Open Access Article
Open Access ArticleRational screening of biomineralisation peptides for colour-selected one-pot gold nanoparticle syntheses†
M.
Tanaka
 a,
Y.
Takahashi
a,
L.
Roach
b,
K.
Critchley
b,
S. D.
Evans
b and
M.
Okochi
a,
Y.
Takahashi
a,
L.
Roach
b,
K.
Critchley
b,
S. D.
Evans
b and
M.
Okochi
 *a
*a
aDepartment of Chemical Science and Engineering, Tokyo Institute of Technology, 2-12-1, O-okayama, Meguro-ku, Tokyo 152-8552, Japan. E-mail: okochi.m.aa@m.titech.ac.jp; Fax: +81-3-5734-2116; Tel: +81-3-5734-2116
bSchool of Physics and Astronomy, University of Leeds, Leeds, LS2 9JT, UK
First published on 20th August 2018
Abstract
Biomineralisation peptides that facilitate the one-pot synthesis of gold nanoparticles (AuNPs) with selected optical properties, were screened using a coherent peptide-spotted array consisting of a AuNP binding peptide library. As the biomineralised AuNPs were captured on each peptide spot, analysis of the images provided information on their collective optical properties.
Living organisms produce finely tuned biomineral architectures with the aid of biomineral-associated molecules.1,2 In contrast, syntheses of inorganic nanoparticles (NPs) are generally performed under relatively severe conditions such as high temperature, high pressure, or extreme pH, and require the presence of various harsh reagents, such as strong reducing/oxidising agents and surfactants. Therefore, the use of biomolecules in NP syntheses has received attention as a potential ‘green synthesis’ vehicles for the morphological and functional regulation of inorganic nanomaterials.3–5
Amongst the plethora of available biological molecules, peptides have attracted particular attention as valuable tools in bionanotechnology,6–12 because of their diverse functionalities and, with certain peptides, the potential for specific binding to particular materials (both organic and inorganic).13–15 Various peptides have been utilised in the functionalisation of nanomaterials,13,16,17 the control of NP synthesis,18,19 as biotemplating for morphological control,20 and biocatalysts for the NP crystalisation.6,21–23 The catalytic peptides used for NP synthesis, named biomineralisation peptides, generally demonstrate high specific binding to a target material, and are screened from a peptide library (e.g. phage display library).5,24 However, the properties of NPs synthesised, using such biomineralisation peptides, cannot be predicted from screening process; thus the development of a rational peptide screening method which can be used to synthesise NPs with specific morphological and optical properties would greatly expand the potential of biomineralisation peptides.
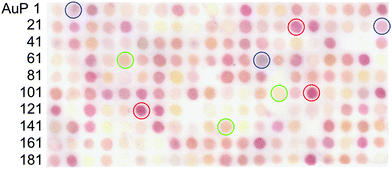
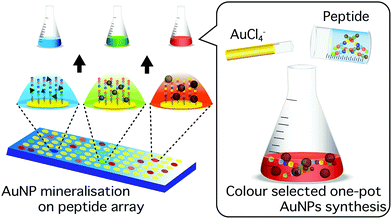
Here, we report a novel approach for the identification of biomineralisation peptides suitable for the synthesis of NPs with selected optical properties. In this study, as a model, gold NPs (AuNPs) were investigated due to their highly stable surface and localized surface plasmon resonance (LSPR) in the visible spectrum; the latter allows colour evaluation of synthesised NPs.25,26 The wavelength of the AuNP LSPR is dependent on the shape, size, and agglomeration state.27–29 The peptide screening technique uses a coherent membrane-supported peptide array library30,31 consisting of AuNP binding peptides.32 Each spot was able to capture synthesised AuNPs and through the accompanying colour change indicate the unique optical properties of bound AuNPs (Fig. 1). The use of this strategy provides a simple and effective technique to screen biomineralisation peptides for the synthesis of AuNPs with selected optical properties in ambient condition.
 | ||
| Fig. 1 Schematic illustration of peptide screening for colour selected one-pot AuNPs biomineralisation. | ||
The schematic flow for AuNP mineralisation peptide screening is shown in Fig. S1.† In a previous study, we developed a technique for the screening of AuNP-binding peptides by designing an array based on varying the amino acid frequency informed by empirical results.32 This simple strategy revealed a list of approximately 1800 peptides with various AuNP binding affinities. Herein, we evaluated the biomineralisation property of the top two hundred high-binding peptide sequences (ESI Table 1†). Various biomineralisation peptides have previously been isolated from peptides strongly bound to target nanocrystals.5,24 Consideration of the chemical equilibrium of metallic ions and their crystalline NPs in solution, suggests that the strong binding of peptides stabilises the crystals, resulting in a shift of equilibrium from ionic state towards that of the crystal; i.e. the mineralisation of target crystal is induced by the presence of the peptide.33 In addition, it is known that crystal binding molecules potentially regulate crystal morphology by enhancing and/or interfering with their growth at random and/or specific crystal facets.24,34–36 Hence, biomineralisation peptides can be screened effectively from a library of AuNP binding peptides, and synthesised particles are expected to show various optical properties derived from different peptide-regulated particle morphologies.
After 1 h incubation of the peptide array in an aqueous solution of 0.5 mM AuCl4− in Tris-buffered saline (pH 7.4), individual peptide array spots were observed to change colour, indicative of AuNP biomineralisation. The observed colours included yellow, red, pink, orange, and purple (Fig. 2). Several spots containing peptides such as AuP90: NLWMQGGIML, AuP94: SMWGTTQGHT, AuP59: MPWESHEISE, AuP80: GGWHKLDESE, and AuP156: GHWQEIGDGV showed no colour change. The characteristic colour of each peptide spot from the scanned image (Canon, LiDE220), was evaluated using ImageJ software, based on their red (R), green (G) and blue (B) intensities. The red chromaticity value (Cr) for each peptide spot was calculated as Cr = R/(R + G + B). The green and blue chromaticity values were also calculated as Cg = G/(R + G + B) and Cb = B/(R + G + B), respectively. After listing the top twenty peptides of each colour for red, green and blue AuNP biomineralisation spots, secondary, the list was sorted by cumulative RGB intensity for the selection of high biomineralisation activity. As (R, G, B) = (0, 0, 0) and (R, G, B) = (255, 255, 255) correspond to zero intensity (black) and maximum intensity (white) respectively. Peptides demonstrating strong biomineralisation are signified by a low summed RGB value. In total, the R, G, and B top ten peptides (re-named to R1–R10, G1–G10 and B1–B10 peptides, respectively) and ten peptides showing the highest RGB value (no or very weak biomineralisation, N1–N10 peptides) were listed with their physicochemical properties in ESI Table 2.† In addition, comparative analyses of amino acid frequency and amino acid sequence in the top ten peptides were shown in Fig. S2 and S3.† Comparison of the amino acid frequency with the average of all 200 peptides in the library showed a significantly higher proportion of tryptophan monomers in R peptides, whilst being notably reduced in the bottom ten. As the number of tryptophan in the top ten G and B peptides was a similar high-level with the average of all 200 peptides, the residue would be prerequisite for all AuNP biomineralisation peptides. It is probable that this results from the indole structure of tryptophan, which can reduce Au(III) to Au(0).37 The large number in R peptides suggests that the biomineralisation of AuNPs showing red colour requires strong Au(III) reducing property. It is curious that histidine is increased in R and G peptides, whilst it is decreased in B peptides. In previous work, the presence of histidine in a biomineralisation peptide (AuP1) was found to reduce AuNP biomineralisation activity.32 As the histidine residue is generally known to chelate metallic ions, the binding between AuCl4− and histidine might restrict Au biomineralisation to form large particles due to steric hindrance, electrostatic interactions, or both. Therefore, the small number of histidine residues may be important in regulating the continuous biomineralisation in the synthesis of larger AuNPs with resonances showing blue colour. In addition, three peptides out of top five G peptide sequence have a unique motif, histidine (H)–isoleucine–tryptophan (HIW; G1, G3 and G4), whilst there are no HIW motif in other top ten sequences (Table S2†). The inclusion of amino acids associated with both the induction and restriction of AuNP biomineralisation in the motif may be important for green colour AuNP syntheses. Acidic amino acids, D (aspartic acid) and E (glutamic acid) were increased in negative and G peptides. As the G peptides biomineralised smaller AuNPs than R and B (see Fig. 4), the electrostatic repulsion between acidic amino acid and AuCl4− may weaken the biomineralisation activity to form small AuNPs (green) or none. This hypothesis is supported by the averaged isoelectric point (pI) and GRAVY (the grand average of the hydropathy value) (Table S2†). The magnitudes of the pI and hydropathy value GRAVY showed the following trends negative < green < blue < red and red < green < blue < negative, respectively. Therefore, peptides with poor biomineralisation properties were relatively acidic and hydrophilic. In addition, regarding to amino acid sequence analysis by logo, R peptides surprisingly showed a unique character that all ten peptides have tryptophan residue at 4th position from N-terminus, while no tryptophan was found at same position in other peptides listed in Table S2.† Although the reason is unknown thus far, the position of a key amino acid, tryptophan, for AuNP biomineralisation appears to regulate NP size producing the observed red colour.
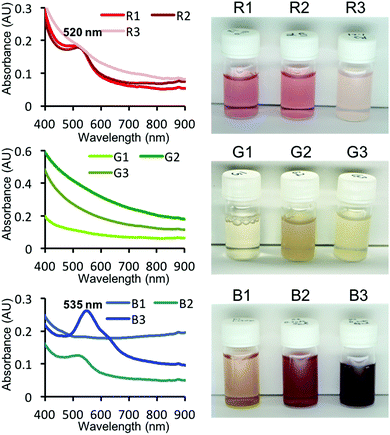
The top three R, G and B peptides that were screened from the peptide array were investigated for use in the one-pot synthesis of AuNPs in free solution. In the presence of each peptide (0.2 mM) and AuCl4− (0.5 mM), AuNPs of different colours were synthesised (Fig. 3). As expected, the observed colours were consistent with the observed colours seen in the peptide array, with the exception of the B1 peptide. The mineralisation by B1 peptide may be affected by the presence of the cellulose membrane (e.g. dispersibility). R peptides showed a peak at 515 nm (red colour) in UV-Vis spectrum. This is well-known peak derived from LSPR, however, R3 showed no peaks – only an extinction curve consistent with scattering. The R3 and G3 peptides revealed white dimmed colour after addition of the AuCl4− solution. This is probably the result of molecular association between the peptide and AuCl4− in this condition, concealing the weak absorbance peak for R3. All G peptides showed no peak, but the colour is clearly different with the sample containing only AuCl4− (no peptide). When AuNP sizes reduce down to <3 nm (called Au nanocluster), the plasmon resonance band is significantly weaker.21,38 Therefore, it seems probable that these G peptides biomineralised ultra small AuNPs which is supported by transmission electron microscope (TEM) observations (Fig. S4†). In addition, B2 and B3 seemed to be purple in colour. In the UV-Vis spectra of these samples showed absorbance peaks (535 nm and weak peak at around 620 nm), especially in the case of peptide B3. As the absorbance peak at around 530 nm and 600 nm shows red and blue colour, the mixed purple colour is derived from a bimodal distribution of AuNPs.
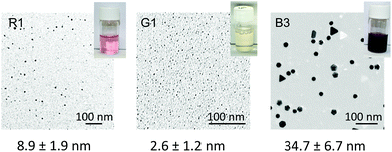
The AuNPs biomineralised by the top three R, G and B were observed by TEM (Fig. S4†). As found in TEM images, the size distribution was different in each sample (green < red < blue). The AuNP sizes synthesised by all three G and R peptides were found to have average diameters of approximately 3 nm and 10 nm, respectively. Some precipitates were found in vials in the presence of R3 and all G peptides (G1, 2, and 3), and were observed unique particle aggregates or coral-like shape, respectively (Fig. S5†). On the other hand, B peptides revealed wider distributions with relatively larger sizes. In the case of B1 peptide, while the size range was similar with other B peptides (B2 and B3), aggregated particles were frequently found. This could explain the unique colour displayed by AuNPs biomineralised by B1. The AuNPs biomineralised by R and G peptides are mostly spherical, while AuNPs biomineralised by B3 showed unique morphologies such as triangular and hexagonal plates and decahedron. The surfaces of such particles are dominated by {111} crystal facets,39,40 suggesting that the peptide is capable of selectively stabilizing individual facets and hence could potentially be used in the controlled formation of complex AuNP morphologies. Such particles will contribute directly to the unique extinction spectra. It is well known, that by comparison with spherical AuNPs, that SPR of such morphologies can be significantly red-shifted.39–41
The time evolution of AuNPs during the synthesis were further characterised by UV-Vis in the presence of different peptide concentrations (Fig. 3 and S6†). R1, G1 and B3 peptides were used due to their high colour intensity (see Fig. 3) and the consideration of peptide solubility (R2 and G3 peptides require DMSO for solubilisation). As found in Fig. S5,† AuNPs biomineralised by G1 showed no plasmon band for whole time at any peptide concentrations (0.25–1.0 mM). In the presence of R1 peptide, the biomineralisation reaction was almost completed within 15 min in all concentrations, whilst the reactions by peptide of B3 proceeded until around 10 h. The difference in reaction kinetics is a key factor in formation of different sizes and morphology of AuNPs.42–44 This phenomenon has been recorded on video (ESI Movie 1†). Whilst the red colours synthesised by peptides of R1 and B3 were similar for approximately the first 30 min, only sample B3 gradually changed colour to purple. The AuNPs synthesised by B3 showed a unique absorbance spectrum, compared to those synthesised by R1. Under high concentration of B3, a near infrared (NIR) peak emerged at 755 nm, which is probably derived from the LSPR of the triangular and hexagonal plates.39,41 The size distributions of AuNPs biomineralised by R1 and G1 at low concentration (0.2 mM) (Fig. S4†) were similar those synthesised at 1 mM (Fig. 4). However, AuNPs synthesised by B3 (1 mM) were larger and well-featured rather than at lower concentration (0.25 and 0.5 mM) (Fig. S4†). Nanoplates are currently attracting increased attention due to their high anisotropy with enhanced cellular uptake45 and a strong plasmon band in the NIR region that has great potential use in plasmonic photothermal therapy, biosensing, cell imaging, and optical coating for solar energy converters.39,41,46,47 Improvement in the shape yield (currently <15%, Fig. 4C) would represent a significant benefit to biomedical applications, because the synthesis process is very simple, safe and environmentally-friendly without the requirement for harsh chemicals such as organic solvents and surfactants.
Conclusions
In this study, we have developed a technique for the screening of biomineralisation peptides for use in the “green” one-pot synthesis of AuNPs with selected optical properties. Using peptide array, the optical property information from biomineralised AuNPs was collectively obtained through image analysis. Subsequently, screened peptides were demonstrated as suitable agents for the one-pot aqueous-phase synthesis of AuNPs. Observed optical properties of synthesised particles were consistent with those observed during peptide screening. Through further optimization of the synthesis conditions, the obtained AuNPs may find use, particularly in biological applications, due to the simple and safe synthetic protocol, free of toxic reagents. For example, green peptides seem to synthesize Au nanoclusters and B3 peptide synthesize AuNPs showing near-infrared absorbance. Materials with such properties are already found and applied in biosensing, bioimaging and therapeutics.21,25,26,47 These techniques are show potential not only in improved AuNP biomineralisations, but also could be used in the synthesis of other inorganic nanomaterials, such as semiconductor and magnetic particles.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported and funded by the Impulsing Paradigm Change through Disruptive Technologies (ImPACT) Program of the Council for Science, Technology, and Innovation (Cabinet Office, Government of Japan) and partially by JSPS KAKENHI (Grant Numbers: 16K14488 and 15H04192). M. T. thanks for funds from Foundation for Promotion of Material Science and Technology (MST) of Japan and the Royal Society UK under the Newton International Fellowships follow on funding scheme. And this work was supported in part by the international collaboration research projects sponsored by the JSPS and the Royal Society (IEC\R3\170038). The authors acknowledge the O-okayama Materials Analysis Division of Tokyo Institute of Technology for their assistance with the TEM and SEM analysis.Notes and references
- S. Mann, Nature, 1993, 365, 499–505 CrossRef CAS.
- L. Addadi and S. Weiner, Angew. Chem., Int. Ed. Engl., 1992, 31, 153–169 CrossRef.
- M. Dickerson, K. Sandhage and R. Naik, Chem. Rev., 2008, 108(11), 4935–4978 CrossRef CAS PubMed.
- G. Mirabello, J. J. M. Lenders and N. A. J. M. Sommerdijk, Chem. Soc. Rev., 2016, 45, 5085–5106 RSC.
- Y. Maeda, O. V. Makhlynets, H. Matsui and I. V. Korendovych, Annu. Rev. Biomed. Eng., 2016, 18, 311–328 CrossRef CAS PubMed.
- W. Yang, W. Guo, J. Chang and B. Zhang, J. Mater. Chem. B, 2017, 5, 401–417 RSC.
- U. O. S. Seker and H. V. Demir, Molecules, 2011, 16, 1426–1451 CrossRef CAS PubMed.
- S. Pavan and F. Berti, Anal. Bioanal. Chem., 2012, 402, 3055–3070 CrossRef CAS PubMed.
- J. M. Galloway, L. Senior, J. M. Fletcher, J. L. Beesley, L. R. Hodgson, R. L. Harniman, J. M. Mantell, J. Coombs, G. G. Rhys, W.-F. Xue, M. Mosayebi, N. Linden, T. B. Liverpool, P. Curnow, P. Verkade and D. N. Woolfson, ACS Nano, 2018, 12, 1420–1432 CrossRef CAS PubMed.
- W. Zhang, X. Yu, Y. Li, Z. Su, K. D. Jandt and G. Wei, Prog. Polym. Sci., 2018, 80, 94–124 CrossRef CAS.
- G. Wei, Z. Su, N. P. Reynolds, P. Arosio, I. W. Hamley, E. Gazit and R. Mezzenga, Chem. Soc. Rev., 2017, 46, 4661–4708 RSC.
- Z. Su, H. Shen, H. Wang, J. Wang, J. Li, G. U. Nienhaus, L. Shang and G. Wei, Adv. Funct. Mater., 2015, 25, 5472–5478 CrossRef CAS.
- M. Tsutsui, M. Tanaka, T. Marui, K. Yokota, T. Yoshida, A. Arima, W. Tonomura, M. Taniguchi, T. Washio, M. Okochi and T. Kawai, Anal. Chem., 2018, 90, 1511–1515 CrossRef CAS PubMed.
- M. Okochi, M. Ogawa, C. Kaga, T. Sugita, Y. Tomita, R. Kato and H. Honda, Acta Biomater., 2010, 6, 2301–2306 CrossRef CAS PubMed.
- M. Okochi, T. Kamiya, T. Omasa, M. Tanaka and H. Honda, Anal. Sci., 2016, 32, 93–97 CrossRef CAS PubMed.
- M. Tanaka, I. H. Harlisa, Y. Takahashi, N. A. Ikhsan and M. Okochi, RSC Adv., 2018, 8, 8795–8799 RSC.
- J. C. Breger, M. Muttenthaler, J. B. Delehanty, D. A. Thompson, E. Oh, K. Susumu, J. R. Deschamps, G. P. Anderson, L. D. Field, S. A. Walper, P. E. Dawson and I. L. Medintz, Nanoscale, 2017, 9, 10447–10464 RSC.
- N. M. Bedford, Z. E. Hughes, Z. Tang, Y. Li, B. D. Briggs, Y. Ren, M. T. Swihart, V. G. Petkov, R. R. Naik, M. R. Knecht and T. R. Walsh, J. Am. Chem. Soc., 2016, 138, 540–548 CrossRef CAS PubMed.
- Y. Li, Z. Tang, P. N. Prasad, M. R. Knecht and M. T. Swihart, Nanoscale, 2014, 6, 3165–3172 RSC.
- D. Gottlieb, S. A. Morin, S. Jin and R. T. Raines, J. Mater. Chem., 2008, 18, 3865–3870 RSC.
- Q. Yuan, Y. Wang, L. Zhao, R. Liu, F. Gao, L. Gao and X. Gao, Nanoscale, 2016, 8, 12095–12104 RSC.
- S. Bolisetty, C. S. Boddupalli, S. Handschin, K. Chaitanya, J. Adamcik, Y. Saito, M. G. Manz and R. Mezzenga, Biomacromolecules, 2014, 15, 2793–2799 CrossRef CAS PubMed.
- W. Zhang, D. Lin, H. Wang, J. Li, G. Ulrich Nienhaus, Z. Su, G. Wei and L. Shang, Bioconjugate Chem., 2017, 28(9), 2224–2229 CrossRef CAS PubMed.
- C. Chen and N. Rosi, Angew. Chem., Int. Ed., 2010, 49, 1924–1942 CrossRef CAS PubMed.
- Z. Miao, Z. Gao, R. Chen, X. Yu, Z. Su and G. Wei, Curr. Med. Chem., 2018, 25(16), 1920–1944 CrossRef CAS PubMed.
- H. Aldewachi, T. Chalati, M. N. Woodroofe, N. Bricklebank, B. Sharrack and P. Gardiner, Nanoscale, 2017, 10, 18–33 RSC.
- L. Roach, S. Ye, S. C. T. Moorcroft, K. Critchley, P. L. Coletta and S. D. Evans, Nanotechnology, 2018, 29, 135601 CrossRef PubMed.
- S. D. E. S. Ye, F. Benz, M. C. Wheeler, J. Oram, J. J. Baumberg, O. Cespedes, H. K. Christenson, P. L. Coletta, L. J. C. Jeuken, A. F. Markham and K. Critchley, Nanoscale, 2016, 8, 14932–14942 RSC.
- S. Ye, G. Marston, J. R. McLaughlan, D. O. Sigle, N. Ingram, S. Freear, J. J. Baumberg, R. J. Bushby, A. F. Markham, K. Critchley, P. L. Coletta and S. D. Evans, Adv. Funct. Mater., 2015, 25, 2117–2127 CrossRef CAS.
- R. Frank, J. Immunol. Methods, 2002, 267, 13–26 CrossRef CAS PubMed.
- R. Frank, Tetrahedron, 1992, 48, 9217–9232 CrossRef CAS.
- M. Tanaka, S. Hikiba, K. Yamashita, M. Muto and M. Okochi, Acta Biomater., 2017, 1, 13–26 Search PubMed.
- T. Hatanaka, A. Matsugami, T. Nonaka, H. Takagi, F. Hayashi, T. Tani and N. Ishida, Nat. Commun., 2017, 8, 15670 CrossRef CAS PubMed.
- A. Yamagishi, M. Tanaka, J. J. M. Lenders, J. Thiesbrummel, N. A. J. M. Sommerdijk, T. Matsunaga and A. Arakaki, Sci. Rep., 2016, 6, 29785 CrossRef CAS PubMed.
- M. Tanaka, E. Mazuyama, A. Arakaki and T. Matsunaga, J. Biol. Chem., 2011, 286, 6386–6392 CrossRef CAS PubMed.
- C. Y. Chiu, Y. Li, L. Ruan, X. Ye, C. B. Murray and Y. Huang, Nat. Chem., 2011, 3, 393–399 CrossRef CAS PubMed.
- P. R. Selvakannan, S. Mandal, S. Phadtare, A. Gole, R. Pasricha, S. D. Adyanthaya and M. Sastry, J. Colloid Interface Sci., 2004, 269, 97–102 CrossRef CAS PubMed.
- A. Baral, K. Basu, S. Ghosh, K. Bhattacharyya, S. Roy, A. Datta and A. Banerjee, Nanoscale, 2017, 9, 4419–4429 RSC.
- L. Chen, F. Ji, Y. Xu, L. He, Y. Mi, F. Bao, B. Sun, X. Zhang and Q. Zhang, Nano Lett., 2014, 14, 7201–7206 CrossRef CAS PubMed.
- P. Yang, H. Portalès and M.-P. Pileni, J. Chem. Phys., 2011, 134, 024507 CrossRef PubMed.
- A. Miranda, E. Malheiro, E. Skiba, P. Quaresma, P. a Carvalho, P. Eaton, B. de Castro, J. a Shelnutt and E. Pereira, Nanoscale, 2010, 2, 2209–2216 RSC.
- Y. Xiong, J. M. McLellan, J. Chen, Y. Yin, Z. Y. Li and Y. Xia, J. Am. Chem. Soc., 2005, 127, 17118–17127 CrossRef CAS PubMed.
- A. Le Beulze, E. Duguet, S. Mornet, J. Majimel, M. Tréguer-Delapierre, S. Ravaine, I. Florea and O. Ersen, Langmuir, 2014, 30, 1424–1434 CrossRef CAS PubMed.
- D. Alloyeau, W. Dachraoui, Y. Javed, H. Belkahla, G. Wang, H. Lecoq, S. Ammar, O. Ersen, A. Wisnet, F. Gazeau and C. Ricolleau, Nano Lett., 2015, 15, 2574–2581 CrossRef CAS PubMed.
- K. Nambara, K. Niikura, H. Mitomo, T. Ninomiya, C. Takeuchi, J. Wei, Y. Matsuo and K. Ijiro, Langmuir, 2016, 32, 12559–12567 CrossRef CAS PubMed.
- H. Daraee, A. Eatemadi, E. Abbasi, S. Fekri Aval, M. Kouhi and A. Akbarzadeh, Artif. Cells, Nanomed., Biotechnol., 2014, 1401, 1–13 Search PubMed.
- S. Ashraf, B. Pelaz, P. del Pino, M. Carril, A. Escudero, W. J. Parak, M. G. Soliman, Q. Zhang and C. Carrillo-Carrion, Top. Curr. Chem., 2016, 370, 169–202 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8na00075a |
| This journal is © The Royal Society of Chemistry 2019 |