Open Access Article
Open Access ArticleA novel method for genetic transformation of C. albicans using modified-hydroxyapatite nanoparticles as a plasmid DNA vehicle
Ketaki
Deshmukh†
a,
Sutapa Roy
Ramanan†
 *b and
Meenal
Kowshik†
*b and
Meenal
Kowshik†
 *a
*a
aBiological Sciences Department, Birla Institute of Technology and Science Pilani, K K Birla Goa Campus, Goa, India. E-mail: ketaki03@gmail.com; meenal@goa.bits-pilani.ac.in
bChemical Engineering Department, Birla Institute of Technology and Science Pilani, K K Birla Goa Campus, Goa, India. E-mail: sutapa@goa.bits-pilani.ac.in
First published on 11th June 2019
Abstract
In modern biological research, genetic transformation is an important molecular biology technique with extensive applications. In this work, we describe a new method for the delivery of plasmid DNA (pDNA) into a yeast species, Candida albicans. This method is based on the use of novel arginine–glucose–PEG functionalized hydroxyapatite nanoparticles (M-HAp NPs) as a vehicle which delivers pDNA into Candida albicans with a high transformation efficiency of 106 cfu μg−1 of pDNA, without the need for preparation of competent cells. A four-fold higher transformation efficiency as compared to that of the electroporation method was obtained. This new method could provide exciting opportunities for the advancement of the applications of yeasts in the field of biotechnology.
Introduction
Yeast has been associated with humans for the preparation of bread, wine, beer, sake, etc., since ancient times and these processes are the basis of modern biotechnology.1,2 Recently, yeast species are being used as experimental models to study signal processes and to characterize molecular events associated with human mitochondrial and neurodegenerative diseases.3–5 To advance the use of yeasts in industries and research, modification of yeast strains by delivering nucleic acids into the cell is crucial.6,7 Several types of yeast species such as Saccharomyces, Aspergillus, Rhizopus, etc. are genetically engineered for the efficient production of various drugs, industrial enzymes, vaccines, and chemicals including insulin, human serum albumin, etc.8,9 Different chemicals like phenolics, isoprenoids, alkaloids and polyketides are also produced by engineered yeast.10 However, delivery of DNA into yeast is a major challenge due to the presence of a tough cell wall which is composed of mannose-containing proteins and glycans. Various approaches like lithium acetate (LiAc)-based methodologies, electroporation, gene gun transformation, protoplast transformation, etc., have been developed to deliver nucleic acids into yeast cells.11–16 However, these methods have limitations such as a low transformation efficiency and high toxicity and many of them need costly reagents or equipment. Hence, there is a need for the development of transformation methods which are more efficient, less toxic and reproducible.Recently, nanotechnology based approaches have received much attention in the field of gene delivery due to factors such as high DNA loading capacity, high transformation efficiency and ease of delivery. Several nanoparticle based transformation methods have been developed for bacterial and eukaryotic cells.17–21 However, to our knowledge, there is only one report on nanoparticle based DNA delivery in yeasts which used oligoelectrolyte polymeric nanoscale carriers for the delivery of DNA into yeasts like Saccharomyces cerevisiae, Hansenula polymorpha and Pichia pastoris.22 Therefore, development of biocompatible nanomaterial based DNA delivery methods for the transformation of yeast cells needs to be actively explored.
Hydroxyapatite nanoparticles (HAp NPs) being biocompatible are good candidates for DNA delivery and have been explored as carriers in animal and plant cells.23–26 HAp also has advantages as it exhibits high affinity to nucleic acids and can be easily transported through cell membranes.23–26 In this work, we have synthesized arginine (R), glucose (G) and polyethylene glycol (PEG) functionalized thin-rod HAp NPs (referred to as M-HAp NPs) which form stable complexes with pDNA. Successful transformation of pDNA in Candida albicans was achieved using these HAp NPs as nanovehicles. This newly developed method exhibited four-fold increase in the transformation efficiency as compared to the electroporation technique. To the best of our knowledge, this is the first report on the transformation of C. albicans using HAp NPs with high efficiency and no toxicity.
Materials and methods
Chemicals
Orthophosphoric acid, dimethyl sulfoxide, acetyl acetone and ammonium hydroxide were procured from Merck India. Calcium chloride di-hydrate, kanamycin and geneticin were obtained from Himedia, India. Kanamycin and geneticin resistance gene containing plasmids and agarose were purchased from Genei, India.Synthesis of modified-HAp NPs
Hydroxyapatite nanoparticles (HAp NPs) were synthesized using a modified sol–gel method developed in our laboratory.27 In brief, 2 M calcium chloride dissolved in DMSO was stirred for 30 min, followed by dropwise addition of orthophosphoric acid. The Ca![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P atomic ratio was maintained at 1.67. This was followed by the addition of acetyl acetone as a stabilizing agent and stirred for 1 h. Liquid ammonia was used for adjusting the pH to 10 and stirring was continued until complete gelation. Subsequently, 0.1 wt% arginine (R), 0.3 wt% glucose (G) and 0.05 wt% polyethylene glycol (PEG) were added and stirred for 2 h to functionalize the HAp NPs. Thorough washing of the final product was carried out using ethanol, followed by dialysis (using a dialysis bag, Bangalore Genei 110) against de-ionized (DI) water for 12 h with frequent change of water for active removal of adsorbed ions. The dialyzed samples were dried at room temperature. Arginine–glucose–PEG modified HAp NPs are referred to as M-HAp NPs in following text.
P atomic ratio was maintained at 1.67. This was followed by the addition of acetyl acetone as a stabilizing agent and stirred for 1 h. Liquid ammonia was used for adjusting the pH to 10 and stirring was continued until complete gelation. Subsequently, 0.1 wt% arginine (R), 0.3 wt% glucose (G) and 0.05 wt% polyethylene glycol (PEG) were added and stirred for 2 h to functionalize the HAp NPs. Thorough washing of the final product was carried out using ethanol, followed by dialysis (using a dialysis bag, Bangalore Genei 110) against de-ionized (DI) water for 12 h with frequent change of water for active removal of adsorbed ions. The dialyzed samples were dried at room temperature. Arginine–glucose–PEG modified HAp NPs are referred to as M-HAp NPs in following text.
Characterization
X-ray diffraction (XRD) studies of HAp and M-HAp NPs were carried out for phase identification using an X-ray diffractometer (Miniflex II Rigaku, Japan) with monochromatic Cu Kα radiation (λ = 1.5405 Å) and a scan range of 2θ = 20° to 80°. The functional groups present in the synthesized compounds were ascertained by Fourier transform infrared spectroscopy (FTIR) (8201 PC Shimadzu, Japan), over the 4000–450 cm−1 region. The pellets for FTIR analysis were obtained by mixing 1 mg of the powdered sample with spectroscopic grade KBr (Merck). The particle shape and size of the M-HAp NPs were studied using a transmission electron microscope (TEM) (Phillips CM 200). NP solutions were sonicated for 15–20 min and a drop of the dispersion was put on a copper coated grid and air dried in vacuum desiccators. Dried samples were examined under a TEM at SAIF IIT Bombay. The zeta potential of M-HAp NPs was determined with a zeta potential analyzer (Delsa Nano S, Beckman Coulter, USA) at room-temperature. The samples were prepared by diluting the NP suspension with DI water. HAp and M-HAp NPs were analyzed by X-ray photoelectron spectroscopy (XPS) (AXIS Supra, Kratos Analytical, UK, monochromatic Al K-alpha, 75 W). A survey spectrum from 0–1000 eV and high-resolution spectra for Ca 2p, O 1s, P 2p N 1s and C 1s were recorded.Plasmid isolation
The plasmid was isolated from E. coli and C. albicans using a Qiagen plasmid isolation midi prep kit, following the protocol specified by the manufacturer (Qiagen plasmid midi kit). It was 10 Kb in size and contained kanamycin and geneticin resistance genes. It is represented as pDNA in the following text.Plasmid DNA binding efficiency
M-HAp NPs (1 mg mL−1) were suspended in DI water, sonicated for 10 min and mixed with 100 ng pDNA at various M-HAp NP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) pDNA ratios (0
pDNA ratios (0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (control), 10
1 (control), 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 30
1, 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 50
1, 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 70
1, 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and 100
1, and 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The suspensions were incubated at room temperature for 5 min, centrifuged at 22
1). The suspensions were incubated at room temperature for 5 min, centrifuged at 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g for 3 min and the pellet obtained was suspended in 50 μL of Tris–EDTA buffer; the dissolved pellet is termed the M-HAp NP–pDNA complex. Unbound pDNA was quantified by measuring the optical density (OD) of the supernatant at 260 nm using a UV spectrometer (UV2450, Shimadzu, Japan), using the supernatant of the NPs without pDNA as a blank. The DNA binding efficiencies (B.Es) were determined using the equation:
000×g for 3 min and the pellet obtained was suspended in 50 μL of Tris–EDTA buffer; the dissolved pellet is termed the M-HAp NP–pDNA complex. Unbound pDNA was quantified by measuring the optical density (OD) of the supernatant at 260 nm using a UV spectrometer (UV2450, Shimadzu, Japan), using the supernatant of the NPs without pDNA as a blank. The DNA binding efficiencies (B.Es) were determined using the equation:| % B.E = {[(pDNA)i − (pDNA)f]/[(pDNA)i]} × 100, |
Each of the M-HAp NP–pDNA complexes (20 μL) was loaded on 0.8% agarose gel and was run using an electrophoresis apparatus at 80 mV for 1 h. It was stained with ethidium bromide for 10 min and the results were recorded using a gel imager (Life technologies, India).
Dynamic light scattering of R-G-HAp NPs and R-G-HAp NP–pDNA complexes
DLS studies of M-HAp NPs and M-HAp NP–pDNA complexes were carried out in minimal media (M.M), phosphate buffered saline of pH 7.0 (PBS) and Sabouraud dextrose broth (SB). Samples were sonicated using a probe sonicator for 15 min after dispersion in the respective medium (0.1 mg/3 mL). DLS studies were carried out immediately and again after 30 min of sonication using a DLS analyzer (Nano Plus Micromeritics).Transformation of non-competent C. albicans cells
Candida albicans was chosen as a model yeast organism for transformation studies. This strain was kanamycin and geneticin sensitive, whereas the selected plasmid of size 10 Kb contains kanamycin and geneticin resistance genes. It was streaked on Sabouraud dextrose agar (SDA) plates and incubated at 37 °C for 48 hours. A single colony from the streaked plate was inoculated into 10 mL Sabouraud broth (SB) and incubated at 37 °C for 48 h. The cells (2 × 108) were added to 20 mL SB after a prescribed time period and incubated at 37 °C in a shaking incubator for 6–8 h. The cells were subsequently centrifuged in two sets at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, each set containing 6 tubes with 1 mL culture. The pellets were dissolved in 200 μL M.M for the first set and in 200 μL of SB media for the second set. Afterwards, 20 μL M-HAp NP
000 rpm, each set containing 6 tubes with 1 mL culture. The pellets were dissolved in 200 μL M.M for the first set and in 200 μL of SB media for the second set. Afterwards, 20 μL M-HAp NP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) pDNA of various ratios (0
pDNA of various ratios (0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 50
1, 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 70
1, 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 80
1, 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 90
1, 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 100
1 and 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was added and vortexed for 5 min. Subsequently, the cells were heat-shocked for 30–60 s at 47 °C, chilled on ice for 2 min, mixed with the respective 1 mL SB/M.M broth, and incubated at 37 °C for 1 h on a rotary shaker. 100 μL of the cell suspension was transferred onto kanamycin (100 μg mL−1) and geneticin (G418) (150 μg mL−1) containing SDA plates and incubated at 37 °C for 36–48 h. The colonies were counted and transformation efficiencies (T.Es) were calculated using the formula T.E = number of colonies on the plate/concentration of pDNA present in 100 μL cell suspension (in μg). A similar protocol was followed to measure the T.E for HAp NP mediated transformation.
1) was added and vortexed for 5 min. Subsequently, the cells were heat-shocked for 30–60 s at 47 °C, chilled on ice for 2 min, mixed with the respective 1 mL SB/M.M broth, and incubated at 37 °C for 1 h on a rotary shaker. 100 μL of the cell suspension was transferred onto kanamycin (100 μg mL−1) and geneticin (G418) (150 μg mL−1) containing SDA plates and incubated at 37 °C for 36–48 h. The colonies were counted and transformation efficiencies (T.Es) were calculated using the formula T.E = number of colonies on the plate/concentration of pDNA present in 100 μL cell suspension (in μg). A similar protocol was followed to measure the T.E for HAp NP mediated transformation.
Electroporation
pDNA of 500 ng concentration was added to 200 μL electrocompetent cells, mixed properly and transferred into an ice cold 1 mm-electroporation cuvette. The cuvette was placed in an electroporator which was set to 2.5 kV; 150 Ω; 22 μF module and press pulsed followed by the addition of 1 mL of SB/M.M media and incubated at 37 °C on a shaking incubator for 1 h. Subsequently, 100 μL of the cell suspension was plated on SB plates with kanamycin and geneticin, and incubated at 37 °C for 36–48 h.Cell toxicity studies
MTT29 and resazurin assays29,30 were used to study the cell viability of M-HAp NPs using 24 well plates. Exponentially growing C. albicans cells were inoculated (5 × 105 cells per well) in SB. The culture was treated with various concentrations (10, 50, 100, 500 and 1000 μg mL−1) of M-HAp NPs dispersed in PBS, incubated for 12, 24 and 36 h, followed by addition of MTT (20 μL of 5 mg mL−1) and further incubated at 37 °C for 4 h. The medium was aspirated out carefully without disturbing the cells after 4 h. The formazan crystals formed were dissolved by adding 150 μL of DMSO, and the absorbance was measured at 570 nm using a UV-visible spectrophotometer (UV-2450, Shimadzu, Japan). All the measurements were carried out in triplicate. The relative cell viability (%) was calculated as ([At]/[Ac]) × 100, where [At] is the absorbance of the test sample and [Ac] is the absorbance of the cells without treatment.For the resazurin assay, a similar protocol was followed, wherein the resazurin dye (10 μL of 5 mg mL−1) was added to the test samples after the respective incubation periods mentioned above followed by incubation at 37 °C for 4 h. The absorbance was measured at 560 nm and the relative cell viability (%) was calculated as mentioned above.
Statistical analysis
All the experiments were carried out in triplicate on different days. The results are expressed as mean ± standard error. Statistical analysis was carried out using Microsoft™ Excel 2007 software. A one-way analysis of variance (ANOVA) test was used for comparison of differences between the multiple tests. Two tailed Student's t-test was used to analyze any significant differences between the control and individual experimental groups. ‘p’ values of less than 0.05 (p < 0.05) were considered significant.Results and discussion
Synthesis and characterization
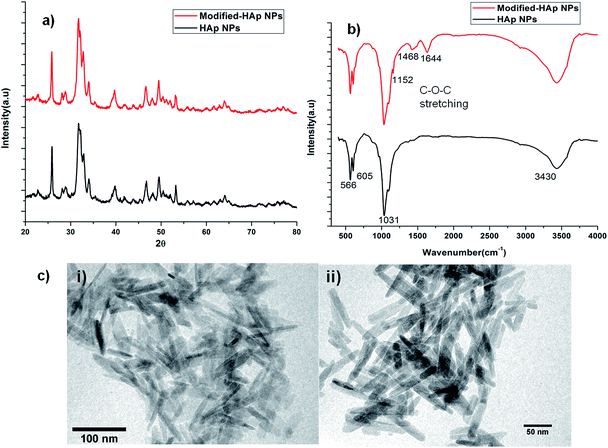
A modified sol gel method was used to synthesize arginine (R), glucose (G) and PEG functionalized HAp NPs (M-HAp NPs) at room temperature.27 Surface functionalization of HAp was carried out with the hypothesis that arginine, a positively charged amino acid could provide a cationic surface to facilitate efficient binding of pDNA, glucose, an essential carbon source for growth, could enhance the uptake of the NP–pDNA complex, and PEG could help pDNA to increase the interaction with the surface of the yeast cells and enhance the transformation efficiency.13,14,16,31The crystalline nature of HAp was confirmed by XRD analysis. The XRD spectra of HAp and M-HAp NPs were similar showing characteristic peaks at 25.8°, 31.34°, 31.95° and 33.97° corresponding to [hkl] values of [002], [211], [112] and [300], respectively (Fig. 1a), confirming the formation of a hexagonal crystalline phase (ICDD 09-0432). FTIR studies were carried out to identify various functional groups associated with HAp wherein signature peaks were noted (Fig. 1b). The bands at ∼600 cm−1 are attributed to the bending modes of P–O bonds in phosphate groups, with contribution from –OH of the apatite group at ∼605 cm−1. The bands in the region of 3400 cm−1 are attributed to –OH bonds and those observed near ∼3571 cm−1 are associated with the –OH stretching vibration of HAp.27,36–38 In M-HAp, additional bands corresponding to glucose, arginine and PEG were also observed. The bands of NH2 and amide I associated with arginine were observed at ∼1448 and ∼1644 cm−1, respectively.26,32,33,39,40 The bands corresponding to –CH of glucose and C–O–C stretching bands of PEG were observed at ∼2943 cm−1 and ∼1100 cm−1 respectively (Fig. 1c).34,35,41
The morphology of HAp NPs and M-HAp NPs was studied using TEM. Modification with PEG increased the average particle length from 62.5 to 72.7 nm and the diameter from 6.2 to 8.3 nm (Fig. 3). The increase in particle size as a result of PEGylation is dependent on the molecular weight of the PEG molecule. PEGylation with high molecular weight PEG contributes towards a larger increase in size as compared to low molecular weight PEG, and this is attributed to the length of the PEG chain.42–45 In this study, PEGylation was carried out using PEG of low molecular weight (6000 kDa) resulting in a minimal increase in size (Fig. 1c).
XPS studies were carried out to determine characteristic elements of HAp and M-HAp NPs. Spectra of HAp and M-HAp NPs were similar and peaks of calcium, phosphate, and oxygen were obtained for both the samples. The presence of arginine, glucose and PEG in M-HAp NPs could not be confirmed by XPS probably due to the low dopant concentration (0.1 wt% arginine, 0.3 wt% glucose (G) and 0.05 wt% PEG).
Zeta potential measurements of HAp and M-HAp NPs were carried out to measure the surface charge. The zeta value for HAp was ∼+13.25 mV which increased to ∼+72 mV for M-HAp NPs. The increase in the zeta value noted after the functionalization of HAp could be attributed to the binding of the α-carboxylate group of arginine and OH− ions of PEG to calcium ions of HAp such that the positively charged amino groups of arginine and H+ ions of PEG are exposed at the NP/solvent interface thereby increasing the zeta potential values.26,40 A decrease in the negative surface charge after functionalization with arginine/PEG has been observed in many NPs such as gold, silica, Fe3O4.etc.42–45
Binding efficiency of M-HAp NPs
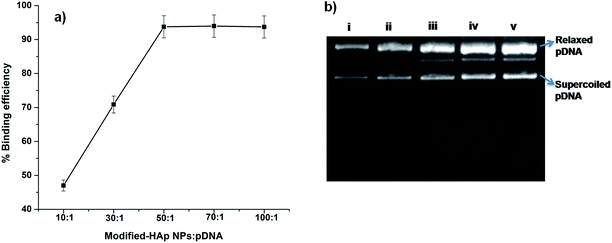
Binding of pDNA to NPs is essential for their proposed application of yeast transformation. The loading capacity of pDNA onto the NPs is dependent on the NP concentration and is critical in controlling the transformation efficiency. Therefore, we examined the binding efficiency (B.E) of pDNA to M-HAp NPs by measuring the absorbance of unbound pDNA at 260 nm at various NP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) pDNA ratios. The B.E of pDNA to M-HAp NPs was <70% when the NP
pDNA ratios. The B.E of pDNA to M-HAp NPs was <70% when the NP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) pDNA ratio was 10
pDNA ratio was 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 30
1 and 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and increased to ∼90% when the ratio was 50
1, and increased to ∼90% when the ratio was 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 70
1, 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 100
1 and 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Fig. 2a). Binding of pDNA to M-HAp NPs was confirmed by agarose gel electrophoresis of the complexes. In Fig. 2b, lanes (i–iv) show pDNA–M-HAp NP complexes with ratios of 10
1 (Fig. 2a). Binding of pDNA to M-HAp NPs was confirmed by agarose gel electrophoresis of the complexes. In Fig. 2b, lanes (i–iv) show pDNA–M-HAp NP complexes with ratios of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 50
1, 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 70
1, 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and 100
1, and 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, demonstrating the binding of pDNA with M-HAp NPs; lane (v) is the control naked pDNA. Calcium phosphate/HAp NPs are being explored for transfection as they exhibit binding affinity for DNA, forming complexes.24,25,28 Increased B.E of DNA with various NPs such as HAp, chitosan and gold has been reported after functionalization with amino acids/PEG/polyethylenimine molecules.39,40,46–48
1, demonstrating the binding of pDNA with M-HAp NPs; lane (v) is the control naked pDNA. Calcium phosphate/HAp NPs are being explored for transfection as they exhibit binding affinity for DNA, forming complexes.24,25,28 Increased B.E of DNA with various NPs such as HAp, chitosan and gold has been reported after functionalization with amino acids/PEG/polyethylenimine molecules.39,40,46–48
Dynamic light scattering
Dynamic light scattering was carried out to study the hydrodynamic radius and aggregation of M-HAp NPs and M-HAp NP–pDNA complexes in various media such as PBS, M.M and S.B (Table 1). M-HAp NPs and M-HAp NP–pDNA complexes dispersed in SB media showed almost 3 times larger size than those in M.M and PBS medium. SB being a complex medium has a higher concentration of bio-micromolecules like peptides, lipids and electrolytes which adsorb onto the NP surface facilitating aggregation due to interparticle interaction.49,50.| Media | Samples | |
|---|---|---|
| M-HAp NPs (nm) | M-HAp NP–pDNA complex (nm) | |
| PBS | 97 | 107 |
| M.M | 100 | 112 |
| S.B | 329 | 337 |
Yeast transformation
M-HAp NPs were used to transform pDNA using C. albicans as the model organism. The transformation process was carried out at various M-HAp NP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) pDNA ratios (0
pDNA ratios (0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 10
1, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 30
1, 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 50
1, 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 70
1, 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 100
1 and 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), using SB and M.M media, to obtain the optimum ratio for the maximum T.E (Table 2). Two different media were used to observe the influence/effect of media components on the transformation efficiency. In M.M media, the T.E at a NP
1), using SB and M.M media, to obtain the optimum ratio for the maximum T.E (Table 2). Two different media were used to observe the influence/effect of media components on the transformation efficiency. In M.M media, the T.E at a NP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) pDNA ratio of 10
pDNA ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 was 103 cfu μg−1, and increased to 106 cfu μg−1 for a ratio of 70
1 was 103 cfu μg−1, and increased to 106 cfu μg−1 for a ratio of 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, beyond which it remained constant. Similarly, in SB media the initial T.E obtained at a ratio of 10
1, beyond which it remained constant. Similarly, in SB media the initial T.E obtained at a ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 was 37 cfu μg−1, and increased to 104 cfu μg−1 for 70
1 was 37 cfu μg−1, and increased to 104 cfu μg−1 for 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and remained constant thereafter. In M.M media, the T.E was higher by two orders of magnitude as compared to that in the complex SB media. This could be attributed to the aggregation of NPs in complex media as observed in DLS studies (Table 1). Moreover, the presence of low concentrations of nutrients in M.M may contribute towards increased uptake of glucose functionalized HAp NP–pDNA complexes resulting in an enhanced T.E. Since the maximum T.E was obtained for a M-HAp NP
1 and remained constant thereafter. In M.M media, the T.E was higher by two orders of magnitude as compared to that in the complex SB media. This could be attributed to the aggregation of NPs in complex media as observed in DLS studies (Table 1). Moreover, the presence of low concentrations of nutrients in M.M may contribute towards increased uptake of glucose functionalized HAp NP–pDNA complexes resulting in an enhanced T.E. Since the maximum T.E was obtained for a M-HAp NP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) pDNA ratio of 70
pDNA ratio of 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, it was considered optimal and used for comparison with the commonly used transformation technique of electroporation. Table 3 shows the comparison between the T.E obtained when transformation was carried out in C. albicans using HAp/M-HAp NP
1, it was considered optimal and used for comparison with the commonly used transformation technique of electroporation. Table 3 shows the comparison between the T.E obtained when transformation was carried out in C. albicans using HAp/M-HAp NP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) pDNA at a ratio of 70
pDNA at a ratio of 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and the electroporation technique. It was observed that the T.E obtained with M-HAp NPs was four and one order of magnitude higher than that obtained with electroporation in M.M and SB media, respectively. To the best of our knowledge, this is the first report on surface modification of HAp NPs with glucose, arginine and PEG as a nanovehicle for the transformation of pDNA in C. albicans (Fig. 3).
1 and the electroporation technique. It was observed that the T.E obtained with M-HAp NPs was four and one order of magnitude higher than that obtained with electroporation in M.M and SB media, respectively. To the best of our knowledge, this is the first report on surface modification of HAp NPs with glucose, arginine and PEG as a nanovehicle for the transformation of pDNA in C. albicans (Fig. 3).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) pDNA ratios
pDNA ratios
M-HAp NP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) pDNA pDNA |
T.E of C. albicans (cfu μg−1) | |
|---|---|---|
| M.M | SB | |
0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
0 | 0 |
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
103 | 37 |
30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
6 × 104 | 7 × 102 |
50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
2 × 105 | 103 |
70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
3 × 106 | 104 |
100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
5 × 106 | 3 × 104 |
| T.E of C. albicans (cfu μg−1) | ||
|---|---|---|
| M.M | SB | |
| HAp NPs | 6 × 103 | 70 |
| M-HAp NPs | 3 × 106 | 104 |
| Electroporation | 5 × 102 | 103 |
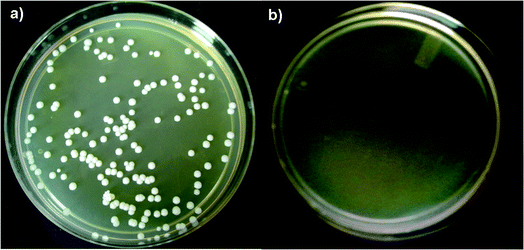 | ||
| Fig. 3 Transformed C. albicans using M-Hap NPs, without the preparation of competent cells, on kanamycin and geneticin containing SB agar plates (a) and the negative control (b). | ||
As hypothesized, the functionalization of HAp with arginine, glucose and PEG molecules enhanced the T.E. In our previous study, the strategy of glucosylation was successfully used to enhance the uptake and T.E of pDNA in bacterial cells.51 The role of glucosylation in drug delivery for improved uptake in the treatment of cancer has been explored earlier.52,53 PEGylation and functionalization of NPs with various amino acids (arginine, lysine, valine, etc.) to increase cellular uptake have also been explored for a variety of applications such as drug delivery, gene delivery and bioimaging.43,54 The potential of PEG to increase the transformation efficiency in yeast/bacteria has been demonstrated. PEG has also been used in the LiAc method to achieve a high transformation efficiency in Saccharomyces cerevisiae.13–15 Fluorescence microscopy studies of pDNA transformation using the LiAc method have shown that pDNA was able to bind to the yeast cell surface only in the presence of PEG, and no transformants were obtained in the absence of PEG.55 In the present study, the positively charged pDNA–M-HAp NP complex possibly attaches to the negatively charged cell surface of C. albicans by electrostatic attraction, facilitating cellular uptake. The possible entry of these complexes through nanopores present on the cell surfaces of the yeast cells could also be a contributing factor.
Plasmid isolation from transformed C. albicans
Fig. 4 shows the agarose gel image of pDNA isolated from transformed C. albicans using M-HAp NPs (lane i) and using the control method of electroporation by growing the cells on SB plates containing kanamycin and geneticin which was similar to that of the control pDNA (lane ii). This confirmed M-HAp NP mediated yeast transformation.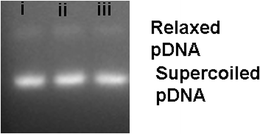 | ||
| Fig. 4 Agarose gel image of pDNA (i) isolated from C. albicans transformed using M-HAp NPs (ii) and the electroporation technique. (ii) Control pDNA. | ||
Cell toxicity
Biocompatibility is an important prerequisite for the successful use of NPs for transformation. To determine the effect of M-HAp NPs on the viability of C. albicans, time and dose dependent studies were carried out using MTT and resazurin assays (Fig. 5a and b). Cell growth was reduced by ∼3–5% at concentrations above 500 μg mL−1. No significant difference (p > 0.05) in cell viability was noted between M-HAp NPs and the untreated control at various concentrations (10, 50, 100, 500 and 1000 μg mL−1), indicating the biocompatibility of M-HAp NPs.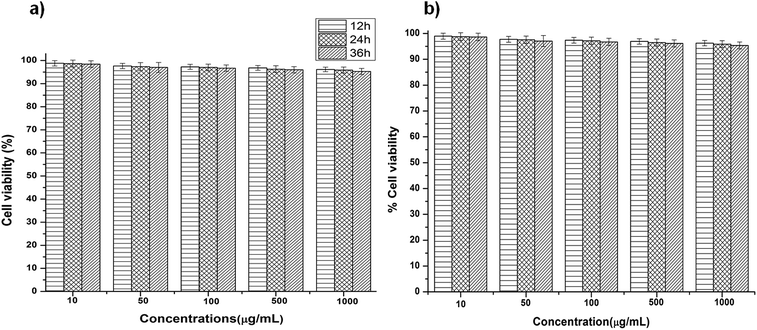 | ||
| Fig. 5 (a) MTT and (b) resazurin data showing the non-toxic nature of modified HAp NPs on C. albicans after 12, 24 and 36 h of growth at various concentrations ranging from 10–1000 μg mL−1. | ||
HAp is a well-studied biocompatible material and its non-toxic nature has been examined on various cell lines.27,32,39,40 It has also been reported that functionalization of NPs with PEG makes them biocompatible by preventing agglomeration and surface oxidation.34,44
Conclusion
This study demonstrated that appropriately functionalized HAp NPs can serve as a vehicle for plasmid transformation in C. albicans. Functionalization of HAp NPs with the positively charged amino acid, arginine, and PEG increased the zeta potential as well as the binding efficiency of the nanovehicle to pDNA. Modification of HAp NPs resulted in a high binding efficiency (93%) and successful transformation in C. albicans with a T.E of 106 cfu μg−1. This nanovehicle delivery system does not require preparation of competent cells and is also non-toxic to yeast cells. This new method could provide exciting opportunities for the advancement of the applications of yeast in the field of biotechnology.Ethical statement
This article does not contain any studies with human participants and animals performed by any of the authors.Conflicts of interest
The authors declare that they have no conflict of interest.Acknowledgements
The authors thank the Department of Science & Technology, India, for the research grant (SR/NM/NS-1354/2014G), and the Sophisticated Analysis Instrumentation Facility and the Central Surface Analytical Facility of the Indian Institute of Technology, Mumbai, for their help with TEM and XPS measurements.References
- J.-L. Legras, D. Merdinoglu, J. Cornuet and F. Karst, Mol. Ecol., 2007, 16, 2091–2102 CrossRef CAS PubMed.
- D. Sicard and J.-L. Legras, C. R. Biol., 2011, 334, 229–236 CrossRef PubMed.
- D. Botstein and G. R. Fink, Genetics, 2011, 189, 695–704 CrossRef CAS PubMed.
- V. Khurana and S. Lindquist, Nat. Rev. Neurosci., 2010, 11, 436–449 CrossRef CAS PubMed.
- S. Tenreiro, M. C. Munder, S. Alberti and T. F. Outeiro, J. Neurochem., 2013, 127, 438–452 CrossRef CAS PubMed.
- A. L. Demain and P. Vaishnav, Biotechnol. Adv., 2009, 27(3), 297–306 CrossRef CAS PubMed.
- V. Meyer, Biotechnol. Adv., 2008, 26, 177–185 CrossRef CAS PubMed.
- K. Ino, Y. Kitagawa, T. Watanabe, H. Shiku, M. Koide, T. Itayama, T. Yasukawa and T. Matsue, Electrophoresis, 2009, 30, 3406–3412 CrossRef CAS PubMed.
- D. Pompon, A. Perret, A. Bellamine, R. Laine, J. C. Gautier and P. Urban, Toxicol. Lett., 1995, 82, 815–822 CrossRef PubMed.
- M. S. Siddiqui, K. Thodey, I. Trenchard and C. D. Smolke, FEMS Yeast Res., 2012, 12(2), 144–170 CrossRef CAS PubMed.
- D. M. Becker and L. Guarente, Methods Enzymol., 1991, 194, 182–187 CAS.
- R. A. Butow, R. M. Henke, J. V. Moran, S. M. Belcher and P. S. Perlman, Methods Enzymol., 1996, 264, 265–278 CAS.
- R. D. Gietz and R. H. Schiestl, Nat. Protoc., 2007, 2, 31–34 CrossRef CAS PubMed.
- R. D. Gietz and R. H. Schiestl, Nat. Protoc., 2007, 2, 38–41 CrossRef CAS PubMed.
- R. D. Gietz and R. A. Woods, Yeast Protoc, 2006, vol. 313, pp. 107–120 Search PubMed.
- R. J. Klebe, J. V. Harriss, Z. D. Sharp and M. G. Douglas, Gene, 1983, 25, 333–341 CrossRef CAS PubMed.
- N. Yoshida, T. Ikeda, T. Yoshida, T. Sengoku and K. Ogawa, FEMS Microbiol. Lett., 2001, 195, 133–137 CrossRef CAS PubMed.
- A. Bozkir and O. M. Saka, J. Drug Targeting, 2004, 12, 281–288 CrossRef CAS PubMed.
- H. Tan, L. Fu and M. Seno, Int. J. Mol. Sci., 2010, 11, 4962–4972 CrossRef CAS PubMed.
- D. R. Radu, C. Y. Lai, K. Jeftinija, E. W. Rowe, S. Jeftinija and V. S. Y. Lin, J. Am. Chem. Soc., 2004, 126, 13216–13217 CrossRef CAS PubMed.
- I. I. Slowing, J. L. Vivero-Escoto, C. W. Wu and V. S. Y. Lin, Adv. Drug Delivery Rev., 2008, 60, 1278–1288 CrossRef CAS PubMed.
- Y. Filyak, N. Finiuk, N. Mitina, O. Bilyk, V. Titorenko, O. Hrydzhuk, A. Zaichenko and R. Stoika, BioTechniques, 2013, 54, 35–43, DOI:10.2144/000113980.
- S. Naqvi, A. Maitra, M. Abdin, M. Akmal, I. Arora and M. Samim, J. Mater. Chem., 2012, 22, 3500–3507 RSC.
- T. N. Tram Do, W.-H. Lee, C.-Y. Loo, A. V. Zavgorodniy and R. Rohanizadeh, Ther. Delivery, 2012, 3(5), 623–632 CrossRef CAS.
- V. Uskoković and D. P. Uskoković, J. Biomed. Mater. Res., Part B, 2011, 96, 152–191 CrossRef PubMed.
- Z. Yan-Zhong, H. Yan-Yan, Z. Jun, Z. Shai-Hong, L. Zhi-You and Z. Ke-Chao, Nanoscale Res. Lett., 2011, 6, 1–8 CrossRef PubMed.
- K. Deshmukh, M. M. Shaik, S. R. Ramanan and M. Kowshik, ACS Biomater. Sci. Eng., 2016, 2(8), 1257–1264 CrossRef CAS.
- D. Olton, J. Li, M. E. Wilson, T. Rogers, J. Close, L. Huang, P. N. Kumta and C. Sfeir, Biomaterials, 2007, 28, 1267–1279 CrossRef CAS PubMed.
- D. Gerlier and N. Thomasset, J. Immunol. Methods, 1986, 94, 57–63 CrossRef CAS PubMed.
- S. D. Sarker, L. Nahar and Y. Kumarasamy, Methods, 2007, 42, 321–324 CrossRef CAS PubMed.
- S. Zhu, B. Huang, K. Zhou, S. Huang, F. Liu, Y. Li, Z. Xue and Z. Long, J. Nanopart. Res., 2004, 6, 307–311 CrossRef CAS.
- R. Gonzalez-McQuire, J.-Y. Chane-Ching, E. Vignaud, A. Lebugle and S. Mann, J. Mater. Chem., 2004, 14, 2277–2281 RSC.
- T. Matsumoto, M. Okazaki, M. Inoue, Y. Hamada, M. Taira and J. Takahashi, Biomaterials, 2002, 23, 2241–2247 CrossRef CAS PubMed.
- G. D. Venkatasubbu, S. Ramasamy, G. Avadhani, V. Ramakrishnan and J. Kumar, Powder Technol., 2013, 235, 437–442 CrossRef CAS.
- X. Zhang, J. Hui, B. Yang, Y. Yang, D. Fan, M. Liu, L. Tao and Y. Wei, Polym. Chem., 2013, 4, 4120–4125 RSC.
- S. Jadalannagari, K. Deshmukh, S. R. Ramanan and M. Kowshik, Appl. Nanosci., 2014, 4, 133–141 CrossRef CAS.
- S. Jadalannagari, K. Deshmukh, A. K. Verma, R. Vohra, M. Kowshik and S. Roy Ramanan, Sci. Adv. Mater., 2014, 6, 312–319 CrossRef CAS.
- S. R. Ramanan and R. Venkatesh, Mater. Lett., 2004, 58, 3320–3323 CrossRef CAS.
- S. Ueno and S. Shimabayashi, Phosphorus Res. Bull., 2009, 23, 57–62 CrossRef CAS.
- G. H. Wang, Y. Z. Zhao, T. Juan, S. H. Zhu and K. C. Zhou, Trans. Nonferrous Met. Soc. China, 2015, 25, 490–496 CrossRef CAS.
- M. Ibrahim, M. Alaam, H. El-Haes, A. F. Jalbout and Ad. Leon, Ecletica Quim., 2006, 31, 15–21 CrossRef CAS.
- M. Anbarasu, M. Anandan, E. Chinnasamy, V. Gopinath and K. Balamurugan, Spectrochim. Acta, Part A, 2015, 135, 536–539 CrossRef CAS PubMed.
- T. Andreani, A. L. R. De Souza, C. P. Kiill, E. N. Lorenzón, J. F. Fangueiro, A. C. Calpena, M. V. Chaud, M. L. Garcia, M. P. D. Gremião and A. M. Silva, Int. J. Pharm., 2014, 473, 627–635 CrossRef CAS PubMed.
- J. Manson, D. Kumar, B. J. Meenan and D. Dixon, Gold Bull., 2011, 44, 99–105 CrossRef CAS.
- B. Pelaz, P. Del Pino, P. Maffre, R. Hartmann, M. Gallego, S. Rivera-Fernandez, J. M. de la Fuente, G. U. Nienhaus and W. J. Parak, ACS Nano, 2015, 9, 6996–7008 CrossRef CAS PubMed.
- B. Layek and J. Singh, Biomacromolecules, 2013, 14(2), 485–494 CrossRef CAS PubMed.
- P. S. Ghosh, C. K. Kim, G. Han, N. S. Forbes and V. M. Rotello, ACS Nano, 2008, 2(11), 2213–2218 CrossRef CAS PubMed.
- L. Wu, J. Xie, T. Li, Z. Mai, L. Wang, X. Wang and T. Chen, R. Soc. Open Sci., 2017, 4(10), 170822 CrossRef PubMed.
- K. Rausch, A. Reuter, K. Fischer and M. Schmidt, Biomacromolecules, 2010, 11(11), 2836–2839 CrossRef CAS PubMed.
- T. L. Moore, L. Rodriguez-Lorenzo, V. Hirsch, S. Balog, D. Urban, C. Jud, B. Rothen-Rutishauser, M. Lattuada and A. Petri-Fink, Chem. Soc. Rev., 2015, 44, 6287–6305 RSC.
- K. Deshmukh, S. R. Ramanan and M. Kowshik, Mater. Sci. Eng., C, 2019, 96, 58–65 CrossRef CAS PubMed.
- X. Jiang, H. Xin, Q. Ren, J. Gu, L. Zhu, F. Du, C. Feng, Y. Xie, X. Sha and X. Fang, Biomaterials, 2014, 35, 518–529 CrossRef CAS PubMed.
- J. Wang, C. Yin, G. Tang, X. Lin and Q. Wu, Int. J. Pharm., 2013, 441, 291–298 CrossRef CAS PubMed.
- J. V. Jokerst, T. Lobovkina, R. N. Zare and S. S. Gambhir, Nanomedicine, 2011, 6, 715–728 CrossRef CAS PubMed.
- P. Chen, H.-H. Liu, R. Cui, Z.-L. Zhang, D. W. Pang, Z. X. Xie, H. Z. Zheng, Z. X. Lu and H. Tong, Talanta, 2008, 77, 262–268 CrossRef CAS PubMed.
Footnote |
| † Present/Permanent Address: Birla Institute of Technology and Science Pilani, K K Birla Goa Campus, Zuarinagar, Goa, India-403726, Phone no: +91-832-2580304, +91-832-2580325; Fax no: +91-832-2517033. |
| This journal is © The Royal Society of Chemistry 2019 |