. non-adhered) versus carbon nanotubes with >90% adherence and 100% tracking for the same protocol. The nanotube-CTC-chip successfully captured CTCs in the peripheral blood of breast cancer patients (stage 1–4) with a range of 4–238 CTCs per 8.5 ml blood or 0.5–28 CTCs per ml. CTCs (based on CK8/18, Her2, EGFR) were successfully identified in 7/7 breast cancer patients, and no CTCs were captured in healthy controls (n = 2). CTC enumeration based on multiple markers using the nanotube-CTC-chip enables dynamic views of metastatic progression and could potentially have predictive capabilities for diagnosis and treatment response.
Introduction
The classical hallmarks of a tumor to become metastatic begin with mobility and invasiveness.1,2 Tumor cells from a primary organ are shed into the vasculature/lymphatics and carried to a distant site to cause metastasis once the conditions become appropriate for their proliferation.1,2 During this process, the circulating epithelial tumor cells (CTCs) change morphology and chemical composition, acquire the ability to overcome the defenses of the immune system, and overcome shear stress in circulation and programmed cell death due to the lack of extracellular interactions in circulation.1,2 CTCs are rare, comprising as few as 1–10 cells per 109 hematological cells, and CTC shedding from a solid tumor into the bloodstream is a highly discontinuous process.3 Thus, the isolation of CTCs with high purity is still a very significant challenge. On top of single CTCs in circulation, some of the rare cells such as CTC clusters, CTCs with micro-tentacles and CTCs of multiple phenotypes are believed to be metastatic initiators and less understood.4,5 Thus, capturing and studying CTCs with biomarker heterogeneity at the single cell level could shed light into the complex biological processes at work and enable dynamic views of cancer metastasis. It could also potentially save lives as identification of a subset of CTCs with metastatic phenotypes among primary tumor cells in early stage cancer can result in customized therapeutic intervention that could result in better outcome (e.g., a small number of EGFR+ CTCs among a group of CK+ CTCs).Technologies for CTC capture and enumeration can be broadly classified into immunoaffinity (antigen-dependent)-based capture and capture based on cellular physical properties (antigen-independent; e.g., size, deformability, cell surface charge, and density).6 Currently, the only FDA-approved technology is the CELLSEARCH® based on immunomagnetic enrichment and is an example of an antigen-dependent capture method.7,8 The sensitivity of CTC capture based on CELLSEARCH® remains poor. Despite a decade of clinical trials, the capture rates are at ∼21.5% (recent SUCCESS trial for breast cancer).8 Filtration technologies such as SCREENCELL,9 MOFF,10 ISET11 and microfabricated filters12,13 isolate CTCs. But size-based isolation of CTCs has challenges in that cells undergoing epithelial–mesenchymal transition (EMT) may not be retained.1,2 Microfluidics has emerged as an active field of research for the isolation of CTCs. Microfluidic technologies such as polymer fluidics,14 CTC-chip,15 Herringbone chip,16 CTC-iChip,17 Vortex,18 Accucyte,19 Fluxion,20 NanoVelcro,21 DEP-Array,22,23 Parsotrix24 and JETTA25 are fluidic devices that have been demonstrated to capture CTCs. Most of the microfluidic devices have challenges in production, imaging, and flow rate. They are inherently flow rate dependent (the faster the flow rate, the lower the capture efficiency), thereby making enrichment slow (0.5–1 ml per hour), and suffer from the large vertical depth of their 3D device features and are difficult to functionalize, making removal of CTCs difficult (e.g. microposts).21 They also require multiple cross-sectional imaging scans and large image files in order to avoid out-of-focus or superimposed images of device-immobilized CTCs.21 Fluidic devices are also prepared using soft lithography, sealing multiple layers of polydimethylsiloxane (PDMS), and CTCs captured inside these chambers are difficult to remove, thereby limiting genomic characterization.25 These methods are therefore highly time-consuming, labor-intensive serial production processes and can enable false positive or negative results, thereby severely restricting their applicability to routine clinical practice.21
In this paper, we have developed a new method of CTC capture based on microarrays of carbon nanotube (CNT) surfaces. This technique is a new type of antigen-independent capture, where the preferential attachment of CTCs to a CNT surface is exploited. Our method presented here has many advantages, such as 1) a microarray format enabling a large volume of blood to be RBC lysed/fractionated into smaller portions that may enable better capture sensitivity from droplets; 2) antigen-independent capture of CTCs enabling isolation of CTCs of variable phenotypes; 3) size-independent capture of CTCs; 4) the preferential adherence of CTCs to the nanotube surface enables 5-log depletion of WBCs; 5) no transfer of CTCs is necessary to do microscopy, eliminating cellular loss; 6) planar surface architecture eliminates out-of-focus problems and large image files associated with imaging CTCs inside a fluidic chamber; 7) surface architecture lends itself to easier CTC downstream analysis, unlike microfluidics, where CTCs may be recovered from sealed chambers; and 8) planar batch manufacturing process resulting in >99% yield of individual devices both in silicon-based and glass based wafers.
Our proof-of-concept results demonstrate the isolation of spiked cancer cells from blood using the preferential attachment at 89–100% capture rate, isolation of CTCs with high purity (5-log depletion of WBCs) and 100% sensitivity (n = 7/7) in breast cancer patients (4 ml and 8.5 ml blood), and capture of single CTCs of multiple phenotypes from the same patient. The microarray format, use of carbon nanotubes for capture based on adherence and the successful isolation of CTCs of different phenotypes suggest that the nanotube-CTC-chip is a versatile platform to capture CTCs in patients. The chip can broadly impact our understanding of the basic metastatic biological processes and clinical decision making, providing dynamic views of metastatic progression at the level of single cells.
Results
The isolation, capture, and enumeration of CTCs of different phenotypes using the nanotube-CTC-chip can be broadly classified into four steps as presented in Fig. 1. In step 1, 8.5 ml blood consisting of approximately 40 billion erythrocytes, 64 million leukocytes and 1–10 CTCs of different phenotypes are collected from a patient. In step 2, the red blood cells (RBCs) are depleted through an RBC lysis protocol.26 The entire contents are centrifuged, and nucleated cell fractions consisting of CTCs and WBCs are pelleted. In step 3, the nucleated cells consisting of CTCs and WBCs are added as standard 10 μl droplets on 6–12 individual nanotube devices (total 60–120 μl; each device is 3 mm × 3 mm). The hypothesis is that CTCs attach to the nanotube substrate and not the other nucleated cells including WBCs. Also called antigen-independent CTC capture, this strategy represents an approach to the enrichment of CTCs with high purity that is not biased by the selection of potentially variably expressed markers on tumor cells. In step 4, the attached CTCs on the nanotube surface are immunostained on-chip using antibodies to identify and enumerate CTCs of different phenotypes. DAPI (4′,6-diamidino-2-phenylindole) is used as the nuclear stain and cytokeratin (CK8/18) and other antibodies (e.g. Her2, EGFR) identify CTCs. CD45 and only DAPI identify WBCs.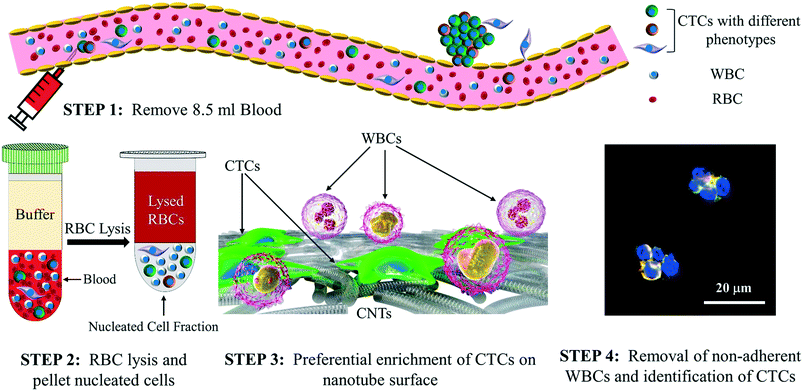 | ||
| Fig. 1 The nanotube-CTC-chip: steps in isolation and enumeration of CTCs using the nanotube-CTC-chip. | ||
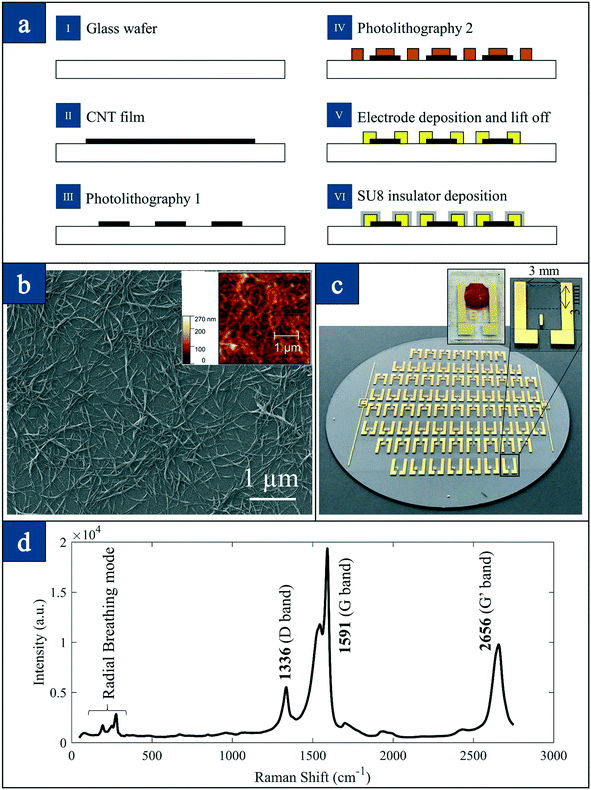
Fig. 2(a) presents the schematic flow chart of the 76-element array fabrication process, which is reported elsewhere.27,28 We have fabricated four generations of devices consisting of 60-element, 76-element and 240-element arrays in silicon27 and 76-element arrays in glass as reported here. With the RBC lysis protocol that we have developed and presented here for isolation of CTCs, one can process 8.5 ml of blood and isolate CTCs based on the nanotube-CTC-chip using the new method of preferential adherence as presented here.
Fig. 2(b) shows the scanning electron micrograph (SEM) of single-walled carbon nanotubes. We used HiPCo carbon nanotubes of about 1 nm in diameter and 1 μm in length. The nanotubes are transferred to the glass surface using a vacuum filtration process reported elsewhere.27 The carbon nanotubes as seen in both SEM (Fig. 2(b)) and AFM (inset, Fig. 2(b)) images show random arrangement. Fig. 2(c) presents the entire wafer consisting of the 76-element array. The inset in this figure shows the single device where the active nanotube film is 3 mm × 3 mm. A single blood droplet is shown on the second inset. All the four sides of the device have a 30 μm thick SU8 layer that enables droplet localization due to hydrophobicity. Fig. 2(d) presents the Raman spectroscopy of carbon nanotube films. The Raman spectroscopy of the carbon nanotube films suggested an RBM mode (275 cm−1), a small D band (1336 cm−1), a large G band (1591 cm−1) and a pronounced 2D band or G′ band (2656 cm−1). The large G/D band ratios suggest high-quality carbon nanotubes.
Tracking single cells using the nanotube-CTC-chip
In our previous work on the nanotube-CTC-chip using spiked blood samples,27 we observed that the cells inside the blood start to settle immediately due to the force of gravity, and density gradients resulting in cells coming in contact and interacting with the base nanotube substrate surface along with RBCs.27 The initial observations on the optical microscope of cancer cell spiked blood sample droplets on top of the devices showed that the spiked cancer cells and RBCs, as a part of the settling process, tend to go to the bottom of the device compared to WBCs which settle on top.27 Knowing this, we decided to track the individual cells from a blood droplet.To track the cells, we used a TNBC cell line (MDA-MB-231; EpCAM−) that was transduced by a lentivirus to actively express a green fluorescent protein (GFP) marker.29 Typically, CTC technologies such as CELLSEARCH® use the epithelial cell adhesion molecule (EpCAM) to identify CTCs from hematological cells. CTCs are highly heterogeneous and actively change their shape and morphology and even downregulate EpCAM during epithelial–mesenchymal transition (EMT).30 Thus, EpCAM-based methods lose CTCs, do not shed light on the subset of metastatic CTCs and therefore are inadequate for clinical decision making. Therefore, alternative methods able to recognize a broader spectrum of CTC phenotypes are needed and are presented here.30 With this in mind, we used an EpCAM− and TNBC basal-like cell line MDA-MB-231 for our spiked cell line studies.
The GFP transduced triple-negative breast cancer cells were spiked in blood and were observed under a fluorescence microscope. For these experiments, blood was diluted to 10% that enabled us to track all the GFP cells in the droplet. Fig. S1 (suppl. (a))† presents the fluorescence image of the GFP tracked cells at a different depth of focus. Fig. S1 (suppl. (b))† shows the number of spiked cells versus some GFP observations. Fig. S1 (suppl. (c))† presents the spiked cell counts in blood for 1, 10, 100, and 1000 spiked GFP cells in the blood. We observed 87% to 100% capture of the GFP cells. A slight error in cell counts is a result of counting the cells using a hemocytometer and is seen in many spiked cell experiments.16 Fig. S2† is the entire image of a droplet with MDA-MB-231-GFP cells marked by arrows.
In our spiking experiments, it was observed that when a droplet of blood was placed on the nanotube device surface, the cancer cells and RBCs went to the bottom as a part of the settling process.27 The RBCs were seen to cover most of the nanosurface, which is not desirable for a preferential cell adherence strategy, and also rare CTCs in patients. Having the cells exposed to the nanosurface is desirable as it enables the conditions for cellular anchorage to the nanotube matrix. In many mechanobiology studies, microfabricated topographic features with specific dimensions have been fabricated to mimic the architecture and orientation of the extracellular matrix (ECM) in vitro.31 The nanotube surface enables topographic anisotropy for cellular attachment due to the collection of nanometer scale tubes on the surface. For CTC isolation based on such topographic features, RBC lysis is necessary as this enables more exposure of the cells to the nanotube surface. While there is a section of the CTC community that is of the understanding that RBC lysis can lead to loss of CTCs and lead to false positives and negatives,1 this work has successfully shown that RBC lysis can be beneficial to obtain viable CTCs of high quality. Further, CTC cultures will not be possible on the nanotube surface without the RBC lysis process. Fig. S3 (suppl. (a))† presents the RBC lysis protocol. Fig. S3 (suppl. (b))† presents the optical image of control blood (from a healthy volunteer) before and after lysis. The WBCs, but none of the RBCs, are observed after the lysis procedure. Fig. S3 (suppl. (c))† presents the cells that are attached to the surface versus non-adhered cells using the RBC lysis protocol.
Preferential adherence of spiked cancer cells on the nanotube-CTC-chip
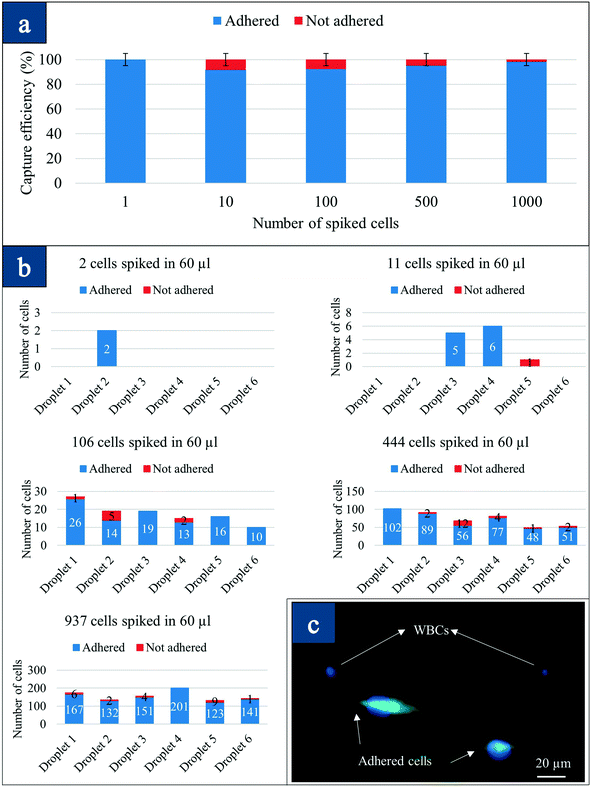
The method of preferential adherence is the hypothesis that CTCs preferentially attach to the nanotube surface and not the other blood components including WBCs. In the recent past, the nano-roughened adhesion-based capture of CTCs with heterogeneous expression and metastatic characteristics has been reported.32 With the nano-roughened glass microfluidic CTC capture device, they were able to achieve capture yields of >80% for both EpCAM+ (MCF-7, SUM-149, A549) and EpCAM− (MDA-MB-231) cancer cell lines spiked in blood samples.32 We have independently discovered this effect on carbon nanotubes in our microarray, and our capture efficiencies are more substantial with very high purity (5-log depletion), partially due to RBC lysis.To determine if all the spiked cancer cells would survive the RBC lysis process, we undertook several experiments. GFP positive, EpCAM−, MDA-MB-231 breast cancer cells were spiked in mice blood, and the lysis protocol was used to determine the adherence/non-adherence of cells on the carbon nanotube surface. The volume of the lysed sample was adjusted to 60 μl to obtain a standard 10 μl droplet on each device, and fluorescence microscopy was done to observe the captured GFP cells. Five samples containing 1, 10, 100, 500, and 1000 MDA-MB-231 cells were spiked into 10 μl blood from wild-type mice in 5 different 1.5 micro-centrifuge tubes. After each sample was lysed, the cells were resuspended in culture medium and were divided into six CNT chips having 10 μl volume each. They were kept inside a sterile culture dish containing PBS to stop the droplets from being dried in a 5% CO2 incubator at 37 °C. After 48 hours, samples were taken out from the incubator, and the droplet was removed and transferred into the second device to count the number of non-adhered cells from the second device. The first device was then washed with PBS, and both the primary and the secondary devices were examined under a fluorescence microscope to count the cells on each device. The number of cells on the primary device was labeled “Adhered” while the ones on the secondary device were labeled “Not Adhered.”
Fig. 3(a) presents the capture efficiency which suggests that 87–100% of the cells adhered on the primary device at all spiked concentrations. By using two devices from the array, we captured both adhered and non-adhered cells or tracked all the spiked cells. Fig. 3(b) presents the number of cells counted in each droplet across all spike concentrations. Our method is a new way to enumerate cells using droplets having a standard volume and a standard number of devices from the array. Since the volumes are quite small, one can ensure highly accurate counts. The standard 6 droplets can also be used for staining cells with 6 different markers, enabling multi-marker analysis of captured cells.
Fig. 3(c) presents the fluorescence image of the captured cells after RBC lysis and preferential attachment in spiked blood experiments. The difference in MDA-MB-231-GFP-Luc cells that are attached versus WBCs (DAPI only) on the same surface is observed. The cells that attached changed shape and morphology, looking elongated/mechanically stretched as presented in the fluorescence images. The stages that characterize the process of static in vitro cell adhesion include the attachment of the cell body to the nanotube matrix, flattening and spreading, and the organization of the actin skeleton network with the formation of focal adhesion between the cell and the nanotubes.33 The deformation, mechanical stretching, and flattening are observed in Fig. 3(c). However, to understand more about the cell adhesion, electron microscopy studies were conducted. Table 1 shows the number of captured cancer cells in spiked blood, number of WBCs and log10 depletion. We obtained almost 4-log depletion in these small volumes, suggesting a high level of purity.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 per microliter in healthy blood
000 per microliter in healthy blood
| Sample | Number of captured CTCs | Number of captured WBCs | % WBC contamination | Log10 depletion |
|---|---|---|---|---|
| 1000 cells spiked in 10 μl mice blood | 937 | 12 | 0.016 | 3.79 |
| 500 cells spiked in 10 μl mice blood | 444 | 14 | 0.018 | 3.72 |
| 100 cells spiked in 10 μl mice blood | 106 | 21 | 0.028 | 3.55 |
| 10 cells spiked in 10 μl mice blood | 12 | 9 | 0.012 | 3.92 |
| 1 cell spiked in 10 μl mice blood | 2 | 16 | 0.021 | 3.67 |
| Patient 1 (8.5 ml) | 8 | 31 | 4.86 × 10−5 | 6.31 |
| Patient 2 (4 ml) | 39 | 637 | 2.12 × 10−5 | 4.67 |
| Patient 3 (4 ml) | 21 | 479 | 1.59 × 10−5 | 4.79 |
| Patient 4 (8.5 ml) | 238 | 277 | 4.34 × 10−6 | 5.36 |
| Patient 5 (8.5 ml) | 27 | 549 | 8.61 × 10−6 | 5.06 |
| Patient 6 (4 ml) | 4 | 151 | 5.03 × 10−6 | 5.29 |
| Patient 7 (8.5 ml) | 9 | 771 | 1.29 × 10−5 | 4.91 |
| Healthy control 1 (8.5 ml) | 0 | 643 | 1.008 × 10−5 | 4.99 |
| Healthy control 2 (8.5 ml) | 0 | 652 | 1.022 × 10−5 | 4.99 |
Electron microscopy of single-cell adhesion on the nanotube surface
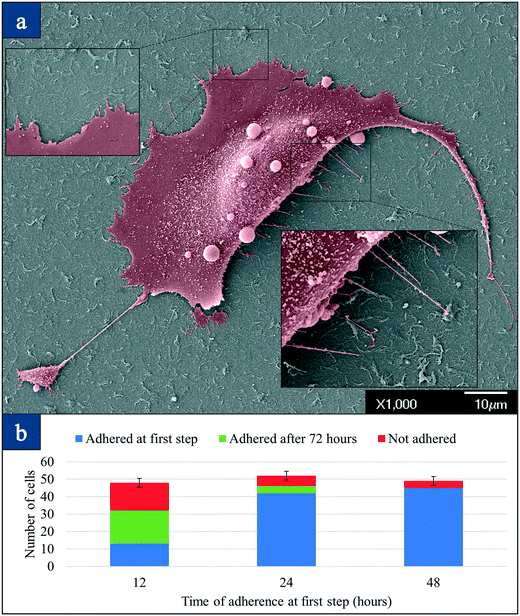
We conducted electron microscopy studies of attached single cells to investigate how cancer cells attach to the nanotube surface. Fig. 4(a) is the SEM image of an attached SKBR3 cell that was incubated for 48 hours on the nanotube surface. The striking image suggests that cancer cells change morphology and spread on the nanotube surface causing strong focal adhesion. The filaments from the main body of the cell extend to the nanotube surface. Many such filaments are observed to attach to the individual nanotubes/bundles directly. The diameter of these filaments is about 150 nm to 200 nm, and they cannot be seen under an optical microscope. The exposure of some of these filaments under an SEM suggests that thousands of such filaments bond to the nanotube matrix from underneath the cell. Integrin receptors are vital in static in vitro cell adhesion and spreading.33 Specific integrin binding provides a mechanical linkage between the intracellular actin cytoskeleton and the nanotube matrix.33 EM studies confirm that tumor-derived epithelial cells attach firmly to the CNT surface including individual CNT bundles and exhibit active dynamics, which confirms our hypothesis.Fig. 4(b) presents the time of adherence versus the captured number of cells. Three different samples containing 50 cells of the MDA-MB-231-GFP-Luc cell line were mixed with 10 μl wild mice blood in a 1.5 ml tube and lysed. Each of these three samples was placed on three separate CNT devices as a 10 μl droplet, and they were given 12, 24, and 48 hours for attachment. After this time, the removed droplet was then placed on another new CNT device surface for another 72 hours to observe if any of the non-attached cells could be attached to a new CNT device surface. It is seen that for the first sample that was incubated for 12 hours, <30% of the cells attached at the first step. An additional 50% attach after 72 hours, and some do not attach. However, when the time of first step adherence is increased to 48 hours, more than 90% of the cells attached, suggesting the increase in adhesion strength with time. The strength of adhesion of the nanotube matrix to the cell is muscular as shown by the increased number of captured cells on the surface with time (grows in number in 48 hours).34 The advantage of small adherence time is that only 7 WBCs (less than 0.005%) adhered to the first CNT device from our observations. For the second sample, 42 cancer cells out of 52 cancer cells (80%) adhered to the CNT surface after 24 hours. 4 cells adhered on the secondary device after 48 hours and 6 cells (11%) were not captured eventually. For the third sample, 45 cells out of 49 cells (92%) adhered to the CNT film. The rest of the cells did not adhere to the second device. The number of WBCs that adhered to the CNT surface for the second and third sample was counted as 21 and 24 (0.02%), respectively, in these small volumes. From these experiments, we concluded that 48 hours is the optimum time to have the highest capture efficiency while the number of WBCs attached to the CNT was negligible.
While all the experiments were done using GFP positive MDA-MB-231 triple negative cells, this technique is not limited to specific cancer types, and therefore could potentially capture any type of epithelial cancer cell which constitutes the four significant cancers (breast, colon, lung, and prostate) using the method of preferential adherence. Further, our method is currently the only one to track both adherent and non-adherent cells on the same chip, thus effectively tracking all the cells, a task that is of high value in CTC capture especially in early-stage cancers where the cell numbers may be meager. Other epithelial cancer cell lines including HeLa (cervix), U-251 (glioblastoma), MCF7 (breast), and LN-291 (brain) were also tested using this method, and the yield of adherence was more than 90% as well. Fig. S4 (suppl. (a))† is a representative image of the brain cancer cells that were stained for DAPI and EGFR. Fig. S4 (suppl. (b))† shows adhered HeLa cells stained with CD59 after adherence to the CNT film. With a suitable functionalization protocol, one can also capture non-epithelial cells such as lymphomas and sarcomas.
Preferential adherence using collagen adhesion matrix on the nanotube-CTC-chip
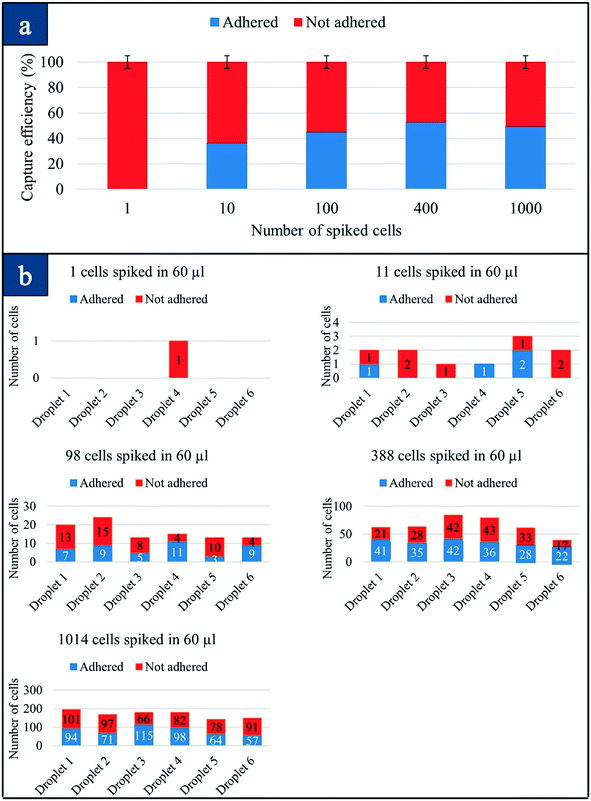
We compared our cancer cell attachment strategy on carbon nanotubes with that of collagen adhesion matrix (CAM) scaffolding for the capture of CTCs. The capture of CTCs based on the CAM strategy (Vita assay) is a unique strategy and is a method of adherence.35 The ability of a tumor cell to invade collagenous matrices is one of the hallmarks of metastasis. In the past, it was hypothesized that populations of CTCs that adhere and invade collagenous matrices would be invasive and would exhibit the natural tendency to undergo metastasis.35Fig. 5 presents the overall capture efficiency and the number of cells (both adhered and non-adhered) for each device for the CAM strategy. The adherence efficiency of the CAM strategy is observed to be only 50%, which is consistent with what has been reported previously.35 Fifty percent of cells did not adhere on the primary CAM device even after 48 hours of incubation, although we were able to track all the cells, both adhered and non-adhered, by counting the number of cells in the secondary device. Repeating these experiments three times enabled us to achieve similar values for the capture rate. It is possible that CAM devices need more time for the cells to digest the collagen, although it should be noted that some of the cells may not attach to the CAM. A study based on CAM coated tubes in stage I–III breast cancer (Vita assay) only was able to detect CTCs in 28/54 patients, which is 52% sensitivity.35 The CAM strategy needs optimization regarding collagen type, the deposition technique, and concentration for the nanotube-CTC-chip. This method has a lower yield compared to bare SWCNT films under the same conditions for preferential adherence using our microarrays.
Clinical studies: the capture of CTCs using the nanotube-CTC-chip from breast cancer patients
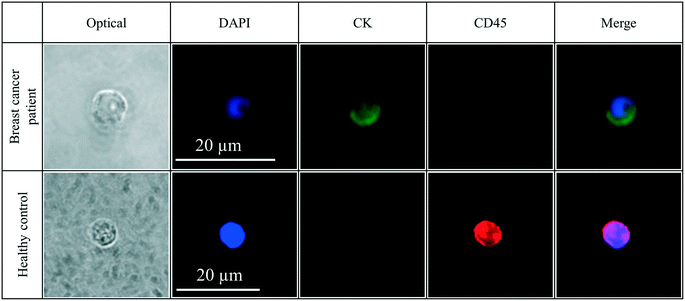
We investigated the ability of the nanotube-CTC-chip to isolate CTCs with high purity from breast cancer patients. De-identified 8.5 ml blood samples were obtained from the University of Louisville under an IRB (IRB#18.0828). De-identified 4 ml blood samples were also obtained from the UMASS tissue bank to determine the numbers of CTCs captured using the nanotube-CTC-chip from these two sources of different volumes.Fig. 6 is a representative image of CTC identification and capture in a breast cancer patient versus no capture of CTCs in healthy control. CTCs were identified and scored as cells that were clearly visible under an optical microscope, possessing cellular morphology, positive for nuclear stain DAPI, positive for CK (8/18), and negative for CD45. Since CTCs are often no larger than WBCs, and since WBCs have a nucleus, it is important to distinguish between CTCs and WBCs for scoring purposes. The lymphocyte common antigen CD45 is expressed in all leukocytes, and therefore WBCs are identified as cells that are clearly visible under an optical microscope, positive for lymphocyte antigen CD45, negative for CK 8/18, and positive for nuclear stain DAPI. It is seen from the image that CTCs do not have to be any larger than leukocytes. Size selective techniques which capture CTCs based on the assumption that epithelial CTCs are much larger than leukocytes may not capture the full range of CTCs.6 The nanotube-CTC-chip enables the capture of CTCs without bias in size and antigen expression, thereby capturing the full range of CTCs.
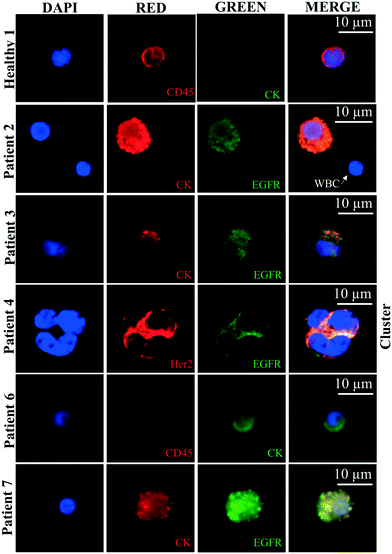
Fig. 7 shows the representative CTCs captured in patients using multiple antigenic markers in 4 ml and 8.5 ml blood. We captured CTCs in n = 7/7 samples in patients, suggesting 100% sensitivity. Healthy controls (n = 2/2) showed no presence of CTCs in the blood. All optical images along with merge images from patients are presented in Fig. S5.† CTCs of different phenotypes were captured based on CK8/18+, Her2+, and EGFR+ cells. Both healthy controls 1 and 2 were CK8/18−, CD45+, and DAPI+. CTCs from patients were CK(8/18)+/Her2+/EGFR+ and CD45− and DAPI+.
Table 2 presents the TNM staging, a number of CTCs and a number of heterogeneous CTCs captured using the nanotube-CTC-chip. Patient 1 (stage 4) was an outlier as the lysis procedure did not work the first time (due to platelet aggregation) and we had to do the lysis more than once. However, we still captured 8 CTCs expressing Her2 and EGFR (Fig. S5†). There was a learning curve associated with processing large volumes of blood for the nanotube-CTC-chip. From the second patient onwards, whole blood stabilization agents (tirofiban; 0.5 μg ml−1) was added before shipping at 4 °C. From the second patient, the protocol was uniform across all the samples. As Table 2 shows, anywhere from 8 to 238 CTCs were captured in 4 ml/8.5 ml blood. CTCs were captured in patients that were lymph node positive and negative. In general, using TNM staging and number of CTCs counted we infer that patients who were staged between stages 1 and 3 (patients 2, 3, 5 and 6) had a lower number of CTCs (4–39 CTCs in 4 ml and 8.5 ml blood or 0.5 to 10 CTCs per ml). Patient 4 had stage 4 breast cancer with an elevated level of CTCs (238 in 8.5 ml blood or 28 CTCs per ml) before treatment. There is an apparent increase in CTC counts between early stage (stage 1–3) and advanced disease (stage 4) using the nanotube-CTC-chip. The CTCs were positive for both Her2 and EGFR, suggesting aggressive disease. Two clusters were also noted in patient 4. Finally, in patient 7, blood was obtained only after radiation therapy (although the patient was chemo naive). Surprisingly, we captured only 9 CTCs in 8.5 ml blood (1 CTC per ml), suggesting that the number of CTCs may have decreased in this stage 4 patient after radiation. Comparing both stage 4 patients, patient 7 (radiation therapy) and patient 4 (treatment naive), we believe the nanotube-CTC-chip may predict the treatment response based on CTC enumeration. More patients need to be tested, but the number of CTCs and the range of CTCs in early stage versus metastatic disease suggest the potential predictive powers of the chip.
| Patient | TNM staging and source | # CTCs captured | # CTCs per ml | # Heterogeneous CTCs | Notes |
|---|---|---|---|---|---|
| Patient 1 | PT4N2M1; treatment naive; stage 4; UofL | 8 in 8.5 ml blood | N/A | 8 based on Her2/EGFR | Lysis used more than once. A criterion cannot be established |
| Patient 2 | PT1CN0; stage 1B; UMASS | 39 in 4 ml blood | 9.75 CTC per ml | 36 CK+ and 3 EGFR+ | Stable disease; lymph node invasion |
| Patient 3 | PT2N0M0; stage 2 UMASS; | 21 in 4 ml blood | 5.25 CTC per ml | 19 CK+, 2 EGFR+ | Stable disease; tumor >20 mm |
| Patient 4 | PT1BN1M1; stage 4; UofL; treatment naive | 238 in 8.5 ml blood | 28 CTC per ml | Her2+ and EGFR+ | 2 CTC clusters; progressive disease |
| Patient 5 | PT1N1M0; treatment naive; stage 2A; UofL | 27 in 8.5 ml blood | 3 CTC per ml | 3 EGFR+ and 26 CK+ | Stable disease |
| Patient 6 | PT2N2A; stage 3A; UMASS | 4 CTCs in 4 ml blood | 1 CTC per ml | Only CK+ | Stable disease |
| Patient 7 | PT4BN1M1; treated with radiation; UofL | 9 CTCs in 8.5 ml blood | 1 CTC per ml | 1 EGFR; 8 CK+ | Metastasis to bone and lung; CTCs still exist after radiation therapy |
CTC purity in patient samples
It is important to capture CTCs of high purity to enable further genomic characterization. Purity describes the ability of the device to capture CTCs within a background of contaminating leukocytes.36 Purity is the one metric that can be measured from clinical samples.36 Using the nanotube-CTC-chip, we established log10-depletion for each patient based on the number of WBCs captured. The log depletion formula can be used to assess CTC purity and is given as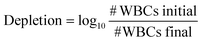 | (1) |
Using this formula, we assessed the log-depletion of WBCs in each of the patient and control samples. Table 1 shows 4- to 5-log depletion of WBCs. Patient 1 is an outlier due to the lysis procedure being done more than once. It can be seen that in both healthy controls we obtained the same log-depletion suggesting the high controllability and uniformity of the process. The range of log-depletion was between 4.6 and 5.3, suggesting that this number could be useful as a calibration marker for suggesting process control in routine clinical practice. However, narrowing this distribution even further in future samples can be highly beneficial to enable comparison across multiple cancer types.
Capture of CTCs of various phenotypes using the nanotube-CTC-chip in breast cancer patients
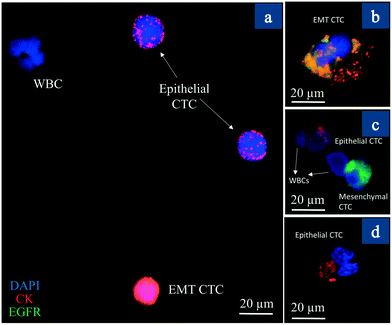
One of the objectives of our study in patients was to investigate the presence of single CTCs of various phenotypes (Her2+/EGFR+ CTC subclones). The EGFR family of receptors is composed of EGFR (ErbB-1, HER1 in humans), HER2 (ErbB-2), HER3 (ErbB-3), and HER4 (ErbB-4).37 There has been a strong interest recently in EGFR and HER2 because of their overexpression in breast carcinomas.37 EGFR (HER1) signalling has been reported to induce EMT through different pathways that results in tumor progression and metastasis.38,39 In our analysis of all patients, apart from CK (8/18)+ cells, we were able to capture 2–3 cells that were strongly EGFR positive in stage 1–3 cancer. This suggests that CTCs of various phenotypes exist in patients even in early-stage cancers. However, advanced stage cancer patients (patient 4) showed both Her2 and EGFR positive CTCs with large numbers of CTCs (238). This may suggest a combination of CTC numbers, and the heterogeneity of CTCs may determine the aggressiveness of the disease in breast cancer. Fig. S6† shows a single CTC at the bottom of the image exhibiting multiple phenotypes (both EGFR and CK(8/18)), while the majority of CTCs were only positive for CK8/18. This suggests that a small clone exists among primary tumor CTCs that can have a metastatic (EMT) phenotype. The question is whether such small subclones (expressing the EGFR family of receptors40) resist chemotherapy and result in progressive disease and metastasis a few years after treatment. One thing is for sure; the presence of CTCs in circulation may be linked negatively to the survival of a patient. Brain metastatic breast cancer (BMBC) can express both Her2 and EGFR,41,42 suggesting that such populations are at high risk for metastatic disease. The high CTC numbers and the presence of both Her2/EGFR pathological features can trigger EMT, metastasis and determine future survival. The nanotube-CTC-chip thus enables us to capture these small subclones and enable better views of disease progression.To distinguish between CTCs of different phenotypes, we further investigated whether different types of CTCs could exist in the same patient sample. Fig. 8 provides dynamic views of epithelial and mesenchymal states of CTCs captured from patient 5. In Fig. 8(a), one can see a WBC (DAPI only), epithelial CTCs (positive only for CK8/18) and an EMT related CTC (CK8/18 and EGFR). Fig. 8(b–d) show the different CTCs from the same sample. In Fig. 8(b), spindle cells with both EGFR and CK8/18 suggest activation of the EMT process. CTCs often change morphology on EMT activation and the presence of EGFR and the morphology of the CTC can be a positive confirmation. Fig. 8(c and d) show the presence of both epithelial and mesenchymal CTCs. Epithelial CTCs were only positive for CK8/18 and not EGFR, but the more aggressive mesenchymal state was also strongly positive for EGFR and lacked complete CK8/18 expression (Fig. 8(c)). Overall, we found 3 EGFR+ CTCs and 26 CK8/18+ CTCs in patient 5, suggesting that the nanotube-CTC-chip can track CTCs of various phenotypes at the single cell level. Further details can be found in Fig. S7† showing the difference between epi+, mesen+, and EMT CTCs. Our analysis of phenotype heterogeneity can also inform decision making as we have focussed mainly on 3 markers, namely CK (8/18), Her2, and EGFR, for which treatment options are available. Overall, the nanotube-CTC-chip enabled a high level of success in analyzing CTCs, CTC enumeration, the capture of heterogeneous CTCs and the ability to distinguish between epithelial, mesenchymal, and EMT related CTCs in breast cancer patients.
Comparison of the nanotube-CTC-chip micro-array with existing CTC capture techniques
Table 3 compares some of the existing CTC capture methods with that of the nanotube-CTC-chip. CELLSEARCH®, a technique based on immunomagnetic enrichment, was the first to arrive in the market based on EpCAM antigen-dependent capture.7 A decade of research on CTC capture based on CELLSEARCH® has yielded only modest results. In the recent German SUCCESS study based on the CELLSEARCH® system involving 2026 breast cancer patients before chemotherapy, CTCs were detected in only 21.5% of patients (n = 435 of 2026) following surgical removal of the primary tumor.8 Another study in comparison with CELLSEARCH® and ISET (filtration system) for circulating tumor cell detection in patients with metastatic carcinomas yielded consistent results only in 55% (11 out of 20) of the patients with breast cancer, in 60% (12 out of 20) of the patients with prostate cancer and in only 20% (4 out of 20) of lung cancer patients.43 Both techniques have discrepancies between the number of CTCs enumerated using both techniques.43| Method/device | Number of CTCs | Number of WBCs | Notes |
|---|---|---|---|
| CELLSEARCH® | 5–1000 CTCs in 7.5 ml blood; >5 CTCs is bad prognosis | No information | 21.5% capture rate for breast cancer; chemically fixed cells7,8 |
| CTC-chip | 5–1281/ml | 50% purity | Micro-post, not surface technique; no clusters15 |
| Herringbone chip | 12–3167 CTCs per ml | No information | 2 clusters each of about 4–12 cells16 |
| CTC-iChip | 1–30 cells/7.5 ml blood in patients | 1188/ml; median, 352/ml; range, 58 to 9249/ml | Has a cutoff for cells larger than 21 μm (ref. 17) |
| NanoVelcro | 1–99 CTCs per ml in patients | No information | CTCs are captured in 1 ml blood21 |
| Vortex | 25–300 CTCs per ml | 57–94% purity,18 >20–500 WBCs per ml | CTC size based collection, deformability of cells, CTC collection depends on aspect ratio18 |
| MOFF, SCREEN CELL, ISET, microfabricated portable filters (filtration) | 1–100 cells, capture of EGFR based cells; 51/57 patients had CTCs | Possibly both CTCs/WBCs will be retained | RBC clogging filters; deformability of cells is an issue. High pressures can damage cells9–13 |
| Collagen adhesion matrix | 10–1000 CTCs per ml | No information | 52% sensitivity in stage1–3 cancers. Not all cancer cells adhere to collagen. Culturing over 33 days is necessary35 |
| Parsortix system | Cell lines (66–92% capture); clinical studies ongoing | 200–800 per mL | Size based capture, 6.5 μm critical gap captures CTCs24 |
| Nanotube-CTC-Chip | 8–238 CTCs per 8.5 ml or 4 ml blood; 7/7 patients with stage 1c to stage 4 cancers had CTCs | 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log to 6 log to 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log depletion of WBCs in patients; 31–652 WBCs in 8.5 mL or 3.6 to 75 per mL log depletion of WBCs in patients; 31–652 WBCs in 8.5 mL or 3.6 to 75 per mL |
Preferential adherence; antigen and size independent capture; 5–6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log depletion of WBCs; CTC of multiple phenotypes. Present work log depletion of WBCs; CTC of multiple phenotypes. Present work |
Microfluidic technologies such as CTC-chip,15 Herringbone chip16 and CTC-iChip17 are essential concepts and fluidic platforms. The CTC-chip with micro-posts is challenging to manufacture and functionalize the surface, and no CTC clusters were captured using this device.16 The Herringbone chip has surface characteristics and also yielded only 2 clusters.16 The CTC-iChip has low WBC contamination and can capture antigen-independent and dependent CTCs,17 but an array with 20 μm gaps on the iChip cannot capture CTC clusters and thus is reduced to size-dependent capture.17 Therefore, newer devices with asymmetry and size-based separation are being developed.44 Most of the filtration/size based techniques such as ScreenCell,9 MOFF10 and ISET11 seem to isolate CTCs. However, RBC saturation and clogging is a problem in these devices. Similarly, microfabricated filters also isolate CTCs.12,13 The problem with most size-based technologies is that the CTCs are highly deformable unless fixed chemically and EMT related CTCs might not be retained. CAM is another unique strategy where only CTCs are captured through digesting the collagen, with 52% sensitivity in stage 1–3 breast cancer.35
Compared to all these essential techniques from the past, the nanotube-CTC-chip has advantages. This is a new antigen-independent and size independent capture technique based on the mechanobiology of tumor cells on nanotube surfaces that has not been described before. The preferential adherence strategy enables 5-log WBC depletion which is one of the best today. The capture yield is 100% at low levels of spiked triple-negative breast cancer cells (1, 10, 100) suggesting that the RBC lysis and preferential attachment of cancer cells to a nanotube surface is a highly competitive strategy. We had high success in capturing CTCs of different heterogeneity in 4 ml and 8.5 ml patient samples of different stages of breast cancer. The clear capture of CTCs in breast cancer patients and no capture of CTCs in healthy controls suggest that this chip is ready for a prospective clinical trial. Many clinical trials today are mainly doing CTC enumeration.6 However, CTC biology, phenotypes, and other pathological features are also relevant in the clinical decision making process. In a single microarray, we have been able to demonstrate the correlation between advanced disease, high CTC numbers, CTC pathological features and ability to track a single CTC with multiple phenotypes.
Discussion
The high mortality in cancer patients is mainly attributable to metastasis and to the fact that tumors in many cancers are generally detected at advanced, inoperable stages of the disease. Early detection of cancer before it metastasizes to other organs is crucial for its treatment and patient survival. Identification of those small subsets of CTCs that can have metastatic potential in early-stage cancer can potentially save lives through suitable intervention. Therefore, minimally invasive liquid biopsies for early detection and diagnosis, especially from a blood sample, have been a significant goal of cancer research for many years. Tissue biopsy is the standard of care in the diagnosis and treatment of cancer. It is possible that the genomic characterization of CTCs based on liquid biopsies may prove useful as a substitute for tissue biopsies, especially when tumor tissues from metastatic breast cancer patients become unavailable for different reasons.45 However, current CTC detection and capture techniques suffer from one or many problems such as sensitivity, specificity, size and deformability of cells through filters, dependence on immunohistochemistry, non-viability of captured fixed cells, WBC contamination and mass production related issues due to complexities.There are several advantages to the nanotube-CTC-chip as mentioned before including size-independent and antigen-independent capture, surface/planar architecture and ability to capture CTCs of different phenotypes. We found that RBC lysis does not affect the target cells and CTCs were found in every patient. We captured 0.5–10 CTCs per ml in early stage (patient 2, 3, 5 and 6) cancer versus >10 CTCs per ml in advanced stages (patient 4; 28 CTCs per ml). The number of CTCs in initially diagnosed advanced disease (patient 7; stage 4 metastasized to lung and bone) was seen to be low with radiation therapy (1 CTC per ml). This suggests that our chip has predictive capabilities and can monitor therapy induced CTC numbers, similar to CELLSEARCH®, except that we capture more CTCs due to the antigen-independent method and our baseline numbers are expected to be higher compared to CELLSEARCH®, which captures CTCs based on EpCAM/CK8/18.
Using this chip, we have overcome many of the technical issues affecting the CTC community, namely the issue of cellular retrieval experienced in most fluidic devices, by using a surface architecture in our microarrays that is amenable to surface chemistry modifications.36 We have overcome the issue of mass production by using semiconductor batch fabrication techniques, with the ability to create 76-element arrays in silicon and glass with >99% yield. We have overcome the issue of WBC contamination by using a combination of RBC lysis and preferential adherence on a nanotube surface that results in the capture of CTCs and 5-log depletion of WBCs. The transparency of glass devices can enable imaging from either side of the device for CTC capture, and that lends itself to manufacturing. The presented nanotube-CTC chip is also gentle and allows for the isolation of viable cells for future CTC culture, whereas magnetic-bead-based approaches such as CELLSEARCH® can isolate only fixed, non-viable cells.46
Over the past decade, researchers have highlighted the importance of matrix stiffness, topography, compressive and shear stresses, and deformation on cells in influencing tumor growth and proliferation.31 CTCs, in order to survive and travel to a distant site, should develop the ability to attach in an environment that is not conducive to attachment. We exploit the ability of CTCs to attach preferentially to a nanotube surface to enrich them. One exciting aspect of our study is that RBC lysis does not affect target cells and nor their clusters. The ability to successfully use RBC lysis along with a method of preferential adherence using a carbon nanotube microarray suggest that our new route is simple and easy to capture CTCs, EMT related cells, and rare clusters.
The most significant aspect of our study is that we successfully identified single CTCs exhibiting multiple phenotypes in early stage (CK8/18, EGFR) and advanced breast cancer patient (Her2, EGFR) samples using our chip. Such dynamic views are not obtained by most CTC technologies currently based on EpCAM and CK8/18 enumeration. While we used immunofluorescence to identify pathological features in captured cells, future capture can directly investigate many other characterization techniques (e.g. FISH, NGS). Dynamic views of cancer genomes to understand evolutionary pathways during the process of metastasis are needed.47–49 For this to happen, high-quality CTCs using elegant and straightforward surface techniques without WBC contamination are needed, which we have shown using this proof-of-concept study in patients. Therefore, the nanotube-CTC-chip is a highly versatile technique for clinical diagnostics and monitoring therapeutic response in human cancers.
Materials and methods
Cell culture
The breast adenocarcinoma cell line luciferase/green fluorescent protein (GFP) dual-labeled MDA-MB-231 was cultured in RPMI-1640 growth medium, MCF7 breast cancer cells were cultured in EMEM growth medium, SKBR-3 breast cancer cells were cultured in McCoy's 5a growth medium, cervical adenocarcinoma cell line HeLa was cultured in low glucose DMEM growth medium, and brain cancer cell lines U251, U-343, LN-229 were cultured in low glucose DMEM growth medium as per their suggested protocol by the manufacturer. All media contain 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin. The cell lines were incubated at 37 °C and 5% CO2. For resuspension of cells, 0.25% EDTA–trypsin solution was used.Five samples containing 1, 10, 100, 400, and 1000 cells were spiked in 10 μl wild mice blood in 5 different 1.5 μl micro-centrifuge tubes. After each sample was lysed, the cells were resuspended in culture medium, and they were divided into six CNT chips each having 10 μl volume. Incubation conditions and time were the same as in previous spiking experiments. The same counting strategy was utilized to count the cells that were adhered to the primary device and not adhered to the secondary devices based on CAM strategy.
Patients' blood sample processing
The collected blood sample was transferred from the original tube to a 15 ml centrifuge tube, and then it was centrifuged at 300g force for 5 minutes. The blood plasma was removed from the supernatant. The cell pellet at the bottom of the tube was resuspended in 12 ml lysis buffer (G-Bioscience #786650). After mixing for 3 minutes, the tube was centrifuged at 130g for 5 minutes. The supernatant of the lysed sample was transferred to another tube (waste tube). Using 1 ml of culture medium, the cells were resuspended at the bottom of the tube and then they were transferred to a 1.5 ml microcentrifuge tube. Cells were centrifuged at 130g for 5 minutes, and then the supernatant was transferred to a waste bottle. The remaining cells were resuspended in 120 μl culture medium, and they were divided into 12 CNT devices. These devices were kept in a larger Petri dish containing PBS for creating a moist environment inside an incubator at 37 °C, 5% CO2. After 48 hours, the droplets on the devices were removed, and the devices were washed once with PBS. The isolated cells on the device are then used for immunofluorescence studies.Immunofluorescence analysis
The cells were fixed with 4% paraformaldehyde for 10 minutes. The sample was washed with PBS and then blocked with Image-iT™ FX Signal Enhancer and immunofluorescence blocking buffer (Cell Signaling #12411) for 1 hour each at room temperature. The primary antibody was diluted based on the suggested concentration by the manufacturer, and then the sample was covered with it and incubated at 4 °C overnight. The sample was washed with PBS 3 times. The secondary antibody was diluted to 1 μg ml−1, and the sample was covered for 1 hour at room temperature in a dark container. The final step was to stain the nucleus with DAPI, wash the devices and mount the sample with a coverslip. Table S1† provides the manufacturer and concentrations used of all the antibodies.Author contributions
MSL and BP developed the idea of the 76-element array in glass for capturing CTCs. MSL, BP, HY, and MDJ had discussions and designed the spiked blood experiments. SMA assisted MSL in the fabrication and testing of the 76-element array chip. FF assisted MSL in EM imaging experiments. VR, an undergraduate intern, helped MSL and SMA. BP, SNR and FA had regular experimental discussions, set up IRB protocols, designed the study in patients and shipped patient samples to BP's laboratory. MM is a breast cancer oncologist who recruited patients at UofL. Informed consent was given by the patients. Melissa Barrouse-Hall was the clinical coordinator at the University of Louisville who coordinated the patient samples. MSL, BP, FA, and SNR analyzed the data together. BP and MSL wrote the manuscript and revised it together.Ethical considerations
The mouse blood was obtained from wild type C57BL/6J mice. All animal studies were conducted under the auspices of an IACUC protocol approved by the University of Massachusetts Medical School Institutional Animal Care and Use Committee (IACUC). De-identified samples from patients were obtained from the University of Louisville (IRB#18.0828). De-identified blood samples were also obtained from the UMass Biorepository and Tissue Bank as a second source to validate the chip. WPI IRB #19.0550.Conflicts of interest
MSL, BP and SNR has financial interest in Strandsmart Inc.Acknowledgements
The authors acknowledge partial funding provided by the Office of the Vice Provost for Research at Worcester Polytechnic Institute. The authors thank the University of Massachusetts Biorepository and Tissue Bank. BP and SNR thank Dr. Jason Chesney, Director, James Graham Brown Cancer Center at the University of Louisville School of Medicine for his support of the project. Partial financial support was provided by Strandsmart Inc., a Silicon Valley start-up company. SNR is supported by the Wendell Cherry Chair in Clinical Trial Research at the University of Louisville. BP, MSL, & SMA thank Dr. Christopher Lambert of WPI for help with cell culture and useful discussions.References
- S. A. Joosse, T. M. Gorges and K. Pantel, Biology, detection, and clinical implications of circulating tumor cells, EMBO Mol. Med., 2015, 7, 1–11 CrossRef CAS PubMed.
- C. Alix-Panabieres and K. Pantel, OPINION Challenges in circulating tumour cell research, Nat. Rev. Cancer, 2014, 14, 623–631 CrossRef CAS PubMed.
- S. Mocellin, U. Keilholz, C. R. Rossi and D. Nitti, Circulating tumor cells: the ‘leukemic phase’ of solid cancers, Trends Mol. Med., 2006, 12, 130–139 CrossRef CAS PubMed.
- E. C. Ory, D. S. Chen, K. R. Chakrabarti, P. P. Zhang, J. I. Andorko and C. M. Jewell, et al., Extracting microtentacle dynamics of tumor cells in a non-adherent environment, Oncotarget, 2017, 8, 111567–111580 CrossRef PubMed.
- S. Watanabe, The metastasizability of tumor cells, Cancer, 1954, 7, 215–223 CrossRef CAS PubMed.
- D. S. Micalizzi, S. Maheswaran and D. A. Haber, A conduit to metastasis: circulating tumor cell biology, Genes Dev., 2017, 31, 1827–1840 CrossRef CAS PubMed.
- M. Cristofanilli, G. T. Budd, M. J. Ellis, A. Stopeck, J. Matera and M. C. Miller, et al., Circulating tumor cells, disease progression, and survival in metastatic breast cancer, N. Engl. J. Med., 2004, 351, 781–791 CrossRef CAS PubMed.
- B. Rack, C. Schindlbeck, J. Juckstock, U. Andergassen, P. Hepp and T. Zwingers, et al., Circulating Tumor Cells Predict Survival in Early Average-to-High Risk Breast Cancer Patients, J. Natl. Cancer Inst., 2014, 106, dju066 CrossRef PubMed.
- I. Desitter, B. S. Guerrouahen, N. Benali-Furet, J. Wechsler, P. A. Janne and Y. A. Kuang, et al., A New Device for Rapid Isolation by Size and Characterization of Rare Circulating Tumor Cells, Anticancer Res., 2011, 31, 427–441 Search PubMed.
- K. A. Hyun, K. Kwon, H. Han, S. I. Kim and H. I. Jung, Microfluidic flow fractionation device for label-free isolation of circulating tumor cells (CTCs) from breast cancer patients, Biosens. Bioelectron., 2013, 40, 206–212 CrossRef CAS PubMed.
- G. Vona, A. Sabile, M. Louha, V. Sitruk, S. Romana and K. Schutze, et al., Isolation by size of epithelial tumor cells - A new method for the immunomorphological and molecular characterization of circulating tumor cells, Am. J. Pathol., 2000, 156, 57–63 CrossRef CAS PubMed.
- H. K. Lin, S. Y. Zheng, A. J. Williams, M. Balic, S. Groshen and H. I. Scher, et al., Portable Filter-Based Microdevice for Detection and Characterization of Circulating Tumor Cells, Clin. Cancer Res., 2010, 16, 5011–5018 CrossRef CAS PubMed.
- S. Zheng, H. Lin, J. Q. Liu, M. Balic, R. Datar and R. J. Cote, et al., Membrane microfilter device for selective capture, electrolysis and genomic analysis of human circulating tumor cells, J. Chromatogr. A, 2007, 1162, 154–161 CrossRef CAS PubMed.
- A. A. Adams, P. I. Okagbare, J. Feng, M. L. Hupert, D. Patterson and J. Gottert, et al., Highly efficient circulating tumor cell isolation from whole blood and label-free enumeration using polymer-based microfluidics with an integrated conductivity sensor, J. Am. Chem. Soc., 2008, 130, 8633–8641 CrossRef CAS PubMed.
- S. Nagrath, L. V. Sequist, S. Maheswaran, D. W. Bell, D. Irimia and L. Ulkus, et al., Isolation of rare circulating tumour cells in cancer patients by microchip technology, Nature, 2007, 450, 1235–1239 CrossRef CAS PubMed.
- S. L. Stott, C. H. Hsu, D. I. Tsukrov, M. Yu, D. T. Miyamoto and B. A. Waltman, et al., Isolation of circulating tumor cells using a microvortex-generating herringbone-chip, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 18392–18397 CrossRef CAS PubMed.
- E. Ozkumur, A. M. Shah, J. C. Ciciliano, B. L. Emmink, D. T. Miyamoto and E. Brachtel, et al., Inertial Focusing for Tumor Antigen-Dependent and -Independent Sorting of Rare Circulating Tumor Cells, Sci. Transl. Med., 2013, 5, 179ra147 Search PubMed.
- E. Sollier, D. E. Go, J. Che, D. R. Gossett, S. O'Byrne and W. M. Weaver, et al., Size-selective collection of circulating tumor cells using Vortex technology, Lab Chip, 2014, 14, 63–77 RSC.
- D. E. Campton, A. B. Ramirez, J. J. Nordberg, N. Drovetto, A. C. Clein and P. Varshavskaya, et al., High-recovery visual identification and single-cell retrieval of circulating tumor cells for genomic analysis using a dual-technology platform integrated with automated immunofluorescence staining, BMC Cancer, 2015, 15, 360 CrossRef PubMed.
- W. Harb, A. Fan, T. Tran, D. C. Danila, D. Keys and M. Schwartz, et al., Mutational Analysis of Circulating Tumor Cells Using a Novel Microfluidic Collection Device and qPCR Assay, Transl. Oncol., 2013, 6, 528–538 CrossRef PubMed.
- Y. T. Lu, L. B. Zhao, Q. L. Shen, M. A. Garcia, D. X. Wu and S. Hou, et al., NanoVelcro Chip for CTC enumeration in prostate cancer patients, Methods, 2013, 64, 144–152 CrossRef CAS PubMed.
- P. R. C. Gascoyne and S. Shim, Isolation of Circulating Tumor Cells by Dielectrophoresis, Cancers, 2014, 6, 545–579 CrossRef PubMed.
- S. Shim, K. Stemke-Hale, A. M. Tsimberidou, J. Noshari, T. E. Anderson and P. R. C. Gascoyne, Antibody-independent isolation of circulating tumor cells by continuous-flow dielectrophoresis, Biomicrofluidics, 2013, 7, 011807 CrossRef PubMed.
- M. C. Miller, P. S. Robinson, C. Wagner and D. J. O'Shannessy, The Parsortix (TM) Cell Separation System-A versatile liquid biopsy platform, Cytometry, Part A, 2018, 93, 1234–1239 CrossRef PubMed.
- R. Riahi, P. Gogoi, S. Sepehri, Y. Zhou, I. Handique and J. Godsey, et al., A novel microchannel-based device to capture and analyze circulating tumor cells (CTCs) of breast cancer, Int. J. Oncol., 2014, 44, 1870–1878 CrossRef PubMed.
- B. L. Khoo, G. Grenci, Y. B. Lim, S. C. Lee, J. Han and C. T. Lim, Expansion of patient-derived circulating tumor cells from liquid biopsies using a CTC microfluidic culture device, Nat. Protoc., 2018, 13, 34–58 CrossRef CAS PubMed.
- F. Khosravi, P. J. Trainor, C. Lambert, G. Kloecker, E. Wickstrom and S. N. Rai, et al., Static micro-array isolation, dynamic time series classification, capture and enumeration of spiked breast cancer cells in blood: the nanotube-CTC chip, Nanotechnology, 2016, 27, 44LT03 CrossRef PubMed.
- F. Khosravi, P. Trainor, S. N. Rai, G. Kloecker, E. Wickstrom and B. Panchapakesan, Label-free capture of breast cancer cells spiked in buffy coats using carbon nanotube antibody micro-arrays, Nanotechnology, 2016, 27, 13LT02 CrossRef PubMed.
- K. Wang, S. M. Xie, Y. Ren, H. B. Xia, X. W. Zhang and J. J. He, Establishment of a bioluminescent MDA-MB-231 cell line for human triple-negative breast cancer research, Oncol. Rep., 2012, 27, 1981–1989 Search PubMed.
- C. Raimondi, C. Nicolazzo and A. Gradilone, Circulating tumor cells isolation: the post-EpCAM era, Chin. J. Cancer Res., 2015, 27, 461–470 CAS.
- P. K. Chaudhuri, B. C. Low and C. T. Lim, Mechanobiology of Tumor Growth, Chem. Rev., 2018, 118, 6499–6515 CrossRef CAS PubMed.
- W. Q. Chen, S. G. Allen, A. K. Reka, W. Y. Qian, S. Han and J. N. Zhao, et al., Nanoroughened adhesion-based capture of circulating tumor cells with heterogeneous expression and metastatic characteristics, BMC Cancer, 2016, 16, 614 CrossRef PubMed.
- A. A. Khalili and M. R. Ahmad, A Review of Cell Adhesion Studies for Biomedical and Biological Applications, Int. J. Mol. Sci., 2015, 16, 18149–18184 CrossRef CAS PubMed.
- S. Hong, E. Ergezen, R. Lec and K. A. Barbee, Real-time analysis of cell-surface adhesive interactions using thickness shear mode resonator, Biomaterials, 2006, 27, 5813–5820 CrossRef CAS PubMed.
- J. Lu, T. Fan, Q. Zhao, W. Zeng, E. Zaslavsky and J. J. Chen, et al., Isolation of circulating epithelial and tumor progenitor cells with an invasive phenotype from breast cancer patients, Int. J. Cancer, 2010, 126, 669–683 CrossRef CAS PubMed.
- M. M. Ferreira, V. C. Romani and S. S. Jeffrey, Circulating tumor cell technologies, Mol. Oncol., 2016, 10, 374–394 CrossRef CAS PubMed.
- S. R. Sirkisoon, R. L. Carpenter, T. Rimkus, L. Miller, L. Metheny-Barlow and H. W. Lo, EGFR and HER2 signaling in breast cancer brain metastasis, Front. Biosci., Elite Ed., 2016, 8, 245–263 CrossRef.
- J. Kim, J. N. Kong, H. Chang, H. Kim and A. Kim, EGF induces epithelial-mesenchymal transition through phospho-Smad2/3-Snail signaling pathway in breast cancer cells, Oncotarget, 2016, 7, 85021–85032 Search PubMed.
- H. W. Lo, S. C. Hsu, W. Y. Xia, X. Y. Cao, J. Y. Shih and Y. K. Wei, et al., Epidermal growth factor receptor cooperates with signal transducer and activator of transcription 3 to induce epithelial-mesenchymal transition in cancer cells via up-regulation of TWIST gene expression, Cancer Res., 2007, 67, 9066–9076 CrossRef CAS PubMed.
- P. M. Navolanic, L. S. Steelman and J. A. McCubrey, EGFR family signaling and its association with breast cancer development and resistance to chemotherapy (Review), Int. J. Oncol., 2003, 22, 237–252 CAS.
- N. L. Grupka, K. C. Lear-Kaul, B. K. Kleinschmidt-DeMasters and M. Singh, Epidermal growth factor receptor status in breast cancer metastases to the central nervous system - Comparison with HER-2/neu status, Arch. Pathol. Lab. Med., 2004, 128, 974–979 CAS.
- J. Gaedcke, F. Traub, S. Milde, L. Wilkens, A. Stan and H. Ostertag, et al., Predominance of the basal type and HER-2/neu type in brain metastasis from breast cancer, Mod. Pathol., 2007, 20, 864–870 CrossRef CAS PubMed.
- F. Farace, C. Massard, N. Vimond, F. Drusch, N. Jacques and F. Billiot, et al., A direct comparison of CellSearch and ISET for circulating tumour-cell detection in patients with metastatic carcinomas, Br. J. Cancer, 2011, 105, 847–853 CrossRef CAS PubMed.
- S. H. Au, J. Edd, A. E. Stoddard, K. H. K. Wong, F. Fachin and S. Maheswaran, et al., Microfluidic Isolation of Circulating Tumor Cell Clusters by Size and Asymmetry, Sci. Rep., 2017, 7, 2433 CrossRef PubMed.
- H. E. Liu, M. Triboulet, A. Zia, M. Vuppalapaty, E. Kidess-Sigal and J. Coller, et al., Workflow optimization of whole genome amplification and targeted panel sequencing for CTC mutation detection, npj Genomic Med., 2017, 2, 34 CrossRef PubMed.
- W. J. Allard, J. Matera, M. C. Miller, M. Repollet, M. C. Connelly and C. Rao, et al., Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases, Clin. Cancer Res., 2004, 10, 6897–6904 CrossRef PubMed.
- M. J. Magbanua, L. Hauranieh, R. Roy, D. Wolf, S. Benz and C. Vaske, et al., Comprehensive genomic characterization of circulating tumor cells (CTCs) in metastatic breast cancer (MBC) sheds light on the biology of blood-borne metastasis, Cancer Res., 2017, 77(4 Suppl), P1-01-04 Search PubMed.
- S. Gkountela, B. Szczerba, C. Donato and N. Aceto, Recent advances in the biology of human circulating tumour cells and metastasis, ESMO Open, 2016, 1, e000078 CrossRef PubMed.
- K. Pantel and M. R. Speicher, The biology of circulating tumor cells, Oncogene, 2016, 35, 1216–1224 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9lc00274j |
| This journal is © The Royal Society of Chemistry 2019 |