Open Access Article
Open Access ArticleControlled disassembly of a DNA tetrahedron using strand displacement†
Arun Richard
Chandrasekaran
 * and
Ken
Halvorsen
*
* and
Ken
Halvorsen
*
The RNA Institute, University at Albany, State University of New York, Albany, NY, USA. E-mail: arun@albany.edu; khalvorsen@albany.edu
First published on 21st December 2018
Abstract
In this study, we assembled a DNA tetrahedron containing single stranded extensions in the middle of the struts. Using these extensions as toeholds, the tetrahedron can be disassembled by nucleic acid triggers via strand displacement. The release mechanism is sequence specific, is functional in biological fluids such as serum and urine, and the kinetics of the disassembly process can be controlled by different molar ratios of the release strand. Such DNA nanostructures that respond to external stimuli have potential use in biosensing and drug delivery, and we demonstrate a proof-of-concept of this approach for microRNA detection.
In a 1999 episode of X-Files, Fox Mulder races to find a cure for his boss' condition, and is told that it is “technology that the world believes is purely theoretical”, to which he exclaims “nanotechnology”. Two decades later, the application of nanotechnology to health and medicine has moved beyond the theoretical. While we are still a far cry from the complex, self-replicating, and autonomous machines presented in science fiction, scientists have created simple nanomachines in the lab. Various versions have been based on metal nanowires, carbon nanotubes, artificial biological assemblies, small molecule motifs and mechanically interlocked molecules.1,2 DNA is one such material that is increasingly used as a building block for the bottom-up construction of a wide variety of nanostructures.3,4 The programmability of these structures allows greater control over defined assembly and response to chemical and molecular cues.5–7 Specifically, hollow or wireframe DNA nanostructures8 can encapsulate guest objects such as proteins,9 fluorescent biopolymers,10 quantum dots11 and nanoparticles,12 with their target application being on-demand cargo delivery.13 To achieve this, DNA nanostructures are designed to respond to chemical or environmental cues such as other biomolecules,14 light,15 pH16 or enzymes.17 These cues affect the self-assembled DNA nanostructures leading to conformational changes such as pore size widening,14 unlinking of cargo,15 or complete dissociation,16,17 all of which result in cargo release. Here we present a DNA tetrahedron that can be controllably disassembled via toehold-based strand displacement process18 that is activated upon binding to a specific nucleic acid trigger.
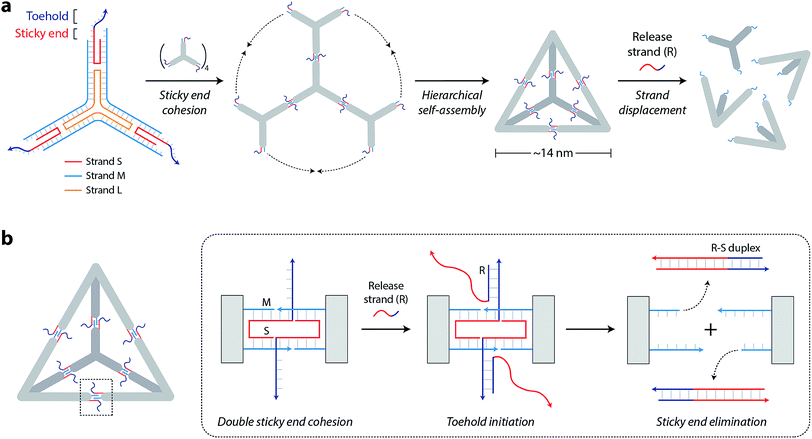
The DNA tetrahedron used in this study is assembled from three-point-star motifs that connect to each other via programmed sticky ends (Fig. 1a).19 The motif contains one long, threefold-repeating strand (L, colored orange in Fig. 1a), three copies of medium strands (M, cyan), and three copies of short, peripheral strands (S, red-and-dark blue). The center portion of the motif contains 5T loops (on strand L) that provide flexibility for the arms to associate into higher order structures (Fig. S1†). Each arm of the motif contains two helices connected by a single crossover. Both helices of the motif are tailed with 7-nt sticky ends (one extending from S and one extending from M) so that motifs can join through sticky end connections to further assemble into the DNA tetrahedron (a tetrahedron contains four units of the motif). The edges of the assembled tetrahedron are four turns of DNA (∼14 nm). The resulting tetrahedron is robust and remains stable at room temperature for a prolonged time (Fig. S2†).
To enable controlled disassembly of our tetrahedron, we modified the design by extending the single-stranded overhang on strand S by 8 nucleotides beyond the sticky end region (dark blue region of strand S in Fig. 1a). Upon assembly, the DNA tetrahedron contains single-stranded tails protruding from the middle of the struts that act as toeholds to recognize external nucleic acid triggers. The release strand (R, dark blue-red) is designed to be complementary to the toehold region and to the remainder of strand S. Addition of the release strand results in strand displacement and elimination of the sticky end interactions that hold the tetrahedron together, leading to disassembly of the DNA tetrahedron into its component motifs (Fig. 1b).
We assembled the DNA tetrahedron by mixing strands L, M, and S in a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
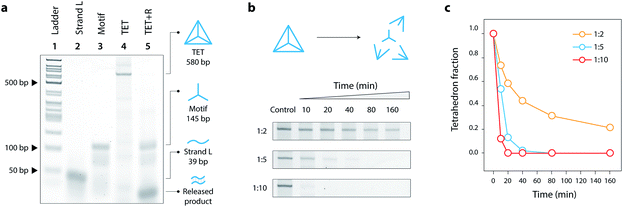
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 in Mg2+-containing tris-acetate-EDTA (TAE) buffer and slowly annealing the solution from 90 °C to 4 °C. This one-step annealing forms the individual three-point-star motifs which further self-assemble to form the DNA tetrahedron. We validated the formation of our toehold modified DNA tetrahedron (TET) using polyacrylamide gel electrophoresis (Fig. 2a, lane 4), a technique well-established to analyze such DNA nanostructures.19 The introduction of single-stranded toeholds does not seem to affect the self-assembly of the tetrahedron from three-point-star motifs (Fig. S3†). By adding the release strand, we confirmed that the tetrahedron is disassembled into its component motifs (Fig. 2a, lane 5). Addition of the release strand did not have any effect on a tetrahedron without a toehold, confirming that disassembly proceeds by toehold-mediated strand displacement (Fig. S4†).
3 in Mg2+-containing tris-acetate-EDTA (TAE) buffer and slowly annealing the solution from 90 °C to 4 °C. This one-step annealing forms the individual three-point-star motifs which further self-assemble to form the DNA tetrahedron. We validated the formation of our toehold modified DNA tetrahedron (TET) using polyacrylamide gel electrophoresis (Fig. 2a, lane 4), a technique well-established to analyze such DNA nanostructures.19 The introduction of single-stranded toeholds does not seem to affect the self-assembly of the tetrahedron from three-point-star motifs (Fig. S3†). By adding the release strand, we confirmed that the tetrahedron is disassembled into its component motifs (Fig. 2a, lane 5). Addition of the release strand did not have any effect on a tetrahedron without a toehold, confirming that disassembly proceeds by toehold-mediated strand displacement (Fig. S4†).
To further investigate the disassembly process, we next measured the progression of disassembly at different time points for different concentrations of the release strand (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, 1
2, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 or 1
5 or 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 molar ratios of tetrahedron
10 molar ratios of tetrahedron![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) release strand) (Fig. 2b, full gels in Fig. S5†). To better visualize the kinetics of disassembly, we calculated the conversion by quantifying the band intensity of the tetrahedron at different time points (Fig. 2c). Results show that the release mechanism can be effective in as little as 10 minutes for 1
release strand) (Fig. 2b, full gels in Fig. S5†). To better visualize the kinetics of disassembly, we calculated the conversion by quantifying the band intensity of the tetrahedron at different time points (Fig. 2c). Results show that the release mechanism can be effective in as little as 10 minutes for 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 molar ratio. The kinetics of the release is also affected by the toehold length, with 8 nucleotides providing the fastest disassembly compared to shorter toeholds (Fig. S6†).
10 molar ratio. The kinetics of the release is also affected by the toehold length, with 8 nucleotides providing the fastest disassembly compared to shorter toeholds (Fig. S6†).
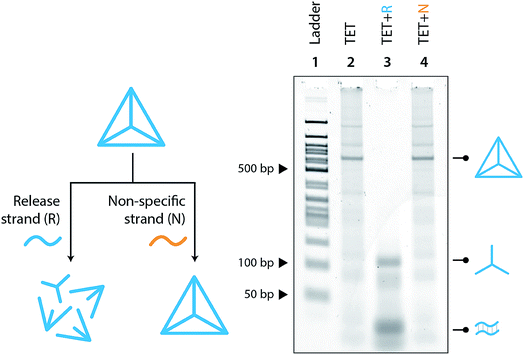
Moving toward biological applications, we investigated the specificity of the release mechanism and compatibility with biological fluids. To test the specificity, we used a non-specific release strand at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
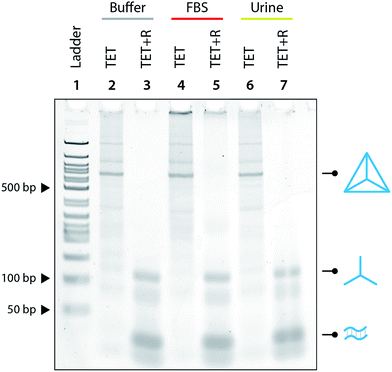
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 ratio and showed that the tetrahedron is not disassembled under those conditions (Fig. 3). We further tested the specificity by using a release strand that contains a single nucleotide mismatch in the toehold region (Fig. S7†). Results showed that the tetrahedron was not disassembled by a release strand with even a single nucleotide mismatch. Extending this concept beyond a single off-target release strand, we challenged the tetrahedron disassembly process by testing in Fetal Bovine Serum (FBS) and in synthetic urine. In both cases, the tetrahedra maintained stability in these fluids and only disassembled in the presence of the release strand (Fig. 4).
10 ratio and showed that the tetrahedron is not disassembled under those conditions (Fig. 3). We further tested the specificity by using a release strand that contains a single nucleotide mismatch in the toehold region (Fig. S7†). Results showed that the tetrahedron was not disassembled by a release strand with even a single nucleotide mismatch. Extending this concept beyond a single off-target release strand, we challenged the tetrahedron disassembly process by testing in Fetal Bovine Serum (FBS) and in synthetic urine. In both cases, the tetrahedra maintained stability in these fluids and only disassembled in the presence of the release strand (Fig. 4).
The features of our tetrahedron disassembly approach could be useful for various nucleic acid sensing applications. One such area of interest is detection of microRNAs, small RNA fragments (sometimes similar in sequence) that are increasingly recognized for their roles in gene regulation and as disease biomarkers.20,21 To demonstrate this as a potential biological application of our approach, we redesigned the tetrahedron to be disassembled upon recognition of a specific microRNA. The resultant tetrahedron had a lower yield (Fig. S8†) than our native tetrahedron, likely due to sensitivity to component sequences that were optimized for self-assembly in the native structure. Even with this lower yield, however, we showed that the tetrahedron was successfully disassembled in the presence of the correct microRNA. Disassembly was progressively reduced but not completely eliminated when presented with microRNA variants that differ by 1 nt and 2 nt.
Stimuli-responsive DNA nanostructures are increasingly shown to be useful in biosensing and drug delivery applications. The inclusion of a toe-hold is a programmable feature, and via strand displacement, allows reconfiguration of small DNA objects,22 nanodevices23 or larger DNA origami structures.24 In this work, we show that designed single-stranded extensions in DNA tetrahedra can be used as toeholds to controllably disassemble the nanostructure into its component units. This concept could be extended to other DNA polyhedra19 and possibly combined with macromolecular sensing strategies. Our preliminary data shows that tetrahedra can be designed to recognize microRNAs, adding to the existing suite of DNA nanotechnology biosensors.25–28 Since the tetrahedron and other polyhedra contain internal cavities that could be loaded with cargo, our strategy also has the potential to be used for drug delivery platforms. Considering the demonstrated stability in biological fluids and the ability to be triggered open on recognizing biologically relevant nucleic acids, these structures show promise in the creation of “sense-and-act” nanomachines.
Conflicts of interest
The authors have no conflicts to declare.Acknowledgements
Research reported in this publication was supported by the NIH through NIGMS under award R35GM124720 to K. H.References
- G. A. Ozin, I. Manners, S. Fournier-Bidoz and A. Arsenault, Adv. Mater., 2005, 17, 3011–3018 CrossRef CAS.
- S. Erbas-Cakmak, D. A. Leigh, C. T. McTernan and A. L. Nussbaumer, Chem. Rev., 2015, 115, 10081–10206 CrossRef CAS PubMed.
- P. L. Xavier and A. R. Chandrasekaran, Nanotechnology, 2018, 29, 062001 CrossRef PubMed.
- A. R. Chandrasekaran, N. Anderson, M. Kizer, K. Halvorsen and X. Wang, ChemBioChem, 2016, 17, 1081–1089 CrossRef CAS PubMed.
- X. Qu, D. Zhu, G. Yao, S. Su, J. Chao, H. Liu, X. Zuo, L. Wang, J. Shi, L. Wang, W. Huang, H. Pei and C. Fan, Angew. Chem., Int. Ed., 2017, 56, 1855–1858 CrossRef CAS PubMed.
- X. Qu, S. Wang, Z. Ge, J. Wang, G. Yao, J. Li, X. Zuo, J. Shi, S. Song, L. Wang, L. Li, H. Pei and C. Fan, J. Am. Chem. Soc., 2017, 139, 10176–10179 CrossRef CAS PubMed.
- W. Lai, L. Ren, Q. Tang, X. Qu, J. Li, L. Wang, L. Li, C. Fan and H. Pei, ACS Nano, 2018, 12, 7093–7099 CrossRef CAS PubMed.
- A. R. Chandrasekaran and O. Levchenko, Chem. Mater., 2016, 28, 5569–5581 CrossRef CAS.
- C. Zhang, C. Tian, F. Guo, Z. Liu, W. Jiang and C. Mao, Angew. Chem., Int. Ed., 2012, 51, 3382–3385 CrossRef CAS PubMed.
- D. Bhatia, S. Surana, S. Chakraborty, S. P. Koushika and Y. Krishnan, Nat. Commun., 2011, 2, 339 CrossRef PubMed.
- D. Bhatia, S. Arumugam, M. Nasilowski, H. Joshi, C. Wunder, V. Chambon, V. Prakash, C. Grazon, B. Nadal, P. K. Maiti, L. Johannes, B. Dubertret and Y. Krishnan, Nat. Nanotechnol., 2016, 11, 1112–1119 CrossRef CAS PubMed.
- Y. Li, Z. Liu, G. Yu, W. Jiang and C. Mao, J. Am. Chem. Soc., 2015, 137, 4320–4323 CrossRef CAS PubMed.
- B. R. Madhanagopal, S. Zhang, E. Demirel, H. Wady and A. R. Chandrasekaran, Trends Biochem. Sci., 2018, 43, 997–1013 CrossRef CAS PubMed.
- C. Zhang, C. Tian, X. Li, H. Qian, C. Hao, W. Jiang and C. Mao, J. Am. Chem. Soc., 2012, 134, 11998–12001 CrossRef CAS PubMed.
- R. E. Kohman, S. S. Cha, H. Y. Man and X. Han, Nano Lett., 2016, 16, 2781–2785 CrossRef CAS PubMed.
- Z. Liu, Y. Li, C. Tian and C. Mao, Biomacromolecules, 2013, 14, 1711–1714 CrossRef CAS PubMed.
- Y. Ma, Z. Wang, Y. Ma, Z. Han, M. Zhang, H. Chen and Y. Gu, Angew. Chem., Int. Ed., 2018, 57, 5389–5393 CrossRef CAS PubMed.
- B. Yurke, A. J. Turberfield, A. P. Mills Jr, F. C. Simmel and J. L. Neumann, Nature, 2000, 406, 605–608 CrossRef CAS PubMed.
- Y. He, T. Ye, M. Su, C. Zhang, A. E. Ribbe, W. Jiang and C. Mao, Nature, 2008, 452, 198–201 CrossRef CAS PubMed.
- H. Dong, J. Lei, L. Ding, Y. Wen, H. Ju and X. Zhang, MicroRNA: function, detection, and bioanalysis, Chem. Rev., 2013, 113, 6207–6233 CrossRef CAS PubMed.
- D. P. Bartel, MicroRNAs: genomics, biogenesis, mechanism, and function, Cell, 2004, 116, 281–297 CrossRef CAS PubMed.
- R. P. Goodman, M. Heilemann, S. Doose, C. M. Erben, A. N. Kapanidis and A. J. Turberfield, Nat. Nanotechnol., 2008, 3, 93–96 CrossRef CAS PubMed.
- A. R. Chandrasekaran, O. Levchenko, D. S. Patel, M. MacIsaac and K. Halvorsen, Nucleic Acids Res., 2017, 45, 11459–11465 CrossRef CAS PubMed.
- E. S. Andersen, M. Dong, M. M. Nielsen, K. Jahn, R. Subramani, W. Mamdouh, M. M. Golas, B. Sander, H. Stark, C. L. Oliveira, J. S. Pedersen, V. Birkedal, F. Besenbacher, K. V. Gothelf and J. Kjems, Nature, 2009, 459, 73–76 CrossRef CAS PubMed.
- A. R. Chandrasekaran, M. MacIsaac, P. Dey, O. Levchenko, L. Zhou, M. Andres, B. K. Dey and K. Halvorsen, Sci. Adv., 2018 DOI:10.1126/sciadv.aau9443.
- M. Xiao, A. R. Chandrasekaran, W. Ji, F. Li, T. Man, C. Zhu, X. Shen, H. Pei, Q. Li and L. Li, ACS Appl. Mater. Interfaces, 2018, 10, 35794–35800 CrossRef CAS PubMed.
- M. Rana, M. Balcioglu, M. Kovach, M. S. Hizir, N. M. Robertson, I. Khan and M. V. Yigit, Chem. Commun., 2016, 52, 3524–3527 RSC.
- M. Lin, P. Song, G. Zhou, X. Zuo, A. Aldalbahi, X. Lou, J. Shi and C. Fan, Nat. Protoc., 2016, 11, 1244–1263 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8na00340h |
| This journal is © The Royal Society of Chemistry 2019 |